Abstract
We identified the ubiquitin-conjugating enzyme E2-EPF mRNA as differentially expressed in breast tumors relative to normal tissues and performed studies to elucidate its putative role in cancer. We demonstrated that overexpression of E2-EPF protein correlated with estrogen receptor (ER) negativity in breast cancer specimens and that its expression is cell cycle-regulated, suggesting a potential function for E2-EPF in cell cycle progression. However, reduction of E2-EPF protein levels by > 80% using RNAi had no significant effects on the proliferation of HeLa cervical cancer cells or ER- MDA-MB-231 or MDA-MB-453 breast cancer cells. Because E2-EPF protein levels were elevated during the G2/M phase of the cell cycle and because E2-EPF mRNA in tumor specimens was frequently coexpressed with genes involved in cell cycle control, spindle assembly, and mitotic surveillance, the possibility that E2-EPF might have a function in the cellular response to agents that induce a G2 checkpoint or an M checkpoint was investigated. E2-EPF knockdown sensitized HeLa cells to the topoisomerase (topo) II inhibitors etoposide and doxorubicin and also increased topo IIα protein levels. These data suggest that combined administration of topo II-directed drugs and E2-EPF inhibitors may enhance their clinical effectiveness.
Keywords: E2-EPF, etoposide, doxorubicin, topoisomerase II, breast cancer
Introduction
In the past decade, DNA microarrays have been employed to study the gene expression profiles of normal and tumorderived specimens from many different tissues, thereby resulting in an exponential increase in the amount of information available to aid our understanding of tumorigenesis and to provide hypotheses for therapeutic intervention. Using a combination of expressed sequence tag (EST) libraries from normal and tumor tissues (i.e., Incyte LifeSeq Foundation database) and DNA microarray gene expression profiles from normal breast and infiltrating ductal carcinomas, we identified the ubiquitin-conjugating enzyme E2-EPF as differentially expressed in breast cancer. Additionally, we noted that the E2-EPF gene was listed as a component of metasignatures for both neoplastic transformation and undifferentiated cancers in a recent meta-analysis of array hybridization data extracted from 21 published data sets encompassing 12 different cancers [1]. Those data indicated that E2-EPF was significantly differentially expressed in multiple cancers, and they enhanced our interest in E2-EPF as a candidate therapeutic target for cancer drug discovery.
E2-EPF is a 24-kDa protein that is a member of the E2 family of ubiquitin-conjugating enzymes [2], which, together with an E1 ubiquitin-activating enzyme and an E3 ubiquitin ligase, catalyze the addition of ubiquitin to substrate proteins (for review, see Pickart [3]). Multiple rounds of ubiquitinylation result in substrate polyubiquitinylation that can target that protein for proteasome-dependent destruction. Although discovered in 1992 [2], candidate substrates and cognate E3 ligases for E2-EPF were unknown until a recent report from Jung et al. [4], who demonstrated that the stability of a von Hippel-Lindau (VHL) tumor-suppressor gene product is dependent on E2-EPF levels. Their study suggested a role for E2-EPF in the stabilization of hypoxia-inducible factor 1α (HIF-1α) under normoxic conditions and in the proliferation and invasion of renal cancer and melanoma cells, but did not address its role in other cancer types.
Because E2-EPF is a component of the meta-signature for undifferentiated cancer that includes genes in the tumor “proliferation signature” (reviewed in Whitfield et al. [5]) comprising many cell cycle-regulated genes, we searched for and found the E2-EPF gene annotated as an M/G1 phase-expressed gene in a comprehensive HeLa cell cycle gene expression analysis [6]. The observed increase in E2-EPF expression in multiple cancers might, therefore, be due to its importance in cell cycle progression, as has been shown for other genes in the proliferation signature. Here, we report the expression profile for E2-EPF in breast tumors and normal tissue specimens and address the possibility that it has an essential role in cancer cell proliferation using RNAi to reduce E2-EPF expression levels. Despite a substantial reduction of E2-EPF protein levels in HeLa cells, as well as in MDA-MB-231 and MDA-MB-453 breast cancer cells, no effects on cell proliferation were observed. Because E2-EPF protein levels were elevated during the G2/M phase of the cell cycle and because E2-EPF mRNA was frequently coexpressed in tumors with genes involved in mitotic surveillance, the possibility that E2-EPF might have a function in cellular response to agents that induce a G2 checkpoint or an M checkpoint was investigated. E2-EPF knockdown had no effect on the sensitivity of HeLa cells to chemotherapeutic agents such as the microtubule stabilizer Taxol or the topoisomerase (topo) I inhibitor camptothecin, but antiproliferative response to the topo II inhibitors etoposide and doxorubicin was significantly enhanced.
Materials and Methods
Chemicals and Reagents
Camptothecin, paclitaxel (Taxol), doxorubicin, etoposide, and nocodazole were all purchased from Sigma (St. Louis, MO) and solubilized in dimethylsulfoxide. The following antibodies were used: polo kinase-like 1 (PLK1; cat. no. 06-813) antibody (Upstate Cell Signaling Solutions, Lake Placid, NY); topo 2A/B (cat. no. 4734; Cell Signaling, Danvers, MA); and cyclin A (cat. no. H-432; Santa Cruz Biotech, Santa Cruz, CA). SiRNA purchased from Ambion, Inc. (Austin, TX), included those for E2-EPF targeting the following sequences: 5′-GGTCTTTCCCAACGAGGAG-3′ (no. 1), 5′-GGTCTGTTCCGCATGAAAC-3′ (no. 2), and 5′-GGGCTACTTCCTGACCAAG-3′ (no. 3), as well as those for Eg5 (5′-CUGAAGACCUGAAGACAAU-3′) and in irrelevant sequence used as a negative control NC (5′-CCUACAACAUAGCUACUCA-3′). Qiagen (Valencia, CA) was the supplier of Firefly luciferase (FFL) siRNA (5′-AACTTACGCTGAGTACTTC-3′). Dharmacon (Lafayette, CO) was the supplier of siRNA targeting FFL2 E2-EPF sequences: 5′-ATAAGAAGCTGGCGGCCAA-3′ (D1), 5′-ACAAGGAGGTGACGACACT-3′ (D2), 5′-GCCAAGAAAAAGACGGACA-3′ (D3), 5′-CATGCTGGCGAGCGCGATA-3′ (D4), 5′-ATAAGAAGCTGGCGGCCAA-3′ (D5), 5′-ACAAGGAGGTGACGACACT-3′ (D6), GCAAGAAAAAGACGGACA (D7), and CATGCTGGCGAGCGCGATA (D8), with D5 to D8 corresponding to their Plus siRNA. NT and MYOG (cat. no. J-010029-05), nontargeting and against myogenin, respectively, are Plus negative control siRNA. E2-EPFp (i.e., D5 + D6 + D7 + D8), NTp, and MYOGp are Smart Pool Plus siRNA purchased from Dharmacon.
