Abstract
Cellular senescence has been theorized to oppose neoplastic transformation triggered by activation of oncogenic pathways in vitro1–3, but the relevance of senescence in vivo has not been established. The PTEN and p53 tumour suppressors are among the most commonly inactivated or mutated genes in human cancer including prostate cancer4,5. Although they are functionally distinct, reciprocal cooperation has been proposed, as PTEN is thought to regulate p53 stability, and p53 to enhance PTEN transcription6–10. Here we show that conditional inactivation of Trp53 in the mouse prostate fails to produce a tumour phenotype, whereas complete Pten inactivation in the prostate triggers non-lethal invasive prostate cancer after long latency. Strikingly, combined inactivation of Pten and Trp53 elicits invasive prostate cancer as early as 2 weeks after puberty and is invariably lethal by 7 months of age. Importantly, acute Pten inactivation induces growth arrest through the p53-dependent cellular senescence pathway both in vitro and in vivo, which can be fully rescued by combined loss of Trp53. Furthermore, we detected evidence of cellular senescence in specimens from early-stage human prostate cancer. Our results demonstrate the relevance of cellular senescence in restricting tumorigenesis in vivo and support a model for cooperative tumour suppression in which p53 is an essential failsafe protein of Pten-deficient tumours.
‘Cellular senescence’ describes a permanent form of cell cycle arrest in primary cultured cells, which can be triggered by DNA damage or activated oncogenes1–3. Although it has been implicated in mediating the response to anti-tumour treatments11, there is still no evidence that senescence opposes tumorigenesis.
Up to 70% of primary prostate tumours lose one PTEN allele and retain the other copy12–15. Similarly, p53 is found completely lost or mutated almost exclusively in advanced prostate cancer16,17. Because complete loss of Pten in the mouse seems to be crucial for the development of invasive prostate tumours18,19, why human invasive prostate cancer would not also select for complete PTEN-loss is puzzling. Similarly, the observation that p53 is lost not at presentation but in advanced prostate cancer was surprising, because loss of p53 favours tumour initiation in many tissues. A network involving the mutual dependence of PTEN and p53 has emerged that could partly reconcile the paradox. Specifically, PTEN may protect p53 from Mdm2-mediated degradation, whereas p53 can enhance the transcription of PTEN6–10. Inactivation of either gene should result in lower protein levels of the other gene. Thus, a loss of PTEN or p53 could mimic two genetic hits in one. To test the relevance of these observations in a well-defined model for tumour progression, we generated mice in which the Pten and/or Trp53 genes are deleted specifically in the prostate. This analysis led to unforeseen observations that contradict the above hypothesis and show the importance of cellular senescence for tumour suppression in vivo.
For prostate-specific inactivation, we made use of the Cre/loxP technique20 and Probasin-Cre4 (Pb-Cre4) transgenic mice expressing Cre after puberty (at age 7 weeks) in prostatic epithelium21. We obtained PtenloxP/loxP;Pb-Cre4 and Trp53loxP/loxP;Pb-Cre4 mice, hereafter referred to as Ptenpc−/− and Trp53pc−/− (Supplementary Fig. S1). As expected, in the presence of Pb-Cre4, recombination of Pten and Trp53 was restricted to the three prostatic lobes, namely the anterior prostate (AP), ventral prostate (VP) and dorsolateral prostate (DLP), with minor recombination occurring in seminal vesicles (Supplementary Fig. S2a).
To study early effects of Pten and/or Trp53 inactivation in the prostate, mice were killed at 9 weeks of age and histopathological analysis was performed. Wild-type (WT) mice displayed normal prostate histology, whereas age-matched Ptenpc−/− littermates consistently displayed high-grade prostatic intraepithelial neoplasia (HG-PIN) in all three prostate lobes (roughly 50–60% of prostate glands affected: n = 10; Fig. 1a and Supplementary Fig. S2b), showing early onset of tumorigenesis immediately after Pten excision. In Trp53pc−/− mice, no pathological changes (hyperplasia or PIN) were found in any prostate lobes, and their prostate glands were indistinguishable from age-matched WT mice (n = 10; Fig. 1b and Supplementary Fig. S2b). Up to 18 months (Supplementary Fig. S2c; not shown), there were no differences in morphology and no differences detected by immunostaining for androgen receptor (glandular epithelial marker) or p63 (basal cell marker) (Supplementary Fig. S2d).
Figure 1. Loss of Trp53 does not initiate prostate tumours but renders Pten-deficient carcinomas lethal.
