Abstract
Dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) is widely used to evaluate tumor permeability, yet measurements have not been directly validated in brain tumors. Our purpose was to compare estimates of forward leakage Ktrans derived from DCE-MRI to the estimates K obtained using [14C]aminoisobutyric acid quantitative autoradiography ([14C]AIB QAR), an established method of evaluating blood-tumor barrier permeability. Both DCE-MRI and [14C]AIB QAR were performed in five rats 9 to 11 days following tumor implantation. Ktrans in the tumor was estimated from DCE-MRI using the threeparameter general kinetic model and a measured vascular input function. Ki was estimated from QAR data using regions of interest (ROI) closely corresponding to those used to estimate Ktrans. Ktrans and Ki correlated with each other for two independent sets of central tumor ROI (R = 0.905, P = .035; R = 0.933, P = .021). In an additional six rats, Ktrans was estimated on two occasions to show reproducibility (intraclass coefficient = 0.9993; coefficient of variance = 6.07%). In vivo blood-tumor permeability parameters derived from DCE-MRI are reproducible and correlate with the gold standard for quantifying blood tumor barrier permeability, [14C]AIB QAR.
Keywords: Blood brain barrier, tumor permeability, anti-angiogenesis
Introduction
Tumor angiogenic vessels are larger, are more tortuous, and have large endothelial fenestrations and caveoli that make them more permeable than normal vessels. Such vessels are thought to promote tumor growth by increasing blood flow and nutrient delivery and are, therefore, a target for cancer therapies. Dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) is currently in wide use as a method for monitoring vascular permeability within tumors, as a means of assessing early response to antiangiogenic therapy in clinical trials [1–3].
Despite its widespread use, there is controversy over the physiological meaning of parameters derived from DCE-MRI, as well as concerns over the reproducibility of such measurements. To date, the only attempts to validate DCE-MRI have been either reproducibility studies, simulations, or qualitative comparisons to histology and immunohistochemical markers of tumor angiogenesis such as CD31 and microvascular density measurements [4]. Although measures of reproducibility and qualitative comparisons to immunohistochemistry are helpful, they are not physiologically based and, therefore, do not directly assess vascular permeability. Indeed, there is concern that DCE-MRI-derived parameters are more dependent on tumor blood flow than on vascular permeability. Others have raised concerns that an assumption of slow water exchange between intracellular and extracellular compartments may be incorrect and may lead to gross errors in permeability measurements [5]. Studies using DCE-MRI to monitor tumor response to vascular-disrupting agents have raised concerns about the utility of DCE-MRI biomarkers [6]. Without comparison studies with “true” functional measures of permeability, the validity of DCE-MRI as an index of vascular permeability is unclear, and the true physiological meaning of the influx rate constant (Ktrans) remains unknown.
In this study, we directly compare the DCE-MRI index of vascular permeability (Ktrans) to a standard experimental method of measuring the influx rate constant (Ki) of small molecules leaking from vascular space into extravascular-extracellular space by [14C]aminoisobutyric acid quantitative autoradiography ([14C]AIB QAR) in the RG2 rat brain tumor model [7,8]. Ktrans and Ki are both measures of the permeability-surface area product per unit volume in the RG2 tumor and, thus, should correlate with each other; yet these measures are obtained through markedly divergent techniques [2,8,9]. Ktrans is derived from a two-compartment pharmacokinetic model based on the curve fitting of the time-gadolinium concentration curve in the tumor. Ki is derived from the unique characteristics of AIB, which leaks unidirectionally from the blood into tissue and is then trapped at the site of leakage. Ki can thus be determined from [14C]AIB QAR data with very high spatial resolution using a “unidirectional model” and simple calculation [7]. In addition to examining correlations to this “gold standard” measure, the reproducibility of DCE-MRI Ktrans estimates is examined for repeated RG2 tumor DCE-MRI estimates.
Materials and Methods
Animal Preparation
All studies were performed after obtaining the approval of the institutional Animal Care and Use Committee. The maintenance and care of animals were in compliance with National Institutes of Health guidelines. RG2 tumor cells were grown in culture and implanted (1 x 105 cells in 5 µl of phosphate-buffered saline) into the caudate-putamen of five adult male Fischer-344 rats (weight range, 250–300 g; Harlan Laboratories, Indianapolis, IN) using previously described techniques [10]. Cells were grown in vivo for 9 to 11 days before injection. On the day of implantation, animals were anesthetized with isoflurane, and one venous and two arterial polyethylene microtubing cannulations (PE-50; Becton Dickinson, Franklin Lakes, NJ) were placed for venous gadolinium diethylenetriamine pentaacetic acid (Gd- DTPA) infusion, venous [14C]AIB injection, arterial [14C]AIB plasma concentration sampling, and arterial blood pressure monitoring.
DCE-MRI Scanning
Animals were placed on a Philips Intera 3.0-T MR scanner (Philips Medical Systems, Andover, MA) with a dedicated 7-cm rat solenoid radiofrequency coil (Philips Research Laboratories, Hamburg, Germany) with maintenance isoflurane anesthesia from 9 to 11 days following tumor implantation. Scout images were obtained to define a coronal plane perpendicular to the dorsum of the rat brain for subsequent imaging. For anatomic imaging, 32 two-dimensional fast spin-echo T2 slices were acquired. For T1 mapping, a low-flip-angle (FA = 3°) three-dimensional fast field echo (FFE) sequence with parameters otherwise matching the DCE-MRI scan sequence was performed immediately before dynamic scans.
DCE-MRI
Thirty DCE-MRI three-dimensional T1 FFE sequence volumes were obtained over 10 minutes at a temporal resolution of 20 sec/vol.
Geometric parameters were as follows: slice thickness = 1 mm; slice gap = -0.5 mm (oversampled); slices = 16 (32); field of view (FOV) = 76.8 mm; matrix = 256 x 244–resulting in an interpolated in-plane resolution of 0.33 mm.
Contrast parameters were: TR = 8.1 milliseconds; TE = 2.3 milliseconds; FA = 12°. Following five baseline scans, 0.5 ml/kg Gd-DTPA (Magnevist; Bayer HealthCare Pharmaceuticals, Wayne, NJ) was injected through the femoral vein using a Hamilton syringe (Hamilton Co., Reno, NV) connected to a polyethylene microtubing (PE-50; Becton Dickinson) at a constant rate of 0.5 ml/kg per minute over 1 minute, with a 2-minute saline flush using a microinfusion pump (PHD 2000; Harvard Apparatus, Holliston, MA).