Cell Lines and Culture Conditions
The human cervical (HeLa) and breast (MDA-MB-453 and MDA-MB-231) cancer cell lines were obtained from the American Type Culture Collection (Manassas, VA). HeLa cells were cultured in Dulbecco's modified Eagle's medium containing 1 mM glutamine and 10% fetal calf serum, and breast cancer cells were cultured as previously described [7]. This study employed the MDA-231-MT-1 derivative of MDA-MB-231 cells (herein referred to as MDA-231) that was established from a rapidly growing athymic nude mouse xenograft [7].
Clinical Specimens
Samples of human breast and breast cancer specimens were supplied by US Biomax, Inc. (Rockville, MD), and the Cooperative Human Tissue Network (CHTN), Western Division (Cleveland, OH). From US Biomax, Inc., we received glass slides of a formalin-fixed paraffin-embedded (FFPE) tissue microarray containing 70 cores (each with a diameter of 1.5 mm) of infiltrating ductal carcinoma (cat. no. BR701). From the CHTN, we received FFPE blocks of 22 normal breast tissues, 11 normal adjacent to tumor (NAT) tissues, and 10 breast carcinoma tissues. Tumor histologic grades, as well as the status of estrogen receptor (ER) and progesterone receptor (PR), were provided by vendors on 65 tissue array cores and 2 breast carcinoma samples from the CHTN.
E2-EPF Polyclonal Antibody Generation
Polyclonal antibodies were generated by immunization of rabbits with a KLH conjugate of peptide GERDKKLAAKKK corresponding to amino acids 201 to 212 of the 222-amino acid protein (Swiss Prot Q16763). Antibodies were purified using protein A affinity chromatography.
E2-EPF mRNA Quantitation by Real-Time Reverse Transcription-Polymerase Chain Reaction (RT-PCR) Analysis
Total RNA from breast tumor specimens was purchased from Biochain (Hayward, CA) or Asterand, Inc. (Detroit, MI), and TaqMan PCR was performed with 50 ng of RNA per sample in duplicate reactions using a TaqMan universal master mix (Applied Biosystems, Foster City, CA) with PCR primers for E2-EPF (forward: 5′-CGACCTCCAGGTCACCAT-3′; reverse: 5′-GGAACAGACCTCCAGCATATGG-3′) and with a probe (5′-CCCCTCAGGGCCCTC-3′) synthesized by Applied Biosystems. Thermal cycling was initiated at 48°C for 30 minutes for cDNA synthesis, followed by a denaturation step for 10 minutes at 95°C and 40 cycles performed in two steps: 15 seconds at 95°C and 1 minute at 60°C. CT values of samples were used to calculate mRNA amounts, and relative mRNA expression data were normalized to 18S ribosomal RNA (primers and probe from Applied Biosystems).
E2-EPF Protein Immunohistochemistry
Sections were cut into 5 µm thickness, deparaffinized, and rehydrated. Following antigen retrieval, sections were stained using an automated instrument (Autostainer; Dako-Cytomation, Carpinteria, CA). Briefly, sections were treated with a peroxidase block, followed by 10% normal goat serum, and incubated in E2-EPF primary antibody (5 µg/ml) for 45 minutes at room temperature. For negative control siRNA, rabbit IgG was used as the primary antibody. Secondary rabbit antibody (EnVision+ System HRP; DakoCytomation) was applied, followed by diaminobenzidine (Liquid DAB+; DakoCytomation), and counterstained with hematoxylin (DakoCytomation). E2-EPF antibody specificity was confirmed by competition with the corresponding E2-EPF peptide, but not with an irrelevant peptide. Additional confirmation of antibody specificity for E2-EPF was demonstrated by a reduction in E2-EPF staining in siRNA-treated HeLa cells.
For histologic evaluation of the amount of E2-EPF staining, the pathologist was blinded to ER and PR status. Staining intensity was relatively similar in all sections, so it was not used in the evaluation of protein expression. The percentage of tumor cells with positive staining was grouped as follows: 0 (0%); 1 (1–5%); 2 (5–25%); 3 (25–50%); 4 (> 50%). Fisher's exact test was applied to assess the statistical significance of the associations between expressions of E2-EPF and ER using JMP version 5.1 (SAS, Cary, NC).
Identification of E2-EPF Coexpressed Genes (ECGs) from the ONCOMINE Database
The search tools provided with the ONCOMINE database (Compendia Bioscience, Ann Arbor, MI) were employed in the acquisition of studies and lists of coexpressed genes. Each microarray study in ONCOMINE is clustered by standard average linkage hierarchical clustering. For query genes, each study with a cluster intracorrelation of > 0.65 was used in our analysis, with the exclusion of studies focused on hematologic malignancies, normal tissues, or cell lines. The numbers of studies retrieved were as follows: 20 (E2-EPF), 29 (CDC20), 32 (CKS2), 32 (TOP2A), 42 (UBE2C), and are named as in ONCOMINE. For these studies, the top 100 genes from clusters coexpressed with the query gene were analyzed, and the number of studies in which genes appeared with a cluster intracorrelation of at least 0.70 were counted. Genes annotated as ESTs were ignored. For genes appearing multiple times in the top 100 list, the highest cluster intracorrelation value for the gene was used. Because ONCOMINE compiles studies across a variety of microarray platforms, not all genes were measured in each study. Heatmaps were generated with Java Treeview [8].
Preparation of Cytoplasmic and Nuclear Extracts
HeLa cells were collected by trypsinization; washed in phosphate-buffered saline (PBS); lysed for 5 minutes on ice in 0.5% NP40, 5 mM Na4P2O7, 5 mM sodium-β-glycerophosphate, 10 mM NaF, 1 mM Na3VO4, 0.5 mM EDTA, and 10% glycerol containing protease inhibitors (pH 7.5); and centrifuged at 10,000g for 1 minute at 4°C. The supernatants used were cytoplasmic extracts, and pellets were lysed in sodium dodecyl sulfate (SDS) buffer (0.5% SDS, 50 mM NaF, 10 mM Na4P2O7, 10 mM Na2β-glycerophosphate, 5 mM Na3VO4, and 5 mM EDTA containing protease inhibitors, pH 7.5) and sonicated to generate nuclear extracts. Protein quantification was performed using the BCA Protein Assay kit (Pierce, Holmdel, NJ).