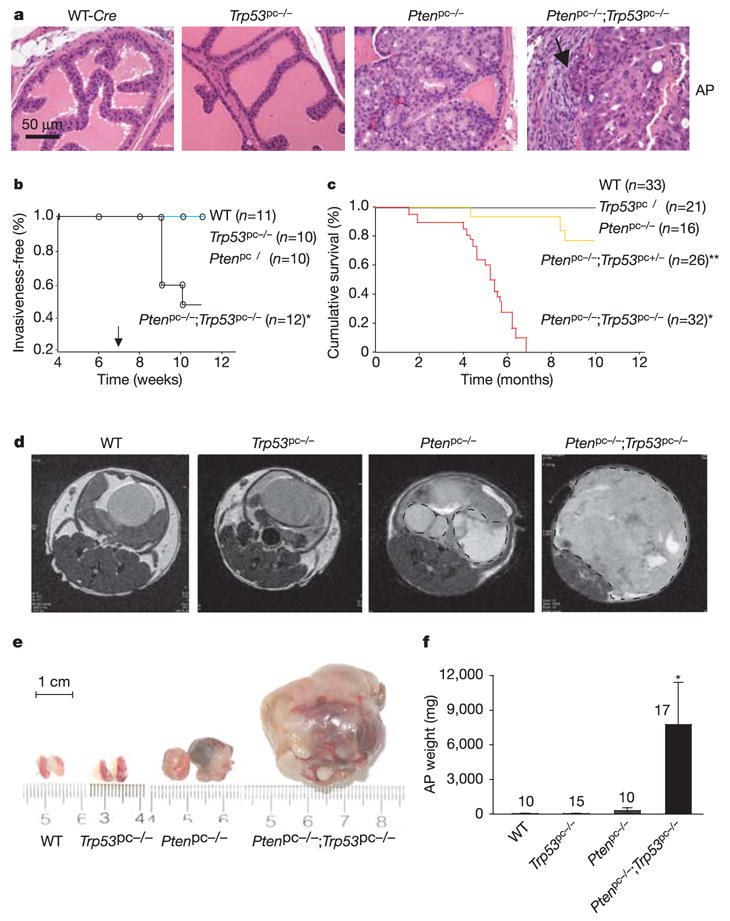
a, Histopathological analysis (haematoxylin/eosin staining) of anterior prostates (AP) in WT, Pten and Trp53 single and double mutants at 9 weeks of age reveals normal glands in WT and Trp53pc−/− mice but PIN lesions in Ptenpc−/− mice and invasion (arrow) in Ptenpc−/−;Trp53pc−/− mice. b, Disease-free survival curve (Kaplan–Meier plot) for prostate cancer. Adenocarcinoma was found only in the Ptenpc−/−;Trp53pc−/− cohort (P < 0.05). The arrow indicates puberty. c, Cumulative survival analysis. A statistically significant decrease in lifespan (P < 0.0001) compared with the Ptenpc−/− cohort was found for the Ptenpc−/−;Trp53pc−/− cohort (asterisk) and for the Ptenpc−/−;Trp53pc+/− cohort (double asterisk). d–f, MRI of AP tumours (dashed circles) at 23–31 weeks (d) and their actual sizes (e) and weights (f) after biopsy. Error bars in f indicate s.d. of AP weight for the numbers of mice indicated above the bars.
In contrast, on combined inactivation of Pten and Trp53, we found HG-PIN in all three lobes from 100% of mice, and invasive prostate cancer in 50% of Ptenpc−/−;Trp53pc−/− mice by age 10 weeks (n = 12) (Fig. 1a, b and Supplementary Fig. S2b). By 11 weeks, invasive adenocarcinoma was restricted to Ptenpc−/−;Trp53pc−/− mutants (Fig. 1b) whereas Ptenpc−/− animals presented with invasive prostate cancer after a 4–6-month latency. Thus, in contrast to loss of Pten, loss of Trp53 does not initiate prostate tumours but accelerates the progression of tumours initiated by loss of Pten.
We followed a cohort of 128 mice over 10 months for survival analysis, with two-weekly magnetic resonance imaging (MRI) analyses19. None of the Ptenpc−/− mutants died of prostate cancer (Fig. 1c), which is consistent with our previous study19. In contrast, reduction of Trp53 in a Pten-null background markedly reduced survival in a dose-dependent manner (Fig. 1c). Notably, all Ptenpc−/−;Trp53pc−/− mice died by 7 months of age (mean survival 5 ± 0.5 months), probably due to bladder obstruction and renal failure. Distant metastases were not observed in these mice or in Ptenpc−/− mice in a 2.5-year follow-up (ref. 19, and Z.C and P.P.P., unpublished observations). By 6 months, MRI revealed masses in Ptenpc−/− prostates but not in Trp53pc−/− or wild-type prostates (Fig. 1d). The average tumour weight in double-null mice was increased 32-fold in comparison with tumours in Pten pc−/− mice (Fig. 1e, f) and tumours predominantly showed poorly differentiated histology (Supplementary Fig. S2c). Tumours arising in Ptenpc−/−;Trp53pc−/− mice encompassed the entire genitourinary tract and pelvis. In contrast, tumours in Ptenpc−/− mice retained clearly distinguishable urogenital organ boundaries. Thus, inactivation of Trp53 leads to massive tumour growth and lethal prostate cancer, but only in combination with Pten deficiency.
