Summary
Hydroxy-naphthoquinones are competitive inhibitors of the cytochrome bc1 complex that bind to the ubiquinol oxidation site between cytochrome b and the iron-sulfur protein and presumably mimic a transition state in the ubiquinol oxidation reaction catalyzed by the enzyme. The parameters that affect efficacy of binding of these inhibitors to the bc1 complex are not well understood. Atovaquone®, a hydroxy-naphthoquinone, has been used therapeutically to treat Pneumocystis carinii and Plasmodium infections. As the pathogens have developed resistance to this drug, it is important to understand the molecular basis of the drug resistance and to develop new drugs that can circumvent the drug resistance. We previously developed the yeast and bovine bc1 complexes as surrogates to model the interaction of atovaquone with the bc1 complexes of the target pathogens and human host.
As a first step to identify new cytochrome bc1 complex inhibitors with therapeutic potential and to better understand the determinants of inhibitor binding, we have screened a library of 2-hydroxy-naphthoquinones with aromatic, cyclic, and non-cyclic alkyl side-chain substitutions at carbon-3 on the hydroxy-quinone ring. We found a group of compounds with alkyl side-chains that effectively inhibit the yeast bc1 complex. Molecular modeling of these into the crystal structure of the yeast cytochrome bc1 complex provides structural and quantitative explanations for their binding efficacy to the target enzyme. In addition we also identified a 2-hydroxy-naphthoquinone with a branched side-chain that has potential for development as an anti-fungal and anti-parasitic therapeutic.
Keywords: Hydroxy-naphthoquinones, cytochrome bc1 complex, malaria, plasmodium, pneumocystis, atovaquone
Introduction
Atovaquone is a substituted hydroxy-naphthoquinone (2-[trans-4-(4′-chlorophenyl) cyclohexyl]-3-hydroxy-1,4-naphthoquinone) that binds tightly and competitively to the ubiquinol oxidation site of the cytochrome bc1 complex (1). This inhibitor was first used therapeutically as an anti-malaria compound with broad spectrum activity against apicomplexan parasites (2–4) and later was also shown to prevent and clear Pneumocystis jirovecii pneumonia (5,6). However, the pathogens soon developed resistance to the drug, and it was shown that the resistance was due to mutations in the gene for cytochrome b, a subunit that forms part of the ubiquinol oxidation site of the cytochrome bc1 complex (7). As the yeast bc1 complex is also inhibited by atovaquone, we have developed the yeast Saccharomyces cerevisiae as a model to explain atovaquone resistance in Pneumocystis jirovecii (8), Plasmodium falciparum (9) and Toxoplasma gondii (10).
In the current study, we have extended the use of the yeast bc1 complex model by combining inhibitor titrations and computer modeling calculations to understand the structural parameters that affect efficacy of binding of 2-hydroxy-naphthoquinones to the bc1 complex. We first screened a randomly chosen library of 2-hydroxy-naphthoquinones with aromatic, cyclic, and non-cyclic alkyl side-chains at position 3 on the hydroxy-quinone ring for inhibition of bc1 complex activity. We then tested a series of 2-hydroxynaphthoquinones with linear alkyl side-chains of varying length in order to obtain a quantitative structure/activity relationship based on side-chain length. We also evaluated the relative efficacy of two stereoisomers with a branched side-chain. The results of these comparisons provide a starting point to synthesize new hydroxy-naphthoquinone inhibitors of the bc1 complex with potential therapeutic uses.
Experimental Procedures
Materials
Dodecylmaltoside was obtained from Roche Applied Science. DEAE-Biogel A was obtained from Bio-Rad. Diisopropylfluorophosphate, decyl ubiquinone and dithionite were purchased from Sigma. Stigmatellin was purchased from Fluka Biochemica. Atovaquone was a gift from GlaxoSmithKline. A collection of 2-hydroxy-1,4-naphthoquinones with aromatic, cyclic, and non-cyclic alkyl side-chains was obtained from the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis of the National Cancer Institute. The hydroxy-1,4-naphthoquinones were dissolved in dimethyl sulfoxide at 2 mM concentration and stored at -20° C.