Immunoblot Analyses
MDA-231 and MDA-MB-453 cell lysates were prepared, electrophoresed, and transferred for immunoblot analysis as previously described [9]. HeLa cell lysates were prepared in SDS buffer and sonicated. SDS-polyacrylamide gel electrophoresis (PAGE; 4–12% acrylamide/Bis-Tris for all, except for topo II, which was 3–8% acrylamide/Tris-acetate) and blotting onto PVDF membranes were performed using the Invitrogen NuPAGE system (Invitrogen, Carlsbad, CA). After blotting, filters were stained for 5 minutes (room temperature) in 0.1% amido black (Sigma) in 25% isopropanol/10% acetic acid/65% water, followed by two washes in 25% isopropanol/10% acetic acid/65% water. Primary antibodies (1 µg/ml) were incubated for 1 hour at room temperature or at 4°C, and secondary anti-rabbit IgG-conjugated horseradish peroxidase (HRP; 1:5000; Pierce) was incubated for 1 hour at room temperature. Blots were processed for the detection of HRP by incubating with SuperSignal West Pico or Femto (Pierce). Serial dilutions of untreated or negative control cell lysates were used in each experiment to estimate the degree of E2-EPF protein knockdown and to monitor detection limits for the anti-E2-EPF antibody. Quantity One software (BioRad, Hercules CA)was used for densitometric quantification of immunoblot band intensity.
RNAi Transfection and Proliferation Assays
Delivery of siRNA was performed according to the forward transfection protocols of the manufacturers, where siRNA and lipid reagents are mixed with cells before plating. Conditions for HeLa cells were 1 to 2 nM siRNA and 2.5 to 5 µl/ml Hiperfect (Qiagen); conditions for MDA-231 and MDA-MB-453 cells were 2 nM siRNA with 1.0 µl/ml Lipofectamine RNAiMAX (Invitrogen). For proliferation assays with image analysis readouts, transfected cells were plated into 96-well plates (HeLa: 800 per well; MDA-231: 2500 per well; MDA-MB-453: 5000 per well). For WST-1 viability assay in HeLa cells, 105 cells/60 cm dish were transfected with siRNA and harvested after 3 days by trypsinization. One percent of each sample was replated in duplicate into a 96-well plate and allowed to attach for 3 hours, WST-1 reagent was added, and the assay was performed according to the instructions of the supplier (Roche Molecular Biochemicals, Indianapolis, IN).
Chemotherapeutic Drug Treatments
Drugs were diluted from dimethyl sulfoxide stock solutions into culture medium working solutions and added at the following concentrations to the cells 24 hours after transfection (doxorubicin: 2.5–20 nM; etoposide: 62.5–500 nM; camptothecin: 2–16 nM; Taxol: 0.5–8 nM). After 3 days of drug treatment, cells were processed for nuclei count by image analysis. Statistical analysis of significance was performed using Student's t test, assuming unequal variances. P < .05 was considered significant.
HeLa Cell Synchronization Conditions
HeLa cells were grown to confluency and cultured for an additional 24 hours, then split 1:4 into 10-cm tissue culture plates for immunoblot analyses and into 24-well plates for cell cycle analysis. Cells were cultured in medium without serum for an additional 24 hours, harvested (ss time point) or fed with fetal bovine serum (FBS; 10% final concentration), and harvested at 3-hour intervals. For thymidine arrest and release experiments, ∼ 30% confluent HeLa cells were grown for 24 hours in 2 mM thymidine (Sigma) and either harvested (thymidine-arrested time point) or washed twice, cultured in thymidine-free medium, and harvested at 3-hour intervals. For nocodazole arrest, HeLa cells were cultured for 24 hours in 200 nM nocodazole (Sigma) before mitotic shakeoff was performed. For cell cycle analysis, a fraction of shakeoff cells was plated back into a poly-d-lysine-coated well and allowed to attach for 5 minutes before fixation.
High-Content Image Analysis
For nuclei quantification and cell cycle analysis, cells were fixed (MDA-231 and MDA-MB-453 cells in 3.7% formaldehyde in PBS at room temperature for 10 minutes; HeLa cells in methanol for 5 minutes at room temperature), stained with 1 µg/ml Hoechst 33342 (Invitrogen) for 30 minutes at 37°C or overnight at 4°C, and washed with PBS before image analysis using Cellomics (ThermoFisher Scientific, Pittsburgh, PA) Arrayscan and Bioapplication software.
Results
E2-EPF mRNA and Protein Are Overexpressed in Breast Tumor Specimens
In a search for genes differentially expressed in breast cancers relative to normal tissues, ∼ 30 specimens from normal breast tissues and infiltrating ductal carcinomas were analyzed by oligonucleotide microarray (HG-U133A; Affymetrix, Santa Clara, CA) hybridization. These studies identified E2-EPF as significantly overexpressed in tumors relative to normal tissues and were confirmed by measuring E2-EPF mRNA levels by real-time PCR analysis (Figure 1A). An E2-EPF overexpression of > 2-fold relative to normal breast samples (n = 10) was observed in ∼ 80% of the tumors tested (n = 34, with data for 17 samples shown in Figure 1A). In most cases, E2-EPF mRNA levels were 5-fold to 10-fold greater in tumors than in normal specimens. E2-EPF EST representation in tumor libraries from the breast, ovary, lung, and pancreas was also elevated in comparison to the corresponding normal tissues (Incyte LifeSeq database; data not shown).
Figure 1.
E2-EPF expression in normal breast tissues and primary breast cancers. (A) E2-EPF mRNA levels in normal breast tissue (NB), mammary gland pool (MG), or NAT and breast cancer (BT) specimens were quantified by real-time RT-PCR analysis. (B) Left: Graphical representation of immunohistochemical data for the percentage of E2-EPF+ samples from normal breast tissues, NAT, and all analyzed breast cancers (all BC), and grouping by ER status (ER+ and ER- breast cancer; data from Table W1). *P values, Fisher's exact test. Right: Graphical representation of the percentage of samples in each IHC scoring category for E2-EPF protein expression (data from Table W2).
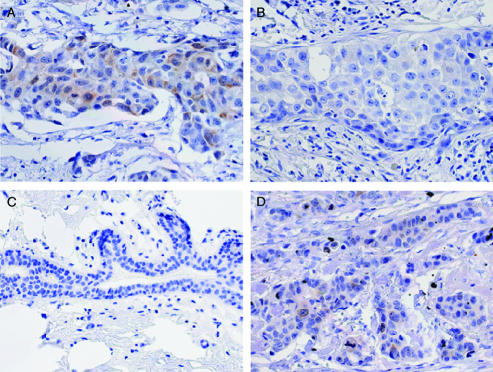
E2-EPF protein expression was evaluated by immunohistochemical analysis in FFPE specimens from normal and cancerous breast tissues. In agreement with observations for mRNA levels, the E2-EPF protein was detectable in the majority of breast cancers (i.e., 67%; 50 of 75) in contrast to only 4.5% (1 of 22) of normal breast samples and 9% (1 of 11) of NAT samples (Figure 1B and Table W1). E2-EPF staining was typically found in scattered epithelial cell clusters within tumors and was rarely found in normal ductal epithelial cells (Figure 2). Both nuclear staining and cytoplasmic staining were observed. When the expression of E2-EPF in this panel of tumor samples was compared with ER and PR status, as well as histologic grade (the majority were grade 2 or 3), a significant positive correlation between ER negativity and E2-EPF expression was observed (Figure 1B and Table W1). Interestingly, tumors with at least 25% of cells expressing detectable levels of E2-EPF (immunohistochemistry score of 3 or 4) were more likely to be ER- than ER+ (Figure 1B and Table W2). No statistically significant associations were noted for E2-EPF positivity and histologic grade or PR expression (Table W1).