We investigated the molecular basis of cooperation between Trp53 and loss of Pten by using primary mouse embryonic fibroblasts (MEFs). To study the effect of acute inactivation of Pten and Trp53 we infected MEFs with retroviruses expressing Cre-PURO-IRES conjugated with green fluorescent protein (GFP) (Fig. 2a and Supplementary Fig. S3a). Virus-mediated expression of Cre led to efficient recombination of loxP alleles and increased Akt activation (Supplementary Fig. S3a). Pten heterozygous (PtenΔ /+) MEFs proliferated more rapidly than Pten WT (Pten+/+) MEFs, but the proliferation of Pten-null (PtenΔ /Δ ) MEFs was similar to that of the WT (Fig. 2a). Pten-null MEFs exhibited the distinctive morphology of senescent cells (flattened large cells; not shown) and were positive for senescence-associated β-galactosidase (SA-β-Gal), a hallmark of senescent cells22 (Fig. 2a and Supplementary Fig. S5a). To determine whether senescence was related to the selection process or to retroviral integration, we made use of transient episomal delivery by means of adenovirus-Cre (Ad-Cre) infection. We found that the loss of one Pten allele resulted in accelerated growth compared with that of wild-type cells, yet the loss of both Pten alleles led to lower proliferation and high β-Gal staining (Supplementary Figs S3b, c and S5d). To ensure that these findings were unrelated to Cre activity, we attempted Pten knockdown by means of RNA interference (RNAi). A decrease in Pten levels by 50% in heterozygous MEFs did indeed result in lowered proliferation and senescence induction, confirming not only the Cre-independence of senescence, but also suggesting that acute loss of Pten below the heterozygosity threshold can elicit a cellular senescence response (Fig. 2b, and Supplementary Figs S3d and S5b).
Figure 2. Acute loss of Pten triggers the p53-dependent senescence pathway in primary mouse embryonic fibroblasts (MEFs).
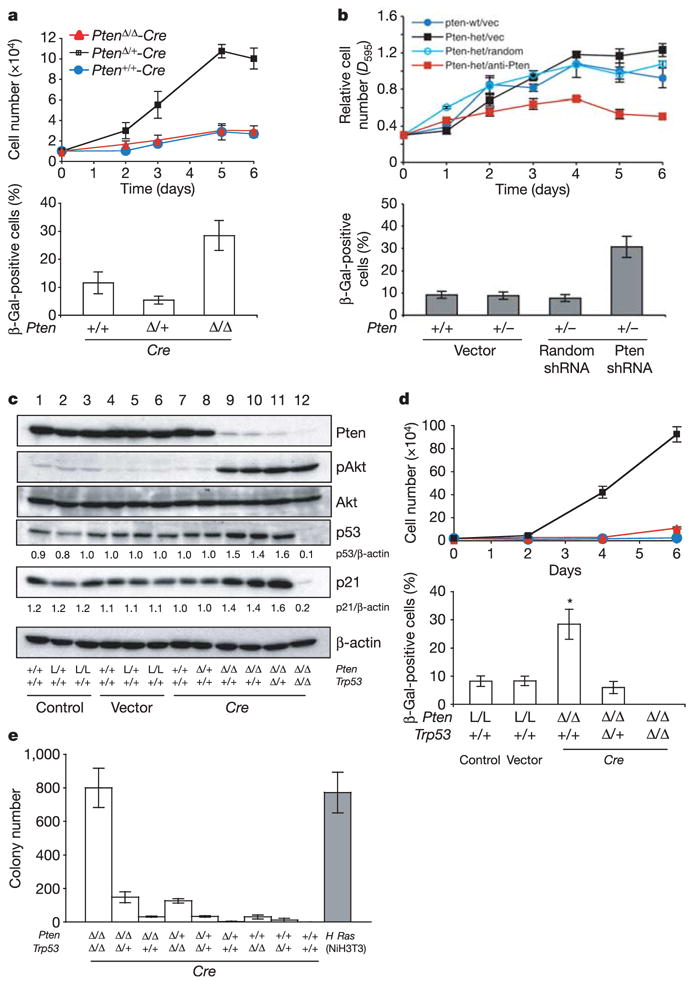
a, Top: growth curves of primary MEFs, infected with retroviral Cre (with selection) and followed over a 6-day period. Bottom: cellular senescence assay of cells from a. b, Top: growth curves of primary MEFs infected with Pten shRNA (with selection) and followed over a 6-day period. Bottom: cellular senescence assay of cells from b. c, Western blots of MEF lysates. Numbers indicate densitometrically determined protein levels relative to β-actin. d, Growth curves (top) and senescence staining (bottom) of PtenΔ /Δ ;Trp53Δ /Δ double-null MEFs (black squares), PtenΔ /Δ ;Trp53Δ /+ MEFs (red triangles) and PtenΔ /Δ MEFs (blue circles). Error bars indicate s.d. L/L indicates PtenloxP/loxP. e, Transformation assay of all combinations of Pten and Trp53 inactivation. H-Ras-infected NIH 3T3 cells served as positive control (grey bar). Error bars indicate s.d. for a representative experiment performed in triplicate.