Synthesis of Naphthoquinones
The synthesis of 3-alkyl-2-hydroxy-1,4-naphthoquinones exploits radical alkylation of commercial 2-hydroxy-1,4-naphthoqinone with alkyl iodides in the presence of tributyltin hydride and 2,2′-azobisisobutyronitrile as radical initiator (11). PhSO2- protection with PhSO2Cl/K2CO3 in DMF gave 66% yield of the O-protected product. Chiral (S)- and (R)- 2-methyloctyl iodides were prepared in accordance with the known sequence (12–14), which started from methyl-(R)- or (S)- 3-hydroxy-2-methylpropionate correspondingly and included copper catalyzed cross coupling of alkyltosylate and Grignard reagent as a key step (15). The synthesized chiral iodides were then used for radical alkylation.
Purification of Cytochrome bc1 Complexes
Cytochrome bc1 complexes from yeast and bovine heart were isolated from mitochondrial membranes as described previously (16,17).
Ubiquinol-Cytochrome c Reductase Activity Measurements
Cytochrome c reductase activity was assayed in 50 mM potassium phosphate, pH 7.0, 250 mM sucrose, 0.2 mM EDTA, 1 mM NaN3, 2.5 mM KCN, 0.01% Tween-20 and 40 μM cytochrome c at 23°C. The cytochrome bc1 complex was diluted to 2.5 nM in the assay buffer, inhibitor was added to the assay mixture and allowed to stir with the enzyme for 1 min, after which the reaction was started by adding 2,3-dimethoxy-5-methyl 6-decyl-1,4- benzoquinol, an analogue of ubiquinol. Reduction of cytochrome c was monitored in an Aminco DW-2a spectrophotometer at 550 versus 539 nm in dual wavelength mode. Data was collected and analyzed using an Online Instrument Systems Inc. computer interface and software. IC50 values for compounds that were inhibitory at concentrations less than 200 nM were determined from titration curves of inhibitor concentration versus activity. Relative amounts of the chiral isomers in the racemic mixture of compound #10576 were calculated from the IC50 values of the two isomers and that of the mixture using the equation X/IC50(S) + Y/IC50(R) = 1/IC50(M) where X + Y = 1.0.
Molecular Modeling
Molecular modeling was carried out on Silicon Graphics O2 and Octane workstations using the commercially available Insight II® software package (Acclerys Inc., San Diego). The starting structure was the energy-minimized atovaquone-liganded yeast cytochrome bc1 complex (1). Briefly, a minimized conformation of atovaquone was docked into a stigmatellin liganded crystal structure (18) that was modified to include the rotation of Glu-272 and a water mediated hydrogen bond observed in the nHDBT liganded crystal structure (19). For the structures shown here, the initial docking was achieved by overlay of the hydroxynaphthoquinone ring with the one from the previously calculated atovaquone structure. The initial position of the linear aliphatic side-chain, prior to the molecular dynamics run, followed the one observed in the nHDBT crystal structure.
For the molecular dynamics calculations, a set of restraints was set up between the hydroxy-naphthoquinones, His-181 of the Rieske protein, and Glu-272 of cytochrome b in order to maintain the ligand in position during the dynamic stages. The naphthoquinones and cytochrome b residues within 4.0 Å were allowed to be flexible. A surrounding 9.5 Å shell of residues in both cytochrome b and the iron-sulfur protein was fixed, and the most distant residues were excluded from the calculation in order to obtain a manageable simulation speed. A 9.5 Å atom-based cut-off for nonbonding interactions was used during the calculations, with the dielectric constant set at 2.0. Eight simulated annealing runs were performed, each from 800 to 298 K, with five temperature steps and a simulation time of 5000 fs/step. The Nose' temperature control method was used with a 0.5 fs/iteration time step. A custom macro was written to select the lowest energy structure from each dynamics run for continued modeling. Between each dynamics run, a minimization of 250 iterations was performed. After the final round of molecular dynamics, the lowest energy structure was minimized to a final convergence criterion of 0.001, using Cauchy's steepest descent method as implemented in the Discover 3® module within the Insight II® software, followed by conjugate gradient and Newton methods in succession. Of the eight minimized results obtained, the three lowest energy structures were chosen for binding energy calculation.