[14C]AIB QAR
[14C]AIB QAR was performed within 30 minutes of MR scanning. Animals were transferred to a radiation-secure facility, placed under maintenance anesthesia, and prepared for constant-withdraw plasma sampling similar to that described in Asotra et al. [11]. A saline-filled PE-90 tubing was attached to the femoral artery line, and a saline-filled (1.0 ml) Hamilton syringe (Hamilton Co.) in a Harvard pump (PHD 22/2000; Harvard Apparatus) was set to constant withdrawal (33 µl/min). A 0.5-ml bolus of 100 µCi (n = 6) or 33 µCi (n = 2) of [14C]AIB was manually injected into the femoral vein over 15 seconds, 1 minute after starting the pump. After approximately 20 minutes, the pump was stopped, PE-90 blood sample was collected, and time at the experimental end point was documented (Te). Blood samples were collected at the time of sacrifice. The animal was immediately guillotined, and the brain was rapidly removed (∼ 5 minutes) and frozen in 2-methylbutane. Guillotining was required in the experimental design to rapidly freeze the brain and to prevent significant diffusion of [14C]AIB away from the initial site of leakage, and was approved as an exception by the institutional Animal Care and Use Committee.
Arterial blood samples were immediately spun, plasma samples were collected, and plasma concentrations for the PE-90 tubing () and the femoral line (Cp(Te)) were measured. At an angle approximating the MR slice plane, frozen brains were sectioned in a cryomicrotome into 40-µm sections per 200-µm interval and photographed. Sections were exposed on film (Fuji Photo Film, Inc., Greenwood, SC) for 3 to 4.5 days for 100-µCi infusions (n = 6), or for 10.5 to 12 days for 33-µCi infusions (n = 2), with 40 µmm [14C]AIB standards (0–35 µCi; American Radiolabeled Chemicals, Inc., St. Louis, MO). The use of standards allowed for changes in the specific radioactivity of [14C]AIB infusions to speed up film development without affecting the accuracy of tissue [14C]AIB concentration determinations or the calculation of Ki. Films were scanned using a BAS-5000 Bio-Imaging Analyzer (Fuji Medical Systems, Stamford, CT).
DCE-MRI Analysis
All DCE-MRI data were processed using an investigational pharmacokinetic analysis package (KinMod plug-in to CINEtool; GE Healthcare, Waukesha, WI) based on IDL (Research Systems, Inc., Boulder, CO). Precontrast T1 maps were generated from data obtained at 3° and 12° flip angles [12,13]. DCE-MRI signal was converted into Gd-DTPA concentration.
A region(s) of interest (ROI) for vascular input function (VIF) selection was drawn around the confluence of dural sinuses, taking care to exclude regions with time-of-flight inflow artifacts. Using the CINEtool program, a semiautomated algorithm based on variance thresholding was used to select an ROI, from which the VIF was calculated. Signals were converted to Gd-DTPA concentration, assuming a normal blood T1 of 1320 milliseconds.
The VIF was convolved with general kinetic model (GKM) transfer function using a first approximation of the three free parameters to each Ct(t) curve according to:
where Cb(t) ibr time data and Hct is the hematocrit of Fischer-344 male rats (0.50). This equation is identical to the bidirectional GKM model equation of Iannotti et al. [14] with a conversion from plasma concentration and fractional plasma volume to blood concentration and fractional blood volume, respectively (Appendix A). The three free parameters (Ktrans, fbv, and ve) were iteratively adjusted to find a best-fit solution that minimized variance between modeled and measured Ct(t) curves using the Levenberg-Marquardt method. Estimates of Ktrans, ve, and fbv were made both on a voxelwise basis and by using ROI in which signal was averaged within the ROI.
Three ROI were hand-drawn on T1-weighted images at peak contrast enhancement, approximately 5 minutes after the injection of contrast media. The mean value of the three regions for contrast-enhancement ratio (CERc), the ratio of enhancement signal to baseline signal, was measured. A CINEtool function automatically generated ROI with boundaries equal to 25%, 50%, and 75% of the CERc for each animal.
[14C]AIB QAR Analysis
Optical density measurements of autoradiographs were converted to tissue concentration using 40-µCi standards and a polynomial fit of the curve of optical density versus tissue radioactivity per volume using MATLAB (MathWorks, Natick, MA). Autoradiograph pixelwise concentration and Ki maps were generated according to:
where Ct is the pixel tissue concentration; the fractional plasma volume (fpv) is assumed to be 2.2% based on prior studies [15]; T is the time of actual sample withdrawal during total experimental time (Te); Cp(Te) is the plasma concentration at Te; and is the concentration of radioactive plasma collected during T. Using a constant rate of sample withdrawal, the integral ∫ Cp(t) dt solves to T.
A radiologist blinded to the study results analyzed anatomic landmarks in gross brain photographs, T2 images, and postcontrast T1 images to select three [14C]AIB QAR section autoradiographs to match MR sections. [14C]AIB QAR ROI were semiautomatically generated using the image processing software MIPAV (http://mipav.cit.nih.gov; National Institutes of Health, Bethesda, MD). The [14C]AIB QAR tumor ROI aimed to match the 25% DCE-MRI ROI were first generated. For each [14C]AIB concentration map, ROI were drawn in the brain region contralateral to the tumor to plot the mean and distribution of normal tissue “background” concentration values, and the value at 3 SD above the mean background value (BG3SD) was recorded. A mask over each tumor section was then created with edges at each individually determined BG3SD threshold. This mask was then used to generate the QAR ROI at the tumor edge, approximating the volume of the 25% DCE-MRI ROI. To generate the QAR ROI matching the 50% and 75% threshold DCE-MRI ROI, concentration threshold values were increased until the volume of the generated QAR mask approximated the volume of the corresponding 50% or 75% DCE-MRI ROI. The final [14C]AIB QAR ROI were then copied and pasted to [14C]AIB QAR pixelwise influx rate constant (Ki) maps. The mean Ki value, standard deviation, and volume were recorded for each QAR Ki map. A final [14C]AIB QAR Ki value corresponding to each DCE-MRI slice was determined as the mean measure of the three selected [14C]AIB QAR sections.