Next we assessed the status of the p53 senescence pathway. Acute loss of Pten resulted in Akt activation and consistent induction of p53 levels (Fig. 2c) but not phosphorylation of p53 serine 15 (Supplementary Fig. S3f). Upregulation of p21 and the senescence marker plasminogen activator inhibitor-1 (PAI-1) was observed in Pten-deficient MEFs but not in Pten/Trp53 double-null MEFs (Fig. 2c and Supplementary Fig. S3g). Furthermore, loss of Trp53 led to a marked increase in proliferation in the Pten-null background (Fig. 2d and Supplementary Fig. S3e). In addition, SA-β-Gal activity in PtenΔ/Δ MEFs decreased sharply in a p53 dose-dependent manner and was undetectable in double-null MEFs. These findings confirm that escape from senescence is essential for PtenΔ /Δ MEFs to realize full proliferative potential (Fig. 2d). Staining by TdT-mediated dUTP nick end labelling (TUNEL) revealed no difference in apoptosis between Pten-null and Pten/Trp53 double-null MEFs (2 and 4 days, not shown). Finally, we tested whether p53 antagonizes cellular transformation of MEFs23. Complete inactivation of Pten and Trp53 resulted in transformation of MEFs, whereas transformation potential decreased in a Pten or Trp53 copy-dependent manner (Fig. 2e). These findings suggested that a p53-mediated senescence response restricts cell growth after loss of Pten.
Next we investigated whether the stability of p53 was altered on loss of Pten as reported previously9. In fact, we observed a prolonged rather than a reduced half-life of p53 protein in Pten-null compared with WT MEFs (Fig. 3a). This could be explained by the fact that complete loss of Pten resulted in increased accumulation of p19Arf (Fig. 3b), which is implicated in both the induction of cellular senescence and p53 stabilization through Mdm2 inhibition1,2. p16INK4a protein levels were also increased (Fig. 3b). In particular we found that the half-life of p21 was prolonged (Fig. 3a), indicating that p53-independent (post-transcriptional) mechanisms might also affect the accumulation of p21. Nevertheless, p53 was essential for p21 induction, because p21 was almost undetectable in p53/Pten double-null cells (Fig. 2c). Moreover, as reported previously24, an activated form of Akt (myristoylated Akt) elicited growth arrest and cellular senescence in primary MEFs, supporting the notion that senescence caused by acute loss of Pten is at least partly due to Akt activation (Fig. 3c and Supplementary Fig. S5c).
Figure 3. Acute loss of Pten results in ARF upregulation and p53/p21 stabilization in primary MEFs.
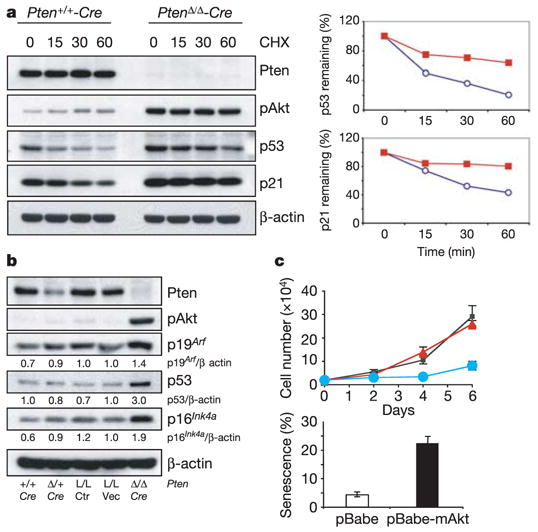
a, Left: inhibition of protein synthesis by cycloheximide (CHX) and western blotting at the indicated times (minutes) shows that the half-life of p53 and p21 proteins is prolonged on acute loss of Pten in MEFs. Right: quantification of p53 and p21 half-life from the western blots, normalized to β-actin. Blue circles, Pten+/+-Cre; red squares, PtenΔ /Δ -Cre. b, Western blotting demonstrates upregulation of p19Arf protein after acute Pten loss in MEFs. Pten+/+-Cre, PtenΔ /+-Cre, PtenloxP/loxP (L/L, Ctrl) and PtenloxP/loxP-MSCV vector (PtenL/L-vec) serve as controls. c, Expression of myristoylated Akt (mAkt) in MEFs induces growth arrest (top) and cellular senescence (bottom). Black squares, control; red triangles, pBabe; blue circles, pBabe-mAkt. Error bars represent s.d. for a representative experiment performed in triplicate.
To determine whether these observations were relevant in vivo to prostate cancer we performed immunohistochemical analysis (IHC). Staining of Pten revealed that Pten was efficiently and specifically deleted and unaffected by Trp53 status (Supplementary Fig. S4a). Akt activation and membrane recruitment were correlated only with loss of Pten in Ptenpc−/− and Ptenpc−/−;Trp53pc−/− mice, and not with p53 status (Supplementary Fig. S4a). In parallel, frozen prostate sections were stained for β-Gal activity. Whereas Pten-null prostates contained large numbers of senescent cells (‘blue glands’), WT prostates had very few (Fig. 4a and Supplementary Fig. S6b). Quantification revealed a 20-fold increase in senescent epithelial cells at 11 weeks compared with WT (Fig. 4b), but senescent cells were markedly decreased in the double-null prostates (Fig. 4a, b and Supplementary Fig. S6b). In addition, no cellular senescence was detected after heterozygous inactivation of Pten in Ptenpc+/− single or compound mutants (Supplementary Fig. S6a), which is consistent with our MEF results (Supplementary Fig. S5a, b, d) and in full agreement with the fact that partial or complete inactivation of Trp53 did not accelerate the PIN phenotype of Ptenpc+/− mutants (not shown). Apoptosis in prostates from various mutants remained comparable (Fig. 4c), whereas senescence of epithelial cells was accompanied by inverse changes in cellular proliferation (Fig. 4d); this was confirmed by double labelling of Pten-mutant and double mutant glands with Ki-67 and β-Gal (Supplementary Fig. S4b).