The binding energy calculation was adapted from a previous method (8) and uses a common subset that included the naphthoquinone and cytochrome b residues within 4.0 Å of the inhibitor. The reported value for each naphthoquinone is an average of the three calculated lowest energy structures and contains non-bonding interactions (van der Waals and electrostatic) as well as internal conformational energies of the ligand and adjacent pocket residues.
Results
Inhibition of Bovine and Yeast bc1 Complexes by Linear Alkyl Naphthoquinones
The molecular target of the hydroxy-naphthoquinone inhibitors is known to be the ubiquinol oxidation pocket at the center P site of the cytochrome bc1 complex (1). We thus tested a library of approximately 60 2-hydroxy-naphthoquinones for inhibition of ubiquinol-cytochrome c reductase activity of both yeast and bovine cytochrome bc1 complexes. The compounds tested included derivatives with aromatic, cyclic non-aromatic, branched and linear side-chains at the carbon-3 position of the hydroxy-quinone ring. The results of this initial survey are summarized in Supplemental Data Table 1, which also includes the structures of the tested compounds. Most of these compounds were found to show no or very poor bc1 complex inhibition and were not studied further.
A small group of alkyl hydroxy-naphthoquinones displayed significant inhibition of enzymatic activity. We thus synthesized the remaining compounds of the series with linear side-chains of varying length for a more systematic study of the structure/activity relationship in this group of inhibitors. All members of the linear series inhibited both yeast and bovine cytochrome bc1 complex activities. As can be seen from the data in Fig. 1, the extent of inhibition of the yeast enzyme was usually twice as great as that of the bovine enzyme with any given compound. The IC50 values with both bc1 complexes show that a linear alkyl chain of at least 8 carbons is required to achieve significant enzymatic inhibition with IC50 values below 100 nM. The best IC50 values obtained with the yeast enzyme were with the C8/C9 side-chains whereas the C9/C10 side-chains were the most effective with the bovine bc1 complex. For both species, a slight decrease in binding was observed when the linear alkyl chain was longer than C10.
Figure 1. Effect of chain length on inhibition of bovine and yeast cytochrome bc1 complexes by 2-hydroxy-naphthoquinones with linear alkyl side-chains.

The IC50 values for inhibition of the bc1 complex activity is shown with light gray bars for the yeast enzyme and dark bars for the bovine enzyme.
Differential Binding of a Branched Alkyl Naphthoquinone to Bovine and Yeast bc1 Complexes
The branched 3-alkyl 2-hydroxy-naphthoquinones included in the library did not show significant inhibition of bc1 activity except for the compound #10576. This hydroxy-naphthoquinone contains an 8-carbon branched side-chain with a methyl group on the second carbon of the side-chain from the quinone ring. The IC50 for inhibition of the yeast enzyme by this branched side-chain hydroxy-naphthoquinone dropped markedly from 80 nM to 15 nM when compared to the linear 8-carbon chain, while the inhibition of the bovine enzyme remained the same with the two compounds (Fig. 2).
Figure 2. Inhibition of bovine and yeast cytochrome bc1 complexes by 2-hydroxy-naphthoquinones with 8-carbon linear alkyl chain and stereoisomers of a methyl substituted 8-carbon branched alkyl chain.

The IC50 values for inhibition of the bc1 complex activity are shown with light gray bars for the yeast enzyme and dark bars for the bovine enzyme. The structure of the branched chain 2-hydroxy-naphthoquinone (compound #10576) is shown above the bar graphs.