Figure 2.
E2-EPF immunohistochemistry. Representative staining with anti-E2-EPF antibody for primary specimens from breast cancers with scores of 4 (A), 0 (B), and 1 (D) and from normal breast (C). Original magnification, x40.
E2-EPF Is Frequently Coexpressed in Breast Tumors with Genes That Function in the G2/M phase of the Cell Cycle
With the aim of providing additional information on which to base exploratory investigations of E2-EPF function, we evaluated gene expression profiles from approximately 160 nonhematologic cancer microarray studies deposited in ONCOMINE [10] for genes that are coordinately expressed with E2-EPF. We extracted the list of coexpressed genes from each of the studies obtained using the search tools provided in ONCOMINE (see Materials and Methods section) and determined the frequency with which each gene was identified as coexpressed with E2-EPF over all the studies. The genes that were most frequently coexpressed with E2-EPF (called ECG) are listed in Table 1. Many have known roles in cell cycle progression and are expressed in G2 or M phase (e.g., CCNB2, TOP2A, CENPA, CDC20, PRC1, STK6, UBE2C. Importantly, E2-EPF was coordinately expressed with a large number of these genes with high correlation in 7 of 20 microarray studies, 5 of which included breast carcinomas (Figure 3A). Not surprisingly, these ECGs were among the coexpressed genes identified for other query genes in the meta-signature for undifferentiated cancers [1], whose mRNA levels are elevated in G2 phase (i.e., UBE2C, TOP2A) or G2/M phase (i.e., CDC20, CKS2) (Table W3). But the ECG were not coexpressed with genes such as ERBB2 (data not shown). In the 20 microarray studies, the strength of the correlation of E2-EPF with ECGs was generally weaker than the correlation of UBE2C, TOP2A, CDC20, and CKS2 with ECGs (Figure 3B, brightness of color, particularly in studies marked with black arrowhead), suggesting greater sample heterogeneity for coexpression with E2-EPF than with TOP2A, CDC20, and UBE2C. Moreover, E2-EPF expression was more correlated with genes other than the ECGs in two studies (i.e., Linn sarcoma and Talantov melanoma), even though ECGs were coexpressed with TOP2A, CDC20, UBE2C, or CKS2 in the same studies (Figure 3B) and E2-EPF was significantly differentially expressed in the tumors in those studies (data not shown). Overall, these data suggest that E2-EPF expression is associated with the proliferating cell population in breast cancers (identified by G2/M gene expression) and also imply that E2-EPF association with genes involved in cell cycle control may not be observed in all cancers or in all tumors of a particular cancer type.
Table 1.
Genes Coexpressed with E2-EPF in Cancer Microarray Studies.
| Gene | # | Description | Cell Cycle* |
| CCNB2 | 9 | cyclin B2 | G2/M |
| TOP2A | 9 | topoisomerase (DNA) II alpha 170kDa | G2 |
| CENPA | 8 | centromere protein A | G2 |
| CDC20 | 7 | cell division cycle 20 homolog (S. cerevisiae) | G2/M |
| MYBL2 | 7 | v-myb myeloblastosis viral oncogene homolog (avian)-like 2 | ND |
| PRC1 | 7 | protein regulator of cytokinesis 1 | M/G1 |
| STK6 | 7 | aurora kinase A | G2 |
| TPX2 | 7 | TPX2, microtubule-associated, homolog (Xenopus laevis) | ND |
| UBE2C | 7 | ubiquitin-conjugating enzyme E2C | G2 |
| BUB1 | 6 | BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) | G2/M |
| CDC2 | 6 | cell division cycle 2, G1 to S and G2 to M | G2 |
| CKS2 | 6 | CDC28 protein kinase regulatory subunit 2 | G2/M |
| DLG7 | 6 | discs, large homolog 7 (Drosophila) | ND |
| FOXM1 | 6 | forkhead box M1 | G2 |
| MAD2L1 | 6 | MAD2 mitotic arrest deficient-like 1 (yeast) | ND |
| MELK | 6 | maternal embryonic leucine zipper kinase | G2/M |
| RRM2 | 6 | ribonucleotide reductase M2 polypeptide | S |
| TACC3 | 6 | transforming, acidic coiled-coil containing protein 3 | G2/M |
| TTK | 6 | TTK protein kinase | G2/M |
| BIRC5 | 5 | baculoviral IAP repeat-containing 5 (survivin) | G2/M |
| C10orf3 | 5 | centrosomal protein 55kDa (CEP55) | ND |
| CCNB1 | 5 | cyclin B1 | G2/M |
| CDCA3 | 5 | cell division cycle associated 3 | ND |
| CDCA8 | 5 | cell division cycle associated 8 | ND |
| H2AFX | 5 | H2A histone family, member X | S |
| MKI67 | 5 | antigen identified by monoclonal antibody Ki-67 | G2 |
| NUSAP1 | 5 | nucleolar and spindle associated protein 1 | ND |
| PTTG1 | 5 | pituitary tumor-transforming 1 | M/G1 |
| ZWINT | 5 | ZW10 interactor | ND |
| AURKB | 4 | aurora kinase B | G2 |
| BUB1B | 4 | BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) | G2/M |
| CCNA2 | 4 | cyclin A2 | G2 |
| DKFZp762E1312 | 4 | hypothetical protein DKFZp762E1312 | G2 |
| EZH2 | 4 | enhancer of zeste homolog 2 (Drosophila) | S |
| HCAP-G | 4 | non-SMC condensin I complex, subunit G | ND |
| KIAA0101 | 4 | p15(PAF) | ND |
| KIF11 | 4 | kinesin family member 11 | ND |
| KIF20A | 4 | kinesin family member 20A | ND |
| KIF4A | 4 | kinesin family member 4A | ND |
| KNTC2 | 4 | kinetochore associated 2 | ND |
| KPNA2 | 4 | karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | G2 |
| NEK2 | 4 | NIMA (never in mitosis gene a)-related kinase 2 | G2/M |
| RAMP | 4 | RA-regulated nuclear matrix-associated protein | ND |
| TRIP13 | 4 | thyroid hormone receptor interactor 13 | ND |
| CDCA1 | 3 | cell division cycle associated 1 | G2/M |
| CDKN3 | 3 | cyclin-dependent kinase inhibitor 3 (CDK2-associated dual specificity phosphatase) | M/G1 |
| CENPE | 3 | centromere protein E, 312kDa | G2/M |
| CENPF | 3 | centromere protein F, 350/400ka (mitosin) | G2/M |
| FLJ10719 | 3 | hypothetical protein FLJ10719 | ND |
| HMMR | 3 | hyaluronan-mediated motility receptor (RHAMM) | G2/M |
| KIF23 | 3 | kinesin family member 23 | ND |
| KIF2C | 3 | kinesin family member 2C | ND |
| KIFC1 | 3 | kinesin family member C1 | G2 |
| LOC146909 | 3 | hypothetical protein LOC146909 | ND |
| MCM10 | 3 | MCM10 minichromosome maintenance deficient 10 (S. cerevisiae) | ND |
| PBK | 3 | PDZ binding kinase | ND |
| RACGAP1 | 3 | Rac GTPase activating protein 1 | ND |
| TYMS | 3 | thymidylate synthetase | S |
The gene name and cellular processes associated with ECGs, as well as their frequency of occurrence in the 20 analyzed cancer microarray studies.