Figure 4. The p53-dependent cellular senescence pathway restricts tumorigenesis in Pten-deficient prostates.
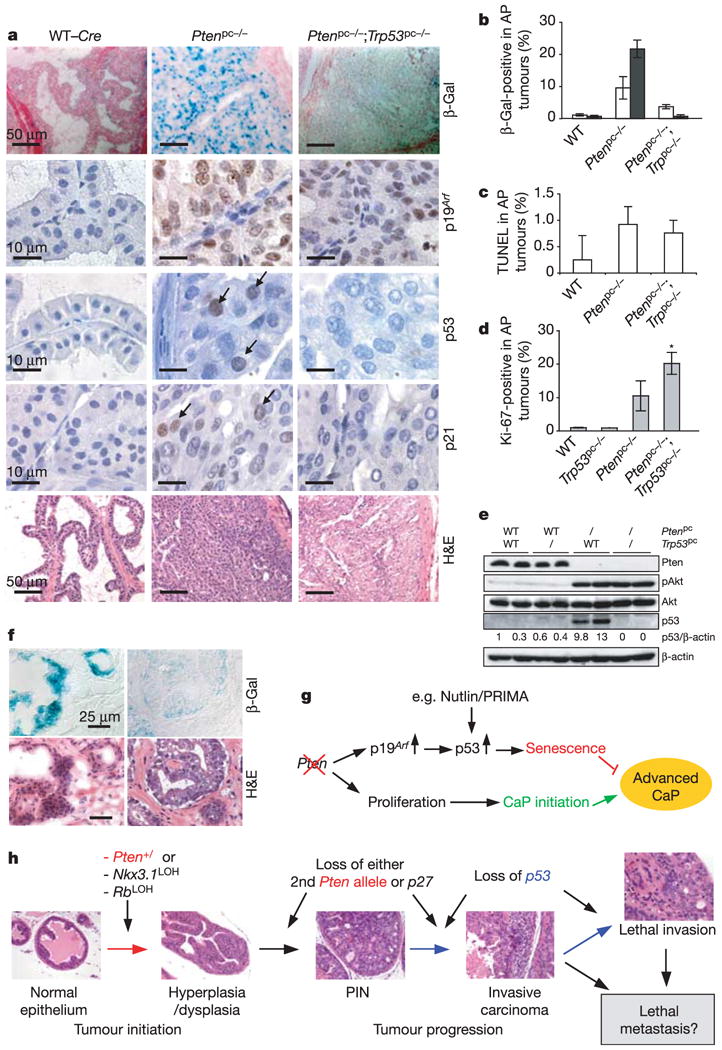
a, Senescence and histopathological analysis of 11-week-old prostates, stained as indicated. H&E, haematoxylin/eosin. Scale bars, 50μm (β-Gal and haematoxylin/eosin stain) and 10μm (p53, p19Arf and p21). b, Quantification of the β-Gal staining seen on AP sections at 8 weeks (open bars) and 11 weeks (filled bars). Representative sections from three mice were counted for each genotype. c, Quantification of TUNEL assay for apoptosis in the AP at 11 weeks (of more than two mice per genotype). d, Quantification of Ki-67 staining of 11-week-old AP done as in b. Error bars in b–d represent s.d. for a representative experiment performed in triplicate. Asterisk indicates statistical significance between Ptenpc−/− and Ptenpc−/−;Trp53pc−/− double mutants (P < 0.05). e, Western blot analysis of AP tissue from each genotype at 11 weeks shows Akt activation and p53 induction in Ptenpc−/− mice. f, Senescence (β-Gal) and histopathological (haematoxylin/eosin stain) analysis of cryosections from human radical prostatectomy samples, showing strong (+++) β-Gal staining from a hyperproliferative (preneoplastic) prostate gland (left) or weak (+) β-Gal staining from a neoplastic gland from a different patient (right). Scale bars, 25μm. g, A model for Pten-deficient tumorigenesis, p53 cooperativity and its therapeutic implications. Pten-deficient tumours may benefit from treatment with drugs that restore the activity of mutated p53 (for example, PRIMA28) or that stabilize WT p53 through the inactivation of MDM2 (for example, Nutlin29). h, A model for prostate tumour initiation, development and progression synergistically contributed by Pten, Trp53 and other genes. Loss of Trp53 accelerates cancer progression by a senescence escape mechanism in Pten-deficient tumours.
We next studied whether p53 is upregulated on Pten inactivation in vivo. Indeed, we found increased accumulation of p53 in the nuclei of prostate epithelial cells from Ptenpc−/− mice but not from WT (or Ptenpc−/−; Trp53pc−/− mice (Fig. 4a). Lysates from Ptenpc−/− prostates showed more than tenfold p53 induction by immunoblotting (Fig. 4e). Importantly, p19Arf and p21 protein levels were also increased in Ptenpc−/− mice (Fig. 4a), confirming in vivo activation of the p19Arf →p53 → p21 senescence pathway.