Statistics
Regression statistics were generated for Ktrans versus Ki for each ROI pair of central tumor slices (SPSS, Chicago, IL).
Reproducibility Studies
In a separate set of experiments on a separate population of six Fischer-344 rats, test-retest repeat DCE-MRI studies were performed between 14 and 24 hours after the first injection of Gd-DTPA. DCE-MRI ROI for each experiment were obtained using hand-drawn ROI to be consistent with commonly used techniques. A user was instructed to select a central slice and to draw ROI (n = 5) within but closely approximating tumor edges to exclude a boundary of approximately 0.5 mm from the tumor edge for all scan MR central slice data. VIFs were then chosen, and parameter estimates were generated as described previously. To reduce variance due to hand-drawn ROI, the value for test-retest scans was calculated as the mean of five Ktrans estimates for five tumor ROI.
Reproducibility Statistics
Reproducibility statistics included the intraclass correlation coefficient (ICC), coefficient of variation (CoV), and within-subject standard deviation (wSD). (Microsoft Excel, Redmond, WA) [16–20].
Results
Tumors were successfully implanted and grown in 11 Fischer-344 rats. [14C]AIB autoradiography and DCE-MRI data were completed and analyzed on five of these animals, whereas repeat DCE-MRI test-retest studies were completed on the remaining six animals.
DCE-MRI Ktrans versus [14C]AIB QAR Ki
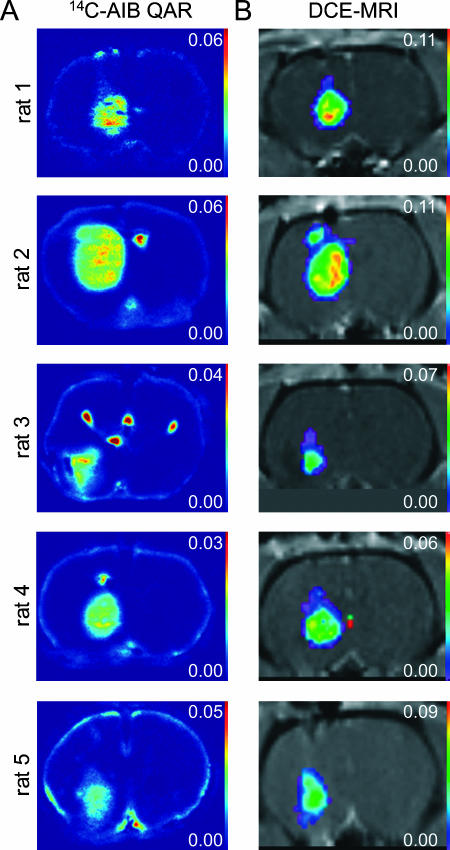
Ki maps generated from [14C]AIB QAR autoradiographs and Ktrans maps generated from DCE-MRI voxelwise data for the central tumor slice of each animal are shown (Figure 1). Similarities in the topographies of Ktrans and Ki maps can be appreciated, with higher influx rate constants in the central tumor and lower influx rate constants peripherally in both Ktrans and Ki maps. Note that distortions from tissue “chatter” caused by displacement during cryomicrotome tissue slicing in [14C]AIB QAR Ki maps are not present in in vivo DCE-MRI Ktrans parameter maps.
Figure 1.
(A) [14C]AIB QAR Ki maps. (B) DCE-MRI Ktrans parameter maps for central tumor slices for each animal. DCE-MRI Ktrans parameter maps are displayed within the 25% threshold ROI.
Tumor ROI, autoradiography Ki measures, and DCE-MRI measures of Ktrans, kep, ve, and fbv are listed for each tumor ROI for each animal (Table 1). The mean percentile differences in transfer constants between autoradiography and DCE-MRI (mean ± SD) were -2.88 ± 9.51%, -1.33 ± 2.28%, and -5.98 ± 26.2% for the 75%, 50%, and 25% thresholds, respectively.
Table 1.
[14C]AIB QAR Influx Rate Constant (Ki) Measures and DCE-MRI Parameter Estimates of the Influx Rate Constant (Ktrans), Efflux Rate Constant (kep), Fractional Leakage Space Volume (ve), and Fractional Blood Volume (fbv) for 75%, 50%, and 25% Threshold ROI and ROI Areas for Central Tumor Slices.
| Animal | Tumor Days | Threshold (%) | Ki (min-1) | Area (mm2) | Ktrans (min-1) | Area (mm2) | Differential Ratio of Tumor Area | kep (min-1) | ve | fbv |
| 1 | 9 | 25 | 0.0272 | 9.1 | 0.0420 | 9.9 | 0.088 | 0.443 | 0.096 | 0.03 |
| 50 | 0.0306 | 7.3 | 0.0660 | 7.2 | -0.014 | 0.546 | 0.123 | 0.02 | ||
| 75 | 0.0346 | 5.4 | 0.0900 | 5.4 | 0.000 | 0.641 | 0.141 | 0.01 | ||
| 2 | 11 | 25 | 0.0208 | 24.3 | 0.0480 | 20.3 | -0.165 | 0.321 | 0.150 | 0.04 |
| 50 | 0.0254 | 15.6 | 0.0690 | 15.6 | 0.000 | 0.387 | 0.174 | 0.03 | ||
| 75 | 0.0273 | 11.3 | 0.0900 | 11.2 | -0.009 | 0.459 | 0.198 | 0.02 | ||
| 3 | 10 | 25 | 0.0126 | 11.6 | 0.0180 | 7.2 | -0.379 | 0.174 | 0.096 | 0.03 |
| 50 | 0.0172 | 3.8 | 0.0360 | 3.6 | -0.053 | 0.268 | 0.132 | 0.04 | ||
| 75 | 0.0185 | 2.6 | 0.0420 | 2.1 | -0.192 | 0.283 | 0.144 | 0.04 | ||
| 4 | 10 | 25 | 0.0121 | 13.0 | 0.0240 | 11.1 | -0.146 | 0.192 | 0.126 | 0.04 |
| 50 | 0.0160 | 6.2 | 0.0420 | 6.2 | 0.000 | 0.249 | 0.168 | 0.03 | ||
| 75 | 0.0171 | 3.9 | 0.0540 | 3.9 | 0.000 | 0.254 | 0.210 | 0.03 | ||
| 5 | 11 | 25 | 0.0055 | 9.9 | 0.0180 | 12.9 | 0.303 | 0.146 | 0.126 | 0.04 |
| 50 | 0.0060 | 7.1 | 0.0270 | 7.1 | 0.000 | 0.168 | 0.162 | 0.05 | ||
| 75 | 0.0072 | 3.5 | 0.0390 | 3.7 | 0.057 | 0.209 | 0.186 | 0.04 | ||
For the central tumor 75% threshold, Ktrans (mean ± SD) measured with DCE-MRI was 0.0630 ± 0.0253, and the corresponding [14C]AIB autoradiography Ki measure (mean ± SD) was 0.0209 ± 0.0104. The fractional extravascular-extracellular space ve (mean ± SD) was 17.6 ± 3.2%. The fractional blood volume (mean ± SD) was 2.80 ± 1.3%. Mean DCE-MRI parameter estimates were comparable to published measures for central tumor regions obtained using radiolabeled serum albumin for ve and fbv (7.5% and 3.2%, respectively) [15,21]. DCE-MRI experimental measures of ve were 2.3-fold higher than published measures for RG2 tumors. For fractional blood volume (fbv), the percentile difference between experimental measures and published measures in RG2 tumors was only 12.5%.