#, frequency with which the indicated gene was identified as correlated with E2-EPF in the 20 microarray studies.
Cell cycle phase determined in Whitfield et al [6]; ND, not cell-cycle regulated or not designated.
Figure 3.
Genes coexpressed with E2-EPF in cancer microarray studies. (A) ECGs were visualized by study to determine patterns distinct to cancer types. Color represents the cluster intracorrelation of each gene with E2-EPF. Note: Chowdary Multicancer includes colon and breast. (B) Cluster intracorrelation of ECGs with specified query gene (i.e., CDC20, CKS2, TOP2A, UBE2C) in the 20 studies. Red arrowhead: Study in which no genes in ECGs were found in the top 100 correlated genes for E2-EPF. Black arrowhead: Study in which E2-EPF is less correlated than CDC20, CKS2, TOP2A, and UBE2C. Genes that were not measured in a study or that did not meet the criteria described in Materials and Methods section are gray in the heatmap. Heatmap bar, correlation values.
E2-EPF Expression Is Cell Cycle-Regulated in HeLa Cervical Carcinoma Cells
Consistent with the observed association of E2-EPF expression with G2/M-expressed genes in multiple cancer microarray studies, we noted that E2-EPF was 1 of 874 cell cycle-regulated genes identified in gene expression analyses of HeLa cells that were synchronized with treatments that arrest cells at the beginning of S phase (i.e., thymidine or hydroxyurea) or in prometaphase (i.e., nocodazole) and subsequently released from those blocks [6]. In that study, E2-EPF was classified as a gene whose expression peaked during the M/G1 phase of the cell cycle, yet its mRNA expression profile was most similar to that of the G2/M phase-expressed BUB1, BIRC5, CCNB2, CDC20, PLK1, and other genes involved in mitotic surveillance (Figure 7 in Whitfield et al. [6]).
We performed similar cell cycle analyses in HeLa cells and found that the E2-EPF protein was detectable under all tested conditions but was significantly elevated in S phase (i.e., thymidine-arrested and released cells 12 hours after serum stimulation) as well as in G2/M phase compared to serum-deprived cells (Figure 4, A and B). In agreement with the data for mRNA levels, E2-EPF protein levels were highest in cells arrested in M phase by nocodazole treatment or in the population with a high fraction of cells in G2/M phase (i.e., Thy + 9–12 hours of release). E2-EPF expression was not as tightly regulated as the phosphorylation of histone H3, which increased markedly in cell populations with a high fraction of cells in M phase (i.e., nocodazole or Thy + 9–12 hours of release). Because cell populations with the highest levels of E2-EPF also have reduced cyclin A expression, which is degraded at the end of prophase, the peak expression of E2-EPF is most likely in middle to late mitosis rather than in early mitosis.
Figure 4.
Cell cycle regulation of E2-EPF gene expression. (A) Top: Immunoblot analysis of E2-EPF protein expression during the HeLa cell cycle. Cell lysates were prepared from cells asynchronously growing (Asy), serum-starved for 24 hours (ss), stimulated with 10% FBS for indicated times (serumstimulated), thymidine-arrested for 24 hours (Thy), released from the thymidine block for indicated times (release), Nocodazole arrested for 24 hours (Noc), (40 µg protein/lane) were resolved by PAGE samples, and immunoblots were processed with indicated antibodies. Amido black stain verified equivalent sample loading. Bottom: Graphical representation of the densitometric quantitation of the above immunoblot normalized for amido black stain. (B) Cell cycle distribution analysis of HeLa cells from the experiment in (A). Histograms from Cellomics Cell Cycle Bioapplication show the number of instances ( y axis) for each DNA content (x axis), with data points binned per 1000 instances in all but Thy and Thy + 12 hours (by 10,000 instances). Arrow: G1 peak.
HeLa Cell Proliferation Is Not Affected By the Reduction in E2-EPF Expression
Having demonstrated cell cycle dependency for E2-EPF expression in HeLa cells, the potential role of E2-EPF in HeLa cell growth was addressed using siRNA directed against E2-EPF mRNA that were introduced into cells by lipid-mediated transfection. Concentrations of lipid reagents and siRNA were optimized for maximal E2-EPF knockdown at the lowest concentration of siRNA (i.e., 0.1–2 fmol/cell or 1–2 nM) and were optimized to have little or no effects on cell proliferation on treatment with multiple “negative” control oligonucleotides (i.e., FFL, FFL2, MYOG). Under these conditions, treatment with five of seven E2-EPF siRNA reduced E2-EPF protein levels to < 25% of control 72 hours after transfection. Following treatment with three of these siRNA (i.e., D1, D2, D4), levels were decreased to < 10% of control (Figure 5A). For effective siRNA, E2-EPF protein levels were reduced by > 75% as early as 24 hours after transfection (Figure 6B; data not shown). In spite of this substantial knockdown of E2-EPF expression with multiple siRNA sequences, including those with oligonucleotide modifications to reduce offtarget effects (i.e., Smart Plus; Dharmacon) (Figure 6B), no specific inhibition of HeLa cell proliferation was observed. In contrast, knockdown of mitotic kinesin 11 (i.e., Eg5; Figure 5A) resulted in a > 90% decrease in cell viability. Furthermore, no alteration in cell cycle distribution was observed in HeLa cells with reduced E2-EPF expression (data not shown).
Figure 5.