Because the methyl group in this position creates an asymmetric carbon, we speculated that the compound obtained from the National Cancer Institute collection would be a racemic mixture and that one of the stereoisomers might display even better inhibition properties. We thus synthesized the R (R-10576) and S (S-10576) stereoisomers of #10576 as described above. Accordingly, a further drop of the IC50 from 15 nM to 6 nM was observed with the S isomer of #10576, as would be expected if the racemic mixture of the original batch contained approximately equal amounts of the two isomers. From the measured IC50 values of the R and S isomers and of the mixture, it can be estimated that the mixture contains approximately 45 ± 5 percent S isomer. The IC50 value for the yeast enzyme was about 28 times lower with the S than with the R stereoisomer and confirmed the strong stereo-selectivity of the binding pocket for this class of compound. Interestingly, the side-chain of R-10576 resulted in weaker inhibition of both yeast and bovine enzymes (with IC50 of 170 nM and 420 nM) than the unbranched 8-carbon chain derivative (80 nM and 190 nM).
In order to compare the efficacy of inhibitors with the bovine and yeast bc1 complexes, we defined the “Selectivity Coefficient” of an inhibitor as the IC50 obtained with the bovine enzyme divided by the IC50 obtained with the yeast enzyme. The Selectivity Coefficients for the linear 8 carbon chain naphthoquinones, atovaquone, myxothiazol, and the R and S stereoisomers of #10576 are show in Fig. 3. Contrary to myxothiazol, the linear 8-carbon chain naphthoquinone, and the R isomer of #10576, which displayed poor enzyme selectivity, atovaquone and the S isomer of #10576 showed high Selectivity Coefficients. Notably, the selectivity of S-10576 was approximately 3 times higher than that recorded for atovaquone.
Figure 3. Selectivity of 2-hydroxy-naphthoquinone inhibitors of the bovine and yeast cytochrome bc1 complexes.

The bar graphs show the Selectivity Coefficients for inhibition of the yeast versus bovine bc1 complex by the 2-hydroxy-naphthoquinone with an 8-carbon linear alkyl side-chain (C8), both R and S stereoisomers of compound #10576 (C8+1R and C8+1S), and atovaquone (ATV). The value for myxothiazol (Myx), a non-quinoid inhibitor, is also shown.
Molecular Modeling of Linear and Branched Alkyl Naphthoquinones Docked in the Yeast Cytochrome bc1 Complex
All members of the linear hydroxy-naphthoquinone series and both stereoisomers of #10576 were docked into the ubiquinol oxidation pocket at center P site of the yeast bc1 complex. We used a previously modeled atovaquone-bound structure (7) as a starting point for calculating the lowest energy conformations of the docked ligands. The results of these docking experiments are overlaid in Figs. 4 and 5. Like atovaquone, the hydroxyl group of the hydroxy-naphthoquinones binds via a hydrogen bond to the nitrogen of His-181 of the Rieske iron-sulfur protein. On the opposite side of the ring system the carbonyl group at position 4 of the quinone ring forms a water-mediated hydrogen bond with Glu-272 of cytochrome b. The bulk of the intermolecular interactions between the naphthoquinones and the cytochrome b are essentially hydrophobic. The side-chains freely interact with a network of aromatic and aliphatic side-chains of the cytochrome b including Leu-282, Leu-275, Ile125, Phe-296, Ile-147, Leu-150 and Phe-151.
Figure 4. Overlaid view of the calculated structures of short (A) and long (B) linear alkyl side-chain naphthoquinones docked into the yeast cytochrome bc1 complex.

Amino acids in the cytochrome b binding pocket are labeled and their carbon atoms are green, nitrogen atoms are blue and oxygen atoms are red. The hydroxy-naphthoquinones with linear alkyl chains containing 4 carbons (white), 6 carbons (yellow), 8 carbons (cyan), 9 carbons (orange), 10 carbons (magenta) and 11 carbons (blue) are shown.
Figure 5. Overlaid view of the calculated structures of the R and S stereoisomers of compound #10576 docked into the yeast cytochrome bc1 complex.