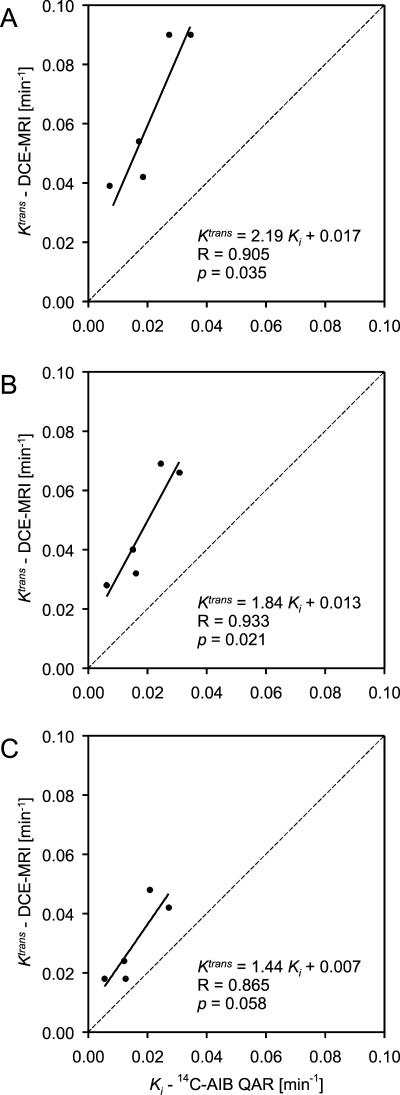
DCE-MRI Ktrans and [14C]AIB QAR Ki maps display the variation and range of influx rate constants measured across the tumors (Figure 1). Ktrans and Ki heterogeneity varied together on a pixel-by-pixel basis. DCE-MRI Ktrans versus [14C]AIB QAR Ki were plotted for the 75%, 50%, and 25% threshold ROI for central tumor slices (Figure 2). For the central 75% threshold, Ktrans and Ki influx rate constants were positively correlated (slope ± SEM = 2.190 ± 0.594; R = 0.905; R2 = 0.819; adjusted R2 = 0.759; F = 13.6; P = .034). For the central 50% threshold, the slope decreased, but R increased (slope ± SEM = 1.838 ± 0.411; R = 0.933; R2 = 0.870; adjusted R2 = 0.826; F = 20.0; P = .021).
Figure 2.
DCE-MRI Ktrans versus [14C]AIB QAR Ki for central tumor slices for the (A) 75%, (B) 50%, and (C) 25% threshold ROI.
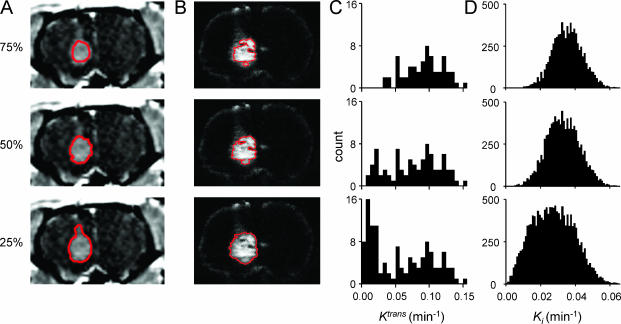
Tumor ROI incorporating increasing proportions of the tumor edge were analyzed to study the impact of tumor periphery on DCE-MRI parameter estimates. Examples of ROI and corresponding histograms of Ktrans voxelwise data and Ki pixelwise data are shown for animal 1 (Figure 3). Despite distortions related to tissue sectioning, the strategy employed for ROI selection was qualitatively accurate in selecting the same tumor tissue in both [14C]AIB QAR autoradiographs and DCE-MRI maps. Lowering the ROI threshold resulted in larger ROI with more tumor periphery. Histograms of Ki from QAR data were unimodal for all three ROI thresholds. Histograms of Ktrans from DCE-MRI data were unimodal for the central tumor 75% threshold, but were bimodal for the 25% and 50% thresholds with more tumor periphery. This “noise,” which is seen as a secondary high-amplitude peak of low Ktrans on histograms of DCE-MRI voxelwise data, can be qualitatively appreciated as a rim of low Ktrans values at tumor edges in Ktrans maps (Figure 1).
Figure 3.
Sample images of tumor ROI and corresponding histograms for animal 1. (A) Images of DCE-MRI ROI (white) for the 25%, 50%, and 75% thresholds. (B) Images of corresponding [14C]AIB QAR ROI (white). (C) Histograms of DCE-MRI Ktrans voxelwise data for the corresponding DCE-MRI ROI threshold with polynomial fit trendlines. (D) Histograms of [14C]AIB QAR Ki pixelwise data for the corresponding [14C]AIB QAR ROI threshold.
A comparison of influx rate constants derived from DCE-MRI and QAR using different ROI demonstrated a systematic decrease in Ktrans relative to Ki as more of the tumor periphery was included in tumor ROI. The slope of regression line (slope ± SEM) for Ktrans versus Ki decreased systematically from 2.190 ± 0.594, to 1.838 ± 0.411, to 1.443 ± 0.483 for the 75%, 50%, and 25% threshold ROI, respectively. For the 25% threshold ROI that included the highest proportion of tumor periphery, the correlation between Ktrans and Ki measures was not statistically significant (R = 0.865; R2 = 0.749; adjusted R2 = 0.665; F = 8.94; P = .058).