Effect of E2-EPF siRNA treatment on cancer cell proliferation. HeLa (A), MDA-231 (B), and MDA-MB-453 (C) cells were transfected with indicated siRNA. HeLa cells were assayed 3 days later for E2-EPF expression (immunoblot) and cell viability (WST-1 assay; duplicate data points shown). Proliferation was assessed at indicated time points (MDA-231) or at 96 hours (MDA-MB-453) by counting nuclei (triplicate data points ± S.D.) and E2-EPF expression from parallel plates by immunoblot analysis. UT = untreated; Lipo = lipid only. FFL, FFL2, MYOG, NC, and NT are negative control siRNA (striped bars). Eg5 and PLK1 are positive control siRNA (open bars). E2-EPF siRNA are nos. 1–3, D1–D6 (black bars). Pooled oligos are NTp, MYOGp, E2-EPFp, and PLK1p. Data are representative of at least two independent experiments from each cell line.
Figure 6.
Effect of chemotherapeutic drugs on HeLa cells treated with E2-EPF siRNA. Cells were transfected with the indicated negative control siRNA (NTp, MYOGp, NT) and E2-EPF siRNA (E2-EPFp, D5, D6) and treated in duplicate (A) or in triplicate (B and C) with the indicated drug concentrations (A and C) or with no drug (B) for 3 days starting 24 hours after transfection. Cell proliferation (nuclei counts) was measured by microscopic imaging analysis. (B) Left: Nuclei counts for “no drug” control transfectants corresponding to the experiment in (C). Right: Samples (40 µg protein/lane, except as noted) on the left were analyzed by immunoblotting for the E2-EPF protein at 24 and 72 hours after transfection. Data are representative of at least three independent experiments. Error bars, S.D. *P < .03 for the comparison of E2-EPF siRNA with negative control siRNA. Note: D6 + dox was not statistically significant (P > .05), but in the repeat experiment, NT + dox versus D6 + dox yielded 63% vs 37% (5 nM) and 28% vs 14% (10 nM) (P = .014). (D) HeLa cells were transfected with the indicated siRNA, harvested 48 hours later, and fractionated into nuclear and cytoplasmic extracts before immunoblot analysis (50 µg protein/lane) for topo IIα (170 kDa) and topo IIβ (180 kDa) expressions. Immunoreactive bands identified in topo IIα and IIβ siRNA-treated cell lysates indicate the specificity of the topo II antibody. Quantification of topo IIα by protein dilution in an independent experiment demonstrated that the increase in topo IIα on E2-EPF knockdown was ∼ 2-fold. Amido black staining verified equivalent sample loading.
E2-EPF Knockdown Has No Effect on the Proliferation of ER- Breast Cancer Cells
Given the positive association of E2-EPF protein expression with ER- breast cancers, the involvement of E2-EPF in the proliferation of ER- MDA-231 and MDA-MB-453 breast cancer cells was addressed by RNAi-based knockdown approaches. In both of these cell lines, transfection with E2-EPF siRNA elicited substantial decreases in E2-EPF protein (Figure 5, B and C) and mRNA levels (data not shown). In MDA-231 cells, E2-EPF protein levels were reduced by > 85% of those in control siRNA-treated cells (i.e., FFL, FFL2) after 24 hours of treatment with multiple E2-EPF siRNA, and similar levels of knockdown were maintained for at least 96 hours (Figure 5B). In contrast to the dramatic inhibition of proliferation observed with Eg5 siRNA, this marked reduction in levels of the E2-EPF protein had no significant effects on MDA-231 cell proliferation (Figure 5B) or cell cycle distribution (data not shown). Similarly, treatment of MDA-MB-453 cells with multiple E2-EPF siRNA substantially decreased the levels of the E2-EPF protein for at least 96 hours (Figure 5C; data not shown) with no effect on their proliferation, whereas treatment with siRNA directed against PLK1 markedly inhibited cell growth.
E2-EPF Knockdown Increased HeLa Cell Sensitivity to Topo II Inhibitors
From the results of E2-EPF knockdown studies in HeLa, MDA-231, and MDA-MB-453 cells, it can be concluded that E2-EPF is not required for the proliferation of these cancer cells under standard culture conditions. The observation that E2-EPF expression in tumors frequently correlated with genes such as TOP2A and CENPA, which have key roles in chromosome condensation and organization, as well as with BUB1, BUB1B, MAD2L1, and TTK, which are involved in spindle assembly and mitotic checkpoint function, prompted us to ask whether G2-phase or M-phase checkpoints might be regulated by E2-EPF under conditions of cellular stress. We used chemotherapeutic drugs to evaluate this hypothesis, selecting agents that damage DNA (i.e., doxorubicin, etoposide, camptothecin) or interfere with spindle assembly (i.e., Taxol). HeLa cells that were transfected with E2-EPF or control siRNA were treated for 72 hours with these drugs at doses spanning their respective IC50 concentrations. E2-EPF knockdown with multiple siRNA [i.e., E2-EPFp, D5, and D6 (Figure 6, A and C); D2 and D4 (data not shown)] resulted in a significantly greater (∼ 2-fold) antiproliferative effect in combination with etoposide [at 250 nM: NTp, 47.5 ± 5.5%; E2-EPFp, 20.1 ± 8.1% (of untreated); n = 3, P < .0001], as well as with doxorubicin (for 10 nM: NTp, 39.4 ± 5.6%; E2-EPFp, 19.2 ± 2.0%; n = 3, P < .0001), than that seen with the drugs alone. In contrast, no significant sensitizing effects of E2-EPF depletion were observed in combination with Taxol or camptothecin (Figure 6C). As previously discussed, no effect of E2-EPF knockdown on cell proliferation in the absence of cotreatment with chemotherapeutic drugs was observed despite the reduction of E2-EPF protein levels by > 75% (Figure 6B).
Both etoposide and doxorubicin damage DNA by forming covalent tertiary complexes with topo II and DNA that result in double-strand breaks (for review, see Giles and Sharma [11]). Topo II has two isoforms, IIα (MW = 170 kDa) and IIβ (MW = 180 kDa), with the former exhibiting cell cycle regulation of its expression that is elevated during S phase and that peaks at G2/M phase [12]. Furthermore, tumor cell sensitivity to topo II inhibitors has been positively correlated with increased nuclear topo IIα levels [13,14], and cytoplasmic relocalization of topo IIα results in drug resistance [15]. Therefore, the effect of E2-EPF knockdown on the levels of nuclear and cytoplasmic topo IIα and IIβ were measured. In HeLa cells, both topo II isoforms were detected in the nuclear compartment, whereas the E2-EPF protein was found in both the cytoplasm and the nucleus (Figure 6D). Following treatment with E2-EPF siRNA for 48 hours, topo IIα protein levels increased by ∼ 2-fold with no change in subcellular localization. Similar increases in topo IIα levels were observed using other E2-EPF siRNA (i.e., D5 and D6; data not shown), but no change in topo IIα levels was detected following treatment with negative control siRNA. As E2-EPF knockdown did not affect HeLa cell cycle distribution, this difference in topo IIα levels is not due to a change in the number of cells in S or G2/M phase (data not shown). No significant difference in topo IIβ levels was observed on treatment with any of the E2-EPF siRNA.