To address the relevance of these findings to human cancer we stained for senescence in specimens from early-stage human prostate cancer (n = 12; Supplementary Table S1). Strongβ-Gal staining was observed specifically in regions of prostate hyperplasia/PIN in some glands (3 of 12 samples, magnification × 100), but never in areas of frank tumour. Weak staining was observed in a further four samples at higher magnification ( × 400), predominantly in areas of prostate hyperplasia/PIN and rarely in areas of carcinoma (Fig. 4f; Supplementary Table S1).
Prostate cancer, the second leading cause of cancer-related deaths in males (after lung cancer), strikes one in six men in the USA25,26 and involves alterations of multiple tumour suppressor genes27. Our data provide insight into the molecular relationship between Pten and p53 in the prostate by demonstrating that loss of Pten induces p53 function (Fig. 4g). Because it has remained unclear whether loss of p53 function is associated with the initiation or progression of prostate cancer, our data firmly associate loss of Trp53 with a crucial role in prostate cancer progression (Fig. 4h). Our results show that acute loss of Pten does not result in decreased, but increased, p53 levels and function. Conversely, loss of Trp53 has no effect on Pten expression in vitro and in prostatic epithelium, whereas in combination with loss of Pten a lethal acceleration of prostate cancer is observed. Thus, regarding the Pten–p53 network, our data lend no support to a ‘two-in-one’ hit model of tumorigenesis, but as both genes need to be ablated for maximal disease progression they suggest a ‘one-by-one’ hit model for the genetic interaction of these major tumour suppressor genes. Our findings have therapeutic implications, because they indicate that PTEN-deficient prostate cancer might benefit from drugs that potentiate p53 activation in favour of a cellular senescence programme28,29 (Fig. 4g). The fact that acute homozygous (not heterozygous) loss of Pten triggers a p53-dependent cellular senescence programme in vivo provides a plausible explanation for why human prostate cancer at presentation does not select for complete loss of PTEN and thus highlights the relevance of PTEN haploinsufficiency for prostate cancer initiation. Furthermore, the finding that complete p53 inactivation alone has no phenotypic consequences in prostate could explain why loss of p53 is preferentially selected for in advanced prostate cancer where in addition it could allow the tumour to reach maximal proliferation through PTEN loss of heterozygosity.
METHODS
Pten and Trp53 mutant mice
PtenloxP/loxP mice were generated as described previously19, and Trp53loxP/loxP mice were generated with a similar strategy (Supplementary Fig. S1; details are available from the authors). Female PtenloxP/loxP;Trp53loxP/loxP mice were crossed with male PB-Cre4 transgenic mice21 for the prostate-specific deletion of Pten and Trp53. For genotyping, tail DNA was subjected to polymerase chain reaction analysis with the following primers. For PtenloxP/loxP, primer 1 (5′-AAAAGTTCCCCTGCTGATGATTTGT-3′ ) and primer 2 (5′-TGTTTTTGACCAATTAAAGTAGGCTGTG-3′ ) were used. To detect the deleted allele, Lpten3 primer (5′-TTCTCTTGAGCA CTGTTTCACAGGC-3′ ) and primer 1 were used. For Trp53loxP/loxP, primer 1 (5′-GAGACGCTGAGTCCGGTTCCCTCC-3′ ) and primer 2 (5′-GCAAGA GGTGACTTTGGGGTGAAGCTC-3′ ) were used.
Constructs
pMSCV-Cre-PIG was constructed by subcloning Cre into pMSCV-PURO-IRES-GFP (MSCV-PIG) vector (a gift from S. W. Lowe). The Nuclear Localization Sequence fusion Cre recombinase (NLS-Cre) was excised from pTZ-CreN vector (a gift from L. Nitschke).
Cell proliferation, transformation, senescence and apoptosis assays
Primary MEFs were prepared as described previously from individual embryos of various genotypes30, and infected with retroviruses expressing Cre-PURO-IRES-GFP, myristoylated Akt (mAkt) or corresponding control viruses. To prepare retro-viral particles, 2 × 106 Phoenix cells were plated per 10-cm culture dish, and then pMSCV-Cre-PURO-IRES-GFP or empty vector was transfected with Lipofectamine 2000 (Invitrogen). For infection, MEFs at passage 2 were plated at a density of (3–4) × 105 cells per 10-cm culture dish and infected by virus from Phoenix cells 48 h after transfection. After selection in the presence of 3μg ml−1 puromycin (Sigma), MEFs at passage 6 were used for growth curves, western blotting and senescence analysis. To determine senescence, MEFs were plated at 104 cells per well of a six-well plate in triplicate, and after 4 days SA-β-Gal activity was revealed with the senescence detection kit (Calbiochem) and quantified (more than 200 cells per sample). For prostate tissue, frozen sections 6μm thick were stained for β-Gal as above. For transformation assay, selected MEFs (3 × 104) at passage 6 were suspended in medium containing 0.3% agar onto solidified 0.6% agar per well of a six-well plate, and the number of colonies was assessed after 21 days. For apoptosis analysis, dewaxed and rehydrated paraffin sections were treated for with the in situ Cell Death Detection Kit (Roche). Apoptotic cells were identified by positive TUNEL staining. A total of 500 cells were counted from five different fields for each section, and the analysis was repeated at least twice for each genotype.