DCE-MRI Ktrans Reproducibility
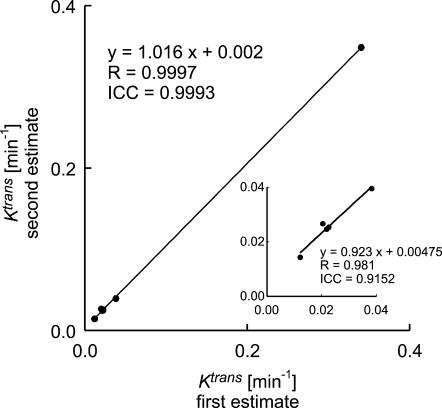
There was positive reproducibility of Ktrans estimates for repeat DCE-MRI experiments on a separate population of animals with implanted RG2 tumors (n = 6) (Figure 4). The mean (± SD) Ktrans for the first test was 0.0759 ± 0.130, and the mean (± SD) Ktrans for the second test was 0.0798 ± 0.139. Test-retest Ktrans estimates had an ICC of 0.9993 and a regression coefficient (R) of 0.9997 (P < .001). The CoV was 10.2%, and the wSD was 0.00323. The 95% confidence interval for measurement error was ± 20.1% or ± 0.00633.
Figure 4.
Test-retest Ktrans estimates from repeat DCE-MRI experiments for all subjects and excluding the highest Ktrans data point (inset).
Given that there was systematic offset in the data, it was alternately analyzed to more closely approximate “true” measurement error. The second Ktrans estimate was consistently higher than the first estimate (mean ± SEM = 0.00385 ± 0.0011). Closely timed studies were required in these fast-growing tumors, and this systematic offset was likely due to the effects of residual contrast in tumors on the second day of Gd-DTPA injection, but could have been caused also by tumor changes. According to accepted statistical methods, systematic changes between scans can be accounted for by subtracting the mean difference between subjects' scans from the second scan values [18]. After adjusting for systematic offset, the CoV was 6.07%, and the wSD was 0.00174. The 95% confidence interval for a given measurement was ± 11.9% or ± 0.00341. The repeatability (r) was 16.8% or 0.00482. Therefore, a difference of 16.8% or 0.00482 in two independent tumor measurements using this method in a single tumor represents a true tumor change in Ktrans.
Reproducibility results were similar for the analysis of data from five animals and excluding the high Ktrans value. The data, excluding this high value, (Figure 4, inset) are on virtually the same line as the data for all six animals. The ICC is lower due to the decreased range in data, with a value of 0.9152. After adjusting for systematic offset, however, the CoV and wSD for five animals were very similar to the results for the six animals. The CoV was 6.63%, and the wSD was 0.00135. The 95% confidence interval for a given measurement was ± 13.0% or ± 0.00265. The repeatability (r) was 18.4% or 0.00374.
Discussion
DCE-MRI is in wide clinical use and has proven useful in detecting early changes in vascularity after angiogenic inhibitor treatment. Wedam et al. [22] used methods similar to this DCE-MRI method, with Gd-DTPA and the same IDL software package, to demonstrate changes in Ktrans in breast cancer patients during treatment with bevacizumab, a vascular endothelial growth factor inhibitor. However, in spite of its wide use, the interpretation of Ktrans values is still uncertain. The results of the current study demonstrate a significant correlation between DCE-MRI-derived Ktrans and [14C]AIB QAR-derived Ki in the RG2 tumor model. [14C]AIB QAR is a long-established quantitative method of measuring vascular leakage for small molecules, and this result is therefore useful in confirming that Ktrans is highly related to vascular permeability, at least in some animal tumor models. Thus, although offset by a factor of roughly 2 in absolute value, DCE-MRI estimates are predictors of “gold standard” measurements of vascular permeability. Differences in the biophysical properties of AIB and DTPA likely account for these small differences. Permeability is a function of the characteristics of the molecule transiting the membrane, such as surface charge, molecular weight, ionization, lipophilicity, and shape. Gd- DTPA (547 Da) is a charged and extremely hydrophilic molecule, whereas AIB (102 Da) is a synthetic amino acid that is neutral and hydrophilic [7,23]. Thus, within practical experimental limits, these two agents produced remarkably similar results.
Although there was a strong correlation between Ktrans and Ki in central tumor regions, the correlation was weaker when the periphery of the tumor was included. There was a measured decrease in [14C]AIB QAR Ki values at RG2 tumor edges consistent with prior results for the RG2 model, and partial volume effects of normal and tumor tissues near the edge are expected to lower DCE-MRI Ktrans [24]. The low Ktrans values seen at the tumor edge were thus likely due to partial volume effects. RG2 tumors are also known to be edematous [25], and it is possible that this effect contributed to the lowering of Ktrans values at tumor edges. The effect of variance in Ktrans relative to Ki as more of the periphery is included in the measurement is reflected in the change in slope in the line of regression (Figure 2). The two-fold decrease in Ktrans values between the 75% threshold and the 25% threshold demonstrates the impact of tumor edge “noise” and the need for strategies of ROI selection that avoid these edge effects. The impact of partial volume effects on tumor edges and necrotic regions, however, is expected to be lower in human studies, which generally include tumors larger than those evaluated in this model.