Discussion
We identified the ubiquitin-conjugating enzyme E2-EPF in DNA microarray and EST database-mining studies as a gene overexpressed in breast tumors relative to normal tissues. Our study demonstrated that the E2-EPF protein is more highly expressed in the majority of breast infiltrating ductal carcinomas than in normal tissues and is regulated during the HeLa cell cycle, achieving the highest levels during mitosis. In tumors, E2-EPF mRNA expression correlated with genes involved in mitotic surveillance, but RNAi-mediated knockdown of the E2-EPF protein did not alter cell cycle distribution or affect the proliferation of HeLa cervical cancer cells or MDA-231 and MDA-MB-453 breast cancer cells. Instead, E2-EPF protein knockdown sensitized HeLa cells to the antiproliferative effects of the topo II inhibitors etoposide and doxorubicin and resulted in elevated topo IIα protein levels.
Our observation that ER- breast tumors are more likely to express the E2-EPF protein and have detectable E2-EPF expression in a greater percentage of cells than ER+ tumors is consistent with the presence of E2-EPF in the metasignature for undifferentiated cancer [1] because ER- tumors are typically poorly differentiated and are of higher mitotic grade than ER+ tumors [16]. Yet, RNAi-mediated reduction of E2-EPF protein levels to < 15% of control levels in HeLa cells or in ER- MDA-231 and MDA-MB-453 breast cancer cells did not affect their proliferation. Our results differ from those of Jung et al. [4], who demonstrated that adenovirus-encoded E2-EPF siRNA that downregulated E2-EPF protein levels inhibited the proliferation of C8161 melanoma and VHL-expressing 786-O renal carcinoma cells by ∼ 1.5-fold to 2-fold 4 days after seeding (approximately two population doublings) and more markedly suppressed their in vivo tumor growth. It is possible that the 80% to 90% reduction in E2-EPF levels achieved in our study is not of sufficient magnitude or duration to impact the proliferation of the cancer cells analyzed or that only certain cancers are dependent on E2-EPF for their growth. In this regard, it is noteworthy that VHL expression was critical for tumor growth inhibition following adenoviral E2-EPF siRNA delivery in Jung et al. [4], as growth of parental 786-O tumors was not affected by E2-EPF downmodulation. Because the cell lines studied here express a wild-type functional VHL protein [17], the lack of an E2-EPF requirement for cell proliferation suggests that the presence of a functional VHL-HIF-1α pathway is not, by itself, responsible for determining E2-EPF dependency.
In our survey of multiple cancer microarray studies using the ONCOMINE database, we found that E2-EPF expression was most frequently correlated with genes such as CCNB2, CENPA, TOP2A, CDC20, and TPX2, which are all implicated in cell cycle progression through G2/M phase. Their coexpression with E2-EPF in tumors may indicate that E2-EPF is expressed in a proliferating tumor cell population identified by the expression of these G2/M genes. The relatively weak correlation of E2-EPF with these G2/M genes, compared with genes such as TOP2A, CDC20, UBE2C, or CKS2, is consistent with the fact that reduction of E2-EPF levels following siRNA treatment had no effect on cell proliferation, but may also suggest that E2-EPF is important for proliferation in only a subset of cancers or may function in a particular tumor microenvironment. Our analysis explored E2-EPF activity in two ER- breast cancer cell lines that may represent different types of breast cancer. MDA-MB-453 is classified as an ERBB2-amplified luminal epithelial cell cancer, whereas MDA-231 is a highly invasive cell line with mesenchymal cell characteristics [18]. Evaluation of additional cell lines representative of other breast cancers, as well as different conditions of in vitro culture (e.g., extracellular matrix, cellular stress), for effects of E2-EPF depletion may therefore be warranted.
The cell cycle-regulated expression of E2-EPF and its coexpression in tumors with genes implicated in chromosome decatenation (i.e., TOP2A), spindle assembly (TPX2), and mitotic checkpoint function (BUB1B, BUB1, and TTK) prompted us to ask whether E2-EPF might have a function in cancer cells that might be revealed only under conditions of cellular stress, modeled by treating cells with agents that induce a G2 checkpoint or a mitotic checkpoint. We discovered that HeLa cells treated with E2-EPF siRNA were approximately two-fold more sensitive to the topo II inhibitors etoposide and doxorubicin than were cells transfected with negative control siRNA. This degree of sensitization is similar to that seen with agents, such as the poly(ADP-Ribose) polymerase inhibitor AG14361, that enhance the potency of topo I inhibitors [19]. Importantly, no sensitization was found for the topo I inhibitor camptothecin, which also induces DNA damage, or for the microtubule-stabilizing drug Taxol. Lack of sensitization to camptothecin suggests that sensitization to topo II inhibitors by E2-EPF depletion was not due to a reduced ability of the cells to sense double-stranded DNA breaks, to trigger the S phase and/or G2 DNA damage checkpoints, or to repair damaged DNA. A possible explanation for the increased antiproliferative effect of topo II inhibitors following E2-EPF knockdown is that topo IIα protein levels were increased. Because topo II inhibitors form tertiary complexes with the enzyme and DNA to enhance strand breakage [11], higher levels of topo II can lead to greater drug sensitivity. Indeed, enhanced sensitivity to topo II inhibitors has been shown in tumors with elevated levels of topo II [13,14,20]. Alternatively, E2-EPF may decrease drug sensitivity by playing a role in the turnover of topo II inhibitor-induced topo II-DNA complexes, thereby enabling repair of DNA damage. Topo IIβ-DNA covalent complexes have been shown to be degraded by the 26S proteasome following ubiquitinylation [21]. In our study, E2-EPF knockdown resulted in an increase in topo IIα, but not topo IIβ, levels in exponentially growing cells that were not treated with drugs. Further studies will be needed to address the effect of topo II inhibitor treatment on topo II protein levels under conditions of E2-EPF depletion, as well as to determine whether ubiquitinylation of topo IIα is affected by knockdown or overexpression of E2-EPF.
Proteasomal activity has also been shown to be important for the regulation of topo IIα protein degradation at the M/G1 transition of the cell cycle [22]. The specific E2 ubiquitin-conjugating enzymes and E3 ubiquitin ligases responsible for topo II turnover have not yet been identified; our E2-EPF depletion, as well as cell cycle studies demonstrating peak levels of E2-EPF during mitosis, suggests but does not prove that E2-EPF is one of the regulators of this process. It is possible that the frequent coexpression of E2-EPF and TOP2A reflects a tumor-specific mechanism that generates adequate levels of E2-EPF to ensure appropriate regulation of topo IIα concentrations and activity.