Short hairpin RNA (shRNA)
Pten-directed shRNA (5′-GATCCCCAGACCA TAACCCACCACAGTTCAAGAGACTGTGGTGGGTTATGGTCTTTTTTA-3′ ) was designed using the Oligoengine RNAi design tool; the resulting oligonucleotides were subcloned into the pSUPER-puro vector (Oligoengine) and trans-fected into Phoenix packaging cells. The random oligonucleotide (5 ′-GATCCCCAAGGAGACGAGCAAGAGAATTCAAGAGATTCTCTTGCTCGTC TCCTTTTTTTA-3′ ) was used as control. Primary Pten WT or heterozygous MEFs at passage 2 were infected, selected (3 days in 2μg ml−1 puromycin) and plated at 2.5 × 104 cells per well in 12-well dishes in duplicates for determination of growth curves by spectroscopic measurement of crystal violet uptake. β-Gal activity was measured on day 4 of the growth curve with cells treated identically and in parallel.
Western blot and immunohistochemistry
MEF lysates were prepared with RIPA buffer (1 × PBS, 1% Nonidet P40, 0.5% sodium deoxycholate, 0.1% SDS and protease inhibitor cocktail (Roche)). The following antibodies were used for western blotting: mouse monoclonal anti-Pten (6H2.1; Cascade BioScience), rabbit polyclonal anti-p19Arf (Ab-1; Calbiochem), rabbit polyclonal anti-p53 (CM5; Novocastra), rabbit polyclonal anti-Akt and anti-phosphoserine 473 of Akt (Cell Signalling), rabbit polyclonal anti-p21 (C-19; Santa Cruz), rabbit polyclonal anti-p16 (M-156; Santa Cruz), rabbit polyclonal anti-PAI-1 (H-135; Santa Cruz) and mouse monoclonal anti-β-actin (AC-74; Sigma). To determine p53/p21 protein half-life, MEFs (Pten+/+-Cre and PtenΔ /Δ -Cre, at 80% con-fluence and equal passage number) were treated with 30μg ml−1 cycloheximide (Sigma) and harvested at the indicated times for western blot analysis. For prostate analysis, protein extracts were prepared by grinding prostate tissues in RIPA buffer at a ratio of 1 mg per 5μl. After brief sonication and centrifugation, the supernatant was collected for western blotting. Antibodies used were rabbit polyclonal anti-PTEN (9552; Cell Signalling). Antibodies against p53, pAkt and Akt were as above. For immunohistochemistry (IHC), tissues were fixed in 10% formalin and embedded in paraffin in accordance with standard procedures. Sections were stained for phospho-Akt (Ser 473) antibody (Cell Signalling), PTEN (Ab-2; NeoMarkers), Ki-67 (Novocastra), p19Arf (ab80-50; Abcam), p21 (F-5; Santa Cruz), p53 (FL-393; Santa Cruz), androgen receptor (N-20; Santa Cruz) and p63 (550025; Becton Dickson Transduction Lab).
MRI
Individual mice were subjected to MRI assessment for the detection of prostate tumours. In brief, mice were anaesthetized with 2% isofluorane, and images were obtained on a Bruker 4.7-T 40-cm bore magnet with a commercial 7-cm inner diameter birdcage coil similar to the protocol described previously19. Low-resolution sagittal and axial scout images were obtained initially, followed by high-spatial-resolution T2-weighted axial images (repetition interval (TR) = 3,800 ms, effective echo time (TE) = 35 ms, eight echoes per phase encoding step, spatial resolution = 1.0 mm slice thickness × 112μm × 112μm in plane resolution, and four repetitions of data acquisition for 8–9 min of imaging time). Invasiveness-free and cumulative survival curves were obtained with Kaplan–Meier analysis as described previously19.