Measures of Ktrans obtained using DCE-MRI proved highly reproducible in this study. A high correlation was observed with two independent repeated measurements. This has implications for the magnitude of the therapeutic effect required to be measurable by DCE-MRI. Galbraith et al. demonstrated that “failed” pixel data had to be removed to reduce interscan variability and that a relatively large variance was required to show statistical changes in human tumors even after data exclusion. Changes of -45% to 83% were needed to overcome the CoV of 24% to 29% in conventional studies [19]. Liu et al. [26] thus used a decrease of > 50% in Ktrans to detect a real change in advanced tumors treated with the angiogenic inhibitor AG-013736. Recent studies of reproducibility in human abdominal tumors, gliomas, and prostate malignancies using an assumed VIF showed similar results requiring changes of > 31% to > 44% (CoV = 11–16%) in one study and a measurement error of > 16% (CoV = 8.2%) in another [20,27]. The ICC, CoV, and 95% confidence interval for measurement error in this study compared favorably to prior results. The improved reproducibility in our model can be traced to improved VIFs, reliable pulse sequences, and a two-compartment model with a sufficient number of parameters to fit most of the pixels within the tumor. Reproducibility results from clinical and preclinical DCE-MRI are, however, highly variable. Such studies often reported pixel “failures” (i.e., inability to fit the data to the model) and reported pixel exclusion ranging between 13% and 74% [19,28]. However, the DCE-MRI method used in this study required no filtering or removal of pixel data within the ROI. This is likely because the modeling allowed better fitting of measured data and because tightly controlled experimental conditions led to robust data sets. Although one study reported a CoV of 7.7% after significant pixel data exclusion (mean = 39%) [28], we were able to achieve a 95% confidence interval of an individual Ktrans measure of ± 0.00341 (± 11.9%) (ICC = 0.9993; CoV = 6.07%) with no pixel data exclusion. The precision of DCE-MRI was predictable from strong correlation results with [14C]AIB QAR Ki measures because the ability to detect a correlation is related to the precision of independent measures. Although a correlation is possible with “noisy” data using a large number of experiments, statistical correlation was demonstrated in this study using only a small number of animals suggesting a relatively high precision of the DCE-MRI method.
The successful use of a VIF certainly contributed to the robustness of this technique. Direct measurement of arterial blood was not possible in this model given the small size of vessels in small animals, and the VIF was instead measured from venous sinus blood. A similar technique using measurements of VIF from the sagittal sinus has been used in human DCE-MRI studies [28]. Although several groups have used slow injections in pharmacokinetic modeling [29], it is argued that a bolus injection is needed for accurate mathematical modeling of Gd-DTPA kinetics [5,28,30]. The results of this study suggest that a slow bolus allows for accurate and precise parameter estimates. Furthermore, a fast bolus requires rapid data acquisition with reduced signal-to-noise ratios to accurately measure the VIF. Finally, a longer acquisition interval allows for higher-resolution images, which are advantageous for clinical studies. Recent simulations and experimental data using this DCE-MRI method suggest that there is no significant difference in Ktrans estimates using a slow injection versus a bolus injection in patient studies (unpublished observations). If correct, a slow injection may be more satisfactory for DCE-MRI in the clinical setting than was originally thought because high resolution is often needed.
In this experiment, it was not possible to correlate other DCE-MRI parameters, such as kep, with [14C]AIB QAR because the latter only measures the forward leakage rate. Two other free parameters were estimated from the best-fit solution of the GKM: fbv and ve. Although parameter estimates of fractional blood (fbv) and leakage space volumes (ve) were comparable to published measures using other techniques for RG2 tumors, leakage space estimates showed greater variation from prior published values. Zhou et al. [31] have shown that measures of ve obtained using a method similar to that used in this study can vary based on the dosing of Gd-DTPA when corrections are not made to account for the effects of transcytolemmal water exchange on tissue relaxivity. Based on these results, the values of ve obtained in this study would be expected to have varied for different doses of Gd-DTPA. It may, therefore, be necessary to make such corrections to obtain accurate DCE-MRI ve measures.
This study has a number of limitations. We cannot discount the possibility that the good correlation between Ktrans and Ki may be related to the nature of the RG2 model. In vessels where flow is high relative to permeability, such as in the RG2 model, the rate of influx of AIB and Gd-DTPA from the vessel to the tumor is a function of vascular permeability [2,8,9]. In tumor regions with lower blood flow, Ktrans may be more dependent on flow than on permeability and, therefore, may not accurately depict vascular permeability. A broader range of tumor models is needed to confirm the validity of DCE-MRI parameters for predicting vascular permeability across a range of tumor blood flow rates. By examining Ktrans estimates in a permeability-limited high-flow tumor model, this study allowed Ktrans estimates to be simplified to indices of permeability—the parameter that is potentially most valuable for monitoring treatment effects on angiogenic tumor vasculature.
Additional limitations of this study include technical challenges in comparing in vivo MRI and invasive QAR methods. There was considerable distortion of tissues using the [14C]AIB QAR technique due to shearing, warping, and tearing, and matching DCE-MRI and [14C]AIB autoradiograph tissue regions was technically difficult. The strategy employed, including the use of large computer-generated ROI and central tumor slices, reduced misregistration error.
In the future, macromolecular contrast agents may be better measures of permeability by separating flow effects from Ktrans indices of permeability [32–34]. This effect would be most prominent in areas with a high permeability-to-flow ratio. Macromolecular agents have been used in preclinical DCE-MRI tumor studies, but these techniques are not yet ready for clinical studies because these macromolecular agents have not yet been approved by regulatory agencies for human use and because techniques used in these studies, such as ex vivo VIF measurements, are not easily applied to clinical studies [35]. QAR validation of macromolecular contrast agents will require a radioisotopically labeled macromolecule of similar hydrodynamic diameter, hydrophilicity, and surface charge as the MR-based macromolecular agent. The advantages of using low-molecular-weight Gd- DTPA versus higher-molecular-weight agents include the increased penetration of smaller compounds in areas of decreased permeability and the general availability and established safety of Gd-DTPA.
In summary, this study demonstrates a statistical correlation between direct measures of the influx (or forward leakage) rate constant obtained using DCE-MRI (K trans) and direct measures of the influx (or forward leakage) rate constant obtained using the “gold standard” method [14C]AIB QAR (Ki).
Acknowledgements
The authors thank Matthew McAuliffe, Paul Morrison, and Kevin O'Brien for their assistance with this study.