The carbon atoms of the cytochrome b binding pocket amino acid residues are green, nitrogen atoms are blue, oxygen atoms are red, and sulfur atom is yellow. The R (yellow) and S (cyan) stereoisomers of compound #10576 are shown, and the modified positions of the side-chains of the residues Met-295 and Phe-296 (purple) are also shown.
With the linear alkyl series of hydroxy-naphthoquinones, we observed that the shortest side-chains, containing 6 or fewer carbons, did not fully optimize the intermolecular hydrophobic interaction within the binding pocket. As can be seen in Fig. 4A, these side-chains remained in an extended, straight position at all stages of the dynamics and minimization process (Fig. 4A). By contrast, the calculated structures obtained with the longer chains, containing 8 or more carbons in the side-chain, displayed bended features (Fig. 4B) that were similar to the bending of the ligand observed in the stigmatellin-bound structure (18).
Docking and energy minimization of the stereoisomers of #10576 into the binding pocket at the center P site of the yeast bc1 complex generated two different calculated structures that are superimposed in Fig. 5. The 8-carbon chain of the S stereoisomer mostly followed the same binding groove as the previously described linear octyl derivative. The side-chain is bent and fully interacts with the cytochrome b (Fig. 5, blue chain). Closer examination of the calculated structure of this stereoisomer reveals that the branched methyl group, by its strong hydrophobic interaction with the Val-146 residue, constrains the rotation of the loose end of the side-chain into the binding groove. The branched methyl group of the R stereoisomer displays the same interaction with the Val-146 residue, but because of steric constraints, the chain cannot bend and shows a straight conformation similar to the short linear derivatives (Fig. 5, yellow chain). In order to accommodate the bulk of this straight chain, the calculated structure has to significantly rotate the position of the side-chains of the cytochrome b residues Met-295 and Phe-296 (Fig. 5, purple residues).
Calculation of Binding Energy of Linear and Branched Alkyl Naphthoquinones to the Yeast Cytochrome bc1 Complex
The energy required for binding of the linear side-chain naphthoquinones was calculated for each of the modeled structures. The calculated binding energies were then compared with the experimentally measured IC50 values. The relative increase in calculated binding energies correlated well with the relative increase in IC50 values for the yeast bc1 complex for each linear alkyl side-chain derivative as shown in Fig. 6. A slight difference was observed for the longest linear chains, for which the calculation predicted an increased binding instead of the observed decrease. Possible reasons for this difference are discussed below.
Figure 6. Experimentally measured IC50 values compared to calculated binding energies of naphthoquinones with linear alkyl side-chains in the yeast cytochrome bc1 complex.

The IC50 values for inhibition of the yeast bc1 complex activity are shown with light gray bars. The dark bars show the calculated changes in binding energy.
A similar graphic comparison was plotted using the calculated binding energies extracted from the modeled structures of the #10576 stereoisomers and the experimentally measured IC50 values obtained with these branched naphthoquinones. As with the linear series, there was a good correlation observed between the two sets of data as shown in Fig. 7.
Figure 7. Experimentally measured IC50 values compared to calculated binding energies of the naphthoquinone with an 8 carbon linear alkyl side-chain and the R and S stereoisomers of compound #10576 in yeast cytochrome bc1 complex.

The IC50 values for inhibition of the yeast bc1 complex activity are shown with light gray bars. The dark bars show the calculated changes in binding energy.
Discussion
Atovaquone is a hydroxy-naphthoquinone that has been used therapeutically against parasites and pathogenic fungi and that acts by inhibiting the cytochrome bc1 complex (1). Spontaneously arising mutations in the cytochrome b gene were genetically linked to drug resistance in the pathogens, but it was not possible to isolate the cytochrome bc1 complex from these organisms to confirm the molecular basis of the drug resistance.