In summary, we have shown that the E2-EPF ubiquitin-conjugating enzyme is overexpressed in the majority of breast cancers, but elevated levels of E2-EPF are not required for cell proliferation under standard culture conditions, as suggested by RNAi-mediated depletion studies with the tested cancer cell lines. However, sensitivity to topo II inhibitors was enhanced in HeLa cells on treatment with E2-EPF-directed siRNA concomitant with an elevation in topo IIα protein levels. Studies that extend these observations to other cancer cells and explore the mechanism(s) by which E2-EPF modulates sensitivity to topo II inhibition are warranted to provide additional rationale for combination therapy with topo II and E2-EPF inhibitors.
Supplementary Material
Acknowledgements
We gratefully acknowledge the assistance of Richard Lin in designing initial siRNA reagents and TaqMan primers and probes; David Vogel and Sara Biancalana for polyclonal antibody generation; and Richard Feldman and Gordon Parry for helpful discussions.
Abbreviations
- topo
topoisomerase
- ER
estrogen receptor
- ECG
E2EPF coexpressed genes
Footnotes
This article refers to supplementary material, which is designated by “W” (i.e., Table W1) and is available online at www.bcdecker.com.
References
- 1.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. Large-scale metaanalysis of cancer microarray data identifies common transcriptional profiles of neoplastic transformation and progression. Proc Natl Acad Sci USA. 2004;101:9309–9314. doi: 10.1073/pnas.0401994101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu Z, Diaz LA, Haas AL, Giudice GJ. cDNA cloning of a novel human ubiquitin carrier protein. An antigenic domain specifically recognized by endemic pemphigus foliaceus autoantibodies is encoded in a secondary reading frame of this human epidermal transcript. J Biol Chem. 1992;267:15829–15835. [PubMed] [Google Scholar]
- 3.Pickart CM. Back to the future with ubiquitin. Cell. 2004;116:181–190. doi: 10.1016/s0092-8674(03)01074-2. [DOI] [PubMed] [Google Scholar]
- 4.Jung CR, Hwang KS, Yoo J, Cho WK, Kim JM, Kim WH, Im DS. E2-EPF UCP targets pVHL for degradation and associates with tumor growth and metastasis. Nat Med. 2006;12:809–816. doi: 10.1038/nm1440. [DOI] [PubMed] [Google Scholar]
- 5.Whitfield ML, George LK, Grant GD, Perou CM. Common markers of proliferation. Nat Rev Cancer. 2006;6:99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
- 6.Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zajchowski DA, Bartholdi MF, Gong Y, Webster L, Liu HL, Munishkin A, Beauheim C, Harvey S, Ethier SP, Johnson PH. Identification of gene expression profiles that predict the aggressive behavior of breast cancer cells. Cancer Res. 2001;61:5168–5178. [PubMed] [Google Scholar]
- 8.Saldanha AJ. Java Treeview—extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 9.Kiewlich D, Zhang J, Gross C, Xia W, Larsen B, Cobb RR, Biroc S, Gu JM, Sato T, Light DR, et al. Anti-EphA2 antibodies decrease EphA2 protein levels in murine CT26 colorectal and human MDA-231 breast tumors but do not inhibit tumor growth. Neoplasia. 2006;8:18–30. doi: 10.1593/neo.05544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giles GI, Sharma RP. Topoisomerase enzymes as therapeutic targets for cancer chemotherapy. Med Chem. 2005;1:383–394. doi: 10.2174/1573406054368738. [DOI] [PubMed] [Google Scholar]
- 12.Woessner RD, Mattern MR, Mirabelli CK, Johnson RK, Drake FH. Proliferation- and cell cycle-dependent differences in expression of the 170 kilodalton and 180 kilodalton forms of topoisomerase II in NIH-3T3 cells. Cell Growth Differ. 1991;2:209–214. [PubMed] [Google Scholar]
- 13.Asano T, Kleinerman ES, Zwelling LA, Zhou Z, Fukunaga Y. Adenovirus-mediated human topoisomerase IIalpha gene transfer increases the sensitivity of etoposide-resistant human and mouse breast cancer cells. Acta Oncol. 2005;44:240–247. doi: 10.1080/02841860510029653. [DOI] [PubMed] [Google Scholar]
- 14.Dingemans AM, Witlox MA, Stallaert RA, van der Valk P, Postmus PE, Giaccone G. Expression of DNA topoisomerase IIalpha and topoisomerase IIbeta genes predicts survival and response to chemotherapy in patients with small cell lung cancer. Clin Cancer Res. 1999;5:2048–2058. [PubMed] [Google Scholar]
- 15.Engel R, Valkov NI, Gump JL, Hazlehurst L, Dalton WS, Sullivan DM. The cytoplasmic trafficking of DNA topoisomerase IIalpha correlates with etoposide resistance in human myeloma cells. Exp Cell Res. 2004;295:421–431. doi: 10.1016/j.yexcr.2004.01.012. [DOI] [PubMed] [Google Scholar]
- 16.Desmedt C, Sotiriou C. Proliferation: the most prominent predictor of clinical outcome in breast cancer. Cell Cycle. 2006;5:2198–2202. doi: 10.4161/cc.5.19.3254. [DOI] [PubMed] [Google Scholar]
- 17.Blancher C, Moore JW, Talks KL, Houlbrook S, Harris AL. Relationship of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha expression to vascular endothelial growth factor induction and hypoxia survival in human breast cancer cell lines. Cancer Res. 2000;60:7106–7113. [PubMed] [Google Scholar]
- 18.Lacroix M, Leclercq G. Relevance of breast cancer cell lines as models for breast tumours: an update. Breast Cancer Res Treat. 2004;83:249–289. doi: 10.1023/B:BREA.0000014042.54925.cc. [DOI] [PubMed] [Google Scholar]
- 19.Smith LM, Willmore E, Austin CA, Curtin NJ. The novel poly(ADP-ribose) polymerase inhibitor, AG14361, sensitizes cells to topoisomerase I poisons by increasing the persistence of DNA strand breaks. Clin Cancer Res. 2005;11:8449–8457. doi: 10.1158/1078-0432.CCR-05-1224. [DOI] [PubMed] [Google Scholar]
- 20.Harris LN, Yang L, Liotcheva V, Pauli S, Iglehart JD, Colvin OM, Hsieh TS. Induction of topoisomerase II activity after ErbB2 activation is associated with a differential response to breast cancer chemotherapy. Clin Cancer Res. 2001;7:1497–1504. [PubMed] [Google Scholar]
- 21.Mao Y, Desai SD, Ting CY, Hwang J, Liu LF. 26S proteasome-mediated degradation of topoisomerase II cleavable complexes. J Biol Chem. 2001;276:40652–40658. doi: 10.1074/jbc.M104009200. [DOI] [PubMed] [Google Scholar]
- 22.Salmena L, Lam V, McPherson JP, Goldenberg GJ. Role of proteasomal degradation in the cell cycle-dependent regulation of DNA topoisomerase IIalpha expression. Biochem Pharmacol. 2001;61:795–802. doi: 10.1016/s0006-2952(01)00580-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.