Abbreviations
- Ktrans
influx (or forward leakage) rate constant DCE-MRI-derived
- Ki
influx (or forward leakage) rate constant, [14C]AIB QAR-derived
- [14C]AIB QAR
[14C]aminoisobutyric acid quantitative autoradiography
- fbv
fractional blood volume
- ve
fractional leakage space (or fractional extravascular-extracellular space) volume
- VIF
curve describing contrast concentration in large vessels as a function of time (or vascular input function)
Appendix
List of Terms
- Ce(t)
Extravascular-extracellular space (or leakage space) tracer concentration
- Ktrans
DCE-MRI influx rate constant (or volume transfer constant)
- Cp
Plasma tracer concentration
- ve
Fractional extravascular-extracellular space (or leakage space) volume
- Ct
Tissue tracer concentration
- ⊗
Convolution
- Cb(t)
Blood tracer concentration per time; VIF
- fbv
Fractional blood volume
- fpv
Fractional plasma volume
- Vrbc
Red blood cell volume
- Vroi
Volume of VIF
Derivation of GKM Bidirectional Pharmacokinetic Model
Convolution equation for bidirectional exchange between plasma and leakage space (extravascular-extracellular space)
| 1 |
| 2 |
| 3 |
Plasma concentration as a function of blood concentration
| 4 |
| 5 |
| 6 |
Substitution of plasma concentration for blood concentration
| 7 |
Balanced equation for the total number of particles in tissue ROI or voxel
| 8 |
| 9 |
Therefore:
References
- 1.Knopp MV, Giesel FL, Marcos H, von Tengg-Kobligk H, Choyke P. Dynamic contrast-enhanced magnetic resonance imaging in oncology. Top Magn Reson Imaging. 2001;12(4):301–308. doi: 10.1097/00002142-200108000-00006. [DOI] [PubMed] [Google Scholar]
- 2.Choyke PL, Dwyer AJ, Knopp MV. Functional tumor imaging with dynamic contrast-enhanced magnetic resonance imaging. J Magn Reson Imaging. 2003;17(5):509–520. doi: 10.1002/jmri.10304. [DOI] [PubMed] [Google Scholar]
- 3.Jordan BF, Runquist M, Raghunand N, Baker A, Williams R, Kirkpatrick L, Powis G, Gillies RJ. Dynamic contrast-enhanced and diffusion MRI show rapid and dramatic changes in tumor microenvironment in response to inhibition of HIF-1alpha using PX-478. Neoplasia. 2005;7(5):475–485. doi: 10.1593/neo.04628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Padhani AR, Husband JE. Dynamic contrast-enhanced MRI studies in oncology with an emphasis on quantification, validation and human studies. Clin Radiol. 2001;56(8):607–620. doi: 10.1053/crad.2001.0762. [DOI] [PubMed] [Google Scholar]
- 5.Yankeelov TE, Rooney WD, Li X, Springer CS., Jr Variation of the relaxographic “shutter-speed” for transcytolemmal water exchange affects the CR bolus-tracking curve shape. Magn Reson Med. 2003;50(6):1151–1169. doi: 10.1002/mrm.10624. [DOI] [PubMed] [Google Scholar]
- 6.McPhail LD, McIntyre DJ, Ludwig C, Kestell P, Griffiths JR, Kelland LR, Robinson SP. Rat tumor response to the vasculardisrupting agent 5,6-dimethylxanthenone-4-acetic acid as measured by dynamic contrast-enhanced magnetic resonance imaging, plasma 5-hydroxyindoleacetic acid levels, and tumor necrosis. Neoplasia. 2006;8(3):199–206. doi: 10.1593/neo.05739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blasberg RG, Fenstermacher JD, Patlak CS. Transport of alpha-aminoisobutyric acid across brain capillary and cellular membranes. J Cereb Blood Flow Metab. 1983;3(1):8–32. doi: 10.1038/jcbfm.1983.2. [DOI] [PubMed] [Google Scholar]
- 8.Groothuis DR, Fischer JM, Pasternak JF, Blasberg RG, Vick NA, Bigner DD. Regional measurements of blood-to-tissue transport in experimental RG-2 rat gliomas. Cancer Res. 1983;43(7):3368–3373. [PubMed] [Google Scholar]
- 9.Groothuis DR, Pasternak JF, Fischer JM, Blasberg RG, Bigner DD, Vick NA. Regional measurements of blood flow in experimental RG-2 rat gliomas. Cancer Res. 1983;43(7):3362–3367. [PubMed] [Google Scholar]
- 10.Kobayashi N, Allen N, Clendenon NR, Ko LW. An improved rat brain-tumor model. J Neurosurg. 1980;53(6):808–815. doi: 10.3171/jns.1980.53.6.0808. [DOI] [PubMed] [Google Scholar]
- 11.Asotra K, Ningaraj N, Black KL. Measurement of blood-brain and blood-tumor barrier permeabilities with [14C]-labeled tracers. Methods Mol Med. 2003;89,:177–190. doi: 10.1385/1-59259-419-0:177. [DOI] [PubMed] [Google Scholar]
- 12.Deoni S, Josseau MJ, Rutt BK, Peters TM. Visualization of thalamic nuclei on high resolution, multi-averaged T1 and T2 maps acquired at 1. 5 T. Hum Brain Mapp. 2005;25(3):353–359. doi: 10.1002/hbm.20117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deoni SC, Rutt BK, Peters TM. Rapid combined T1 and T2 mapping using gradient recalled acquisition in the steady state. Magn Reson Med. 2003;49(3):515–526. doi: 10.1002/mrm.10407. [DOI] [PubMed] [Google Scholar]
- 14.Iannotti F, Fieschi C, Alfano B, Picozzi P, Mansi L, Pozzilli C, Punzo A, Del Vecchio G, Lenzi GL, Salvatore M, et al. Simplified, noninvasive PET measurement of blood-brain barrier permeability. J Comput Assist Tomogr. 1987;11(3):390–397. doi: 10.1097/00004728-198705000-00004. [DOI] [PubMed] [Google Scholar]
- 15.Nakagawa H, Groothuis DR, Owens ES, Patlak CS, Pettigrew KD, Glasberg RR. Dexamethasone effects on vascular volume and tissue hematocrit in experimental RG-2 gliomas and adjacent brain. J Neuro-Oncol. 1988;6(2):157–168. doi: 10.1007/BF02327392. [DOI] [PubMed] [Google Scholar]