Supplementary Material
Acknowledgments
We thank T. Maeda and T. Jacks for helpful suggestions; D. Peeper, C. Schmitt and M. Serrano for exchanging unpublished data and coordinating the submission of manuscripts; N. Hay, U. Greber and S. Hemmi for reagents; L. Cai and L. DiSantis for critical reading and editing of the manuscript; other members of the Pandolfi lab for advice and discussion; K. Manova and C. Farrell from the Molecular Cytology Core Facility for assistance with IHC analysis; and C. Le, C. Matei, D. Procissi and I. Buchanan for MRI analysis. This work was supported by NIH grants to P.P.P.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432:307–315. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 2.Serrano M, Blasco MA. Putting the stress on senescence. Curr Opin Cell Biol. 2001;13:748–753. doi: 10.1016/s0955-0674(00)00278-7. [DOI] [PubMed] [Google Scholar]
- 3.Campisi J. Cellular senescence as a tumour-suppressor mechanism. Trends Cell Biol. 2001;11:S27–S31. doi: 10.1016/s0962-8924(01)02151-1. [DOI] [PubMed] [Google Scholar]
- 4.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 5.Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumour suppression. Cell. 2000;100:387–390. doi: 10.1016/s0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- 6.Stambolic V, et al. Regulation of PTEN transcription by p53. Mol Cell. 2001;8:317–325. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 7.Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci USA. 2001;98:11598–11603. doi: 10.1073/pnas.181181198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou BP, et al. HER-2/neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nature Cell Biol. 2001;3:973–982. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 9.Freeman DJ, et al. PTEN tumour suppressor regulates p53 protein levels and activity through phosphatase-dependent and -independent mechanisms. Cancer Cell. 2003;3:117–130. doi: 10.1016/s1535-6108(03)00021-7. [DOI] [PubMed] [Google Scholar]
- 10.Trotman LC, Pandolfi PP. PTEN and p53: who will get the upper hand? Cancer Cell. 2003;3:97–99. doi: 10.1016/s1535-6108(03)00022-9. [DOI] [PubMed] [Google Scholar]
- 11.Schmitt CA, et al. A senescence program controlled by p53 and p16INK4a contributes to the outcome of cancer therapy. Cell. 2002;109:335–346. doi: 10.1016/s0092-8674(02)00734-1. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki H, et al. Interfocal heterogeneity of PTEN/MMAC1 gene alterations in multiple metastatic prostate cancer tissues. Cancer Res. 1998;58:204–209. [PubMed] [Google Scholar]
- 13.Feilotter HE, Nagai MA, Boag AH, Eng C, Mulligan LM. Analysis of PTEN and the 10q23 region in primary prostate carcinomas. Oncogene. 1998;16:1743–1748. doi: 10.1038/sj.onc.1200205. [DOI] [PubMed] [Google Scholar]
- 14.Muller M, Rink K, Krause H, Miller K. PTEN/MMAC1 mutations in prostate cancer. Prostate Cancer Prostatic Dis. 2000;3:S32. doi: 10.1038/sj.pcan.4500457. [DOI] [PubMed] [Google Scholar]
- 15.Hermans KG, et al. Loss of a small region around the PTEN locus is a major chromosome 10 alteration in prostate cancer xenografts and cell lines. Genes Chromosom Cancer. 2004;39:171–184. doi: 10.1002/gcc.10311. [DOI] [PubMed] [Google Scholar]
- 16.Qian J, et al. Loss of p53 and c-myc overrepresentation in stage T(2–3)N(1–3)M(0) prostate cancer are potential markers for cancer progression. Mod Pathol. 2002;15:35–44. doi: 10.1038/modpathol.3880487. [DOI] [PubMed] [Google Scholar]
- 17.Navone NM, et al. p53 mutations in prostate cancer bone metastases suggest that selected p53 mutants in the primary site define foci with metastatic potential. J Urol. 1999;161:304–308. [PubMed] [Google Scholar]
- 18.Di Cristofano A, De Acetis M, Koff A, Cordon-Cardo C, Pandolfi PP. Pten and p27KIP1 cooperate in prostate cancer tumour suppression in the mouse. Nature Genet. 2001;27:222–224. doi: 10.1038/84879. [DOI] [PubMed] [Google Scholar]
- 19.Trotman LC, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:385–396. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jonkers J, Berns A. Conditional mouse models of sporadic cancer. Nature Rev Cancer. 2002;2:251–265. doi: 10.1038/nrc777. [DOI] [PubMed] [Google Scholar]
- 21.Wu X, et al. Generation of a prostate epithelial cell-specific Cre transgenic mouse model for tissue-specific gene ablation. Mech Dev. 2001;101:61–69. doi: 10.1016/s0925-4773(00)00551-7. [DOI] [PubMed] [Google Scholar]
- 22.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinberg RA. The cat and mouse games that genes, viruses, and cells play. Cell. 1997;88:573–575. doi: 10.1016/s0092-8674(00)81897-8. [DOI] [PubMed] [Google Scholar]
- 24.Miyauchi H, et al. Akt negatively regulates the in vitro lifespan of human endothelial cells via a p53/p21-dependent pathway. EMBO J. 2004;23:212–220. doi: 10.1038/sj.emboj.7600045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stewart SL, King JB, Thompson TD, Friedman C, Wingo PA. Cancer mortality surveillance—United States, 1990–2000. MMWR Surveill Summ. 2004;53:1–108. [PubMed] [Google Scholar]
- 26.Levi F, Lucchini F, Negri E, Boyle P, La Vecchia C. Leveling of prostate cancer mortality in Western Europe. Prostate. 2004;60:46–52. doi: 10.1002/pros.20058. [DOI] [PubMed] [Google Scholar]
- 27.Abate-Shen C, Shen MM. Molecular genetics of prostate cancer. Genes Dev. 2000;14:2410–2434. doi: 10.1101/gad.819500. [DOI] [PubMed] [Google Scholar]
- 28.Bykov VJ, et al. Restoration of the tumour suppressor function to mutant p53 by a low-molecular-weight compound. Nature Med. 2002;8:282–288. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- 29.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 30.Maeda T, et al. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 2005;433:278–285. doi: 10.1038/nature03203. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