- 16.Bland JM, Altman DG. Measurement error. BMJ. 1996;313(7059):744. doi: 10.1136/bmj.313.7059.744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bland JM, Altman DG. Measurement error and correlation coefficients. BMJ. 1996;313(7048):4–42. doi: 10.1136/bmj.313.7048.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bland JM, Altman DG. Measuring agreement in method comparison studies. Stat Methods Med Res. 1999;8(2):135–160. doi: 10.1177/096228029900800204. [DOI] [PubMed] [Google Scholar]
- 19.Galbraith SM, Lodge MA, Taylor NJ, Rustin GJ, Bentzen S, Stirling JJ, Padhani AR. Reproducibility of dynamic contrast-enhanced MRI in human muscle and tumours: comparison of quantitative and semi-quantitative analysis. NMR Biomed. 2002;15(2):132–142. doi: 10.1002/nbm.731. [DOI] [PubMed] [Google Scholar]
- 20.Roberts C, Issa B, Stone A, Jackson A, Waterton JC, Parker GJ. Comparative study into the robustness of compartmental modeling and model-free analysis in DCE-MRI studies. J Magn Reson Imaging. 2006;23(4):554–563. doi: 10.1002/jmri.20529. [DOI] [PubMed] [Google Scholar]
- 21.Nakagawa H, Groothuis DR, Owens ES, Fenstermacher JD, Patlak CS, Blasberg RG. Dexamethasone effects on [125I]albumin distribution in experimental RG-2 gliomas and adjacent brain. J Cereb Blood Flow Metab. 1987;7(6):687–701. doi: 10.1038/jcbfm.1987.123. [DOI] [PubMed] [Google Scholar]
- 22.Wedam SB, Low JA, Yang SX, Chow CK, Choyke P, Danforth D, Hewitt SM, Berman A, Steinberg SM, Liewehr DJ, et al. Antiangiogenic and antitumor effects of bevacizumab in patients with inflammatory and locally advanced breast cancer. J Clin Oncol. 2006;24(5):769–777. doi: 10.1200/JCO.2005.03.4645. [DOI] [PubMed] [Google Scholar]
- 23.Weinmann HJ, Brasch RC, Press WR, Wesbey GE. Characteristics of gadolinium-DTPA complex: a potential NMR contrast agent. AJR Am J Roentgenol. 1984;142(3):619–624. doi: 10.2214/ajr.142.3.619. [DOI] [PubMed] [Google Scholar]
- 24.Aas AT, Brun A, Blennow C, Stromblad S, Salford LG. The RG2 rat glioma model. J Neuro-Oncol. 1995;23(3):175–183. doi: 10.1007/BF01059948. [DOI] [PubMed] [Google Scholar]
- 25.Tjuvajev J, Gansbacher B, Desai R, Beattie B, Kaplitt M, Matei C, Koutcher J, Gilboa E, Blasberg R. RG-2 glioma growth attenuation and severe brain edema caused by local production of interleukin-2 and interferon-gamma. Cancer Res. 1995;55(9):1902–1910. [PubMed] [Google Scholar]
- 26.Liu G, Rugo HS, Wilding G, McShane TM, Evelhoch JL, Ng C, Jackson E, Kelcz F, Yeh BM, Lee FT, Jr, et al. Dynamic contrastenhanced magnetic resonance imaging as a pharmacodynamic measure of response after acute dosing of AG-013736, an oral angiogenesis inhibitor, in patients with advanced solid tumors: results from a phase I study. J Clin Oncol. 2005;23(24):5464–5473. doi: 10.1200/JCO.2005.04.143. [DOI] [PubMed] [Google Scholar]
- 27.Walker-Samuel S, Parker CC, Leach MO, Collins DJ. Reproducibility of reference tissue quantification of dynamic contrastenhanced data: comparison with a fixed vascular input function. Phys Med Biol. 2007;52(1):75–89. doi: 10.1088/0031-9155/52/1/006. [DOI] [PubMed] [Google Scholar]
- 28.Jackson A, Jayson GC, Li KL, Zhu XP, Checkley DR, Tessier JJ, Waterton JC. Reproducibility of quantitative dynamic contrastenhancedMRI in newly presenting glioma. Br J Radiol. 2003;76(903):153–162. doi: 10.1259/bjr/70653746. [DOI] [PubMed] [Google Scholar]
- 29.Brix G, Semmler W, Port R, Schad LR, Layer G, Lorenz WJ. Pharmacokinetic parameters in CNS Gd-DTPA enhanced MR imaging. J Comput Assist Tomogr. 1991;15(4):621–628. doi: 10.1097/00004728-199107000-00018. [DOI] [PubMed] [Google Scholar]
- 30.Henderson E, Rutt BK, Lee TY. Temporal sampling requirements for the tracer kinetics modeling of breast disease. Magn Reson Imaging. 1998;16(9):1057–1073. doi: 10.1016/s0730-725x(98)00130-1. [DOI] [PubMed] [Google Scholar]
- 31.Zhou R, Pickup S, Yankeelov TE, Springer CS, Jr, Glickson JD. Simultaneous measurement of arterial input function and tumor pharmacokinetics in mice by dynamic contrast enhanced imaging: effects of transcytolemmal water exchange. Magn Reson Med. 2004;52,(2):248–257. doi: 10.1002/mrm.20143. [DOI] [PubMed] [Google Scholar]
- 32.Barrett T, Kobayashi H, Brechbiel M, Choyke PL. Macromolecular MRI contrast agents for imaging tumor angiogenesis. Eur J Radiol. 2006;60(3):353–366. doi: 10.1016/j.ejrad.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 33.Turetschek K, Roberts TP, Floyd E, Preda A, Novikov V, Shames DM, Carter WO, Brasch RC. Tumor microvascular characterization using ultrasmall superparamagnetic iron oxide particles (USPIO) in an experimental breast cancer model. J Magn Reson Imaging. 2001;13(6):882–888. doi: 10.1002/jmri.1126. [DOI] [PubMed] [Google Scholar]
- 34.de Lussanet QG, Langereis S, Beets-Tan RG, van Genderen MH, Griffioen AW, van Engelshoven JM, Backes WH. Dynamic contrast-enhanced MR imaging kinetic parameters and molecular weight of dendritic contrast agents in tumor angiogenesis in mice. Radiology. 2005;235(1):65–72. doi: 10.1148/radiol.2351040411. [DOI] [PubMed] [Google Scholar]
- 35.Marzola P, Degrassi A, Calderan L, Farace P, Crescimanno C, Nicolato E, Giusti A, Pesenti E, Terron A, Sbarbati A, et al. In vivo assessment of antiangiogenic activity of SU6668 in an experimental colon carcinoma model. Clin Cancer Res. 2004;10(2):739–750. doi: 10.1158/1078-0432.ccr-0828-03. [DOI] [PubMed] [Google Scholar]