Because the yeast cytochrome bc1 complex is also inhibited by atovaquone, we have previously developed Saccharomyces cerevisiae as a model to study cytochrome b mutations conferring atovaquone resistance in Pneumocystis (8), Plasmodium (9), and Toxoplasma (10) species. In those studies, using structures of the yeast bc1 complex co-crystallized with various inhibitors (18,19) as a starting point, we were able to design a coherent computer model of the interactions between the drug and its target. This virtual model of atovaquone binding was validated by strong correlations between the experimental measurements of atovaquone resistance in the mutated yeast enzymes and the calculated drug binding energies. An overlay of these crystal structures with our modeled structures (Fig. 8) shows how the hydroxy-naphthoquinones share common features with stigmatellin, such as the bended position of the chains, and nHDBT, which has a similar ring geometry as the hydroxy-naphthoquinone ring and a similiar position of the short alkyl side-chain.
Figure 8. Binding of different inhibitors to the ubiquinol oxidation pocket at center P of the yeast cytochrome bc1 complex.

Binding of stigmatellin (STG) and nHDBT are compared by overlays of the two inhibitors in panels A and B, with stigmatellin colored by atom in panel A and nHDBT colored by atom in panel B. Views of stigmatellin (18) and nHDBT (19) were constructed from the coordinates of the crystal structures of the yeast enzyme with the inhibitors bound. Binding of atovaquone (ATV) and the S isomer of #10576 are compared by overlays of the two inhibitors in panels C and D, with atovaquone colored by atom in panel C and #10576 colored by atom in panel D. When colored, oxygen atoms are red, carbons are green, sulfur is yellow and nitrogen is blue. Portions of the Rieske protein and cytochrome b are shown as yellow and blue ribbons, respectively, and His-181 of the Rieske protein is also shown for perspective. In panel A the carboxyl group of Glu-272 of cytochrome b is oriented toward stigmatellin and forms a hydrogen bond directly to the hydroxy-chromone ring of the inhibitor. In panels B, C, and D the carboxyl group of Glu-272 is rotated away from the inhibitor, and a water molecule forms hydrogen bonds between a carbonyl oxygen on the quinone ring of the benzoxythiazole and hydroxynaphthoquinone inhibitors and the backbone nitrogen of Glu-272.
In the current study, we have used this set of tools to screen a small library of hydroxy-naphthoquinones that included a wide variety of linear, branched, saturated, unsaturated and aromatic substitutions in position 3 of the quinone ring (see Supplemental Data Table 1). Most of these compounds, including derivatives known for their anti-tumor (20) and anti-leishmanial (21) activities, were found to have no or very poor bc1 complex inhibition activity and were not studied further. Of the remaining set, approximately half of the compounds were suspected as likely to be toxic for humans and discarded because of a better inhibition of the bovine bc1 complex than of the yeast enzyme.
The linear alkyl side-chain derivatives displayed significant inhibition of the yeast enzyme although they did not exhibit significant species selectivity. Molecular modeling revealed that the longer the side-chain, the greater the hydrophobic interaction between the inhibitor and the binding pocket of the cytochrome b (Fig. 4B). Calculation of the binding energy for these compounds was correlated by titrations of their inhibitory activity with the yeast enzyme, except for the 4-carbon side-chain compound (Fig. 6). This discrepancy can be explained by the set of restraints that was included in the modeling in order to maintain low affinity ligands in the binding pocket during the high temperature annealing runs of the dynamics stage.
The drug atovaquone has been known for poor bioavailability attributed to its poor solubility (22). Although we did not evaluate bioavailability, the size and hydrophobicity of the side-chain impacted the aqueous solubility of the hydroxy-naphthoquinones in the current study (data not shown). We observed that saturated alkyl side-chain derivatives longer than C9 are less effective with both yeast and bovine bc1 complexes (Fig. 1). Because our molecular modeling could not incorporate the solubility effect of these long side-chains, it usually predicted slightly higher binding affinity for the naphthoquinones with longer chains (e.g. C11) than the observed degree of inhibition (Fig. 6).
It was observed that insertion of a methyl group in the beta position of a saturated 8-carbon linear side-chain (compound #10576) significantly impacted the inhibition efficacy, lowering the IC50 more than five times when compared to the linear C8 side-chain. Of the various compounds that we tested during our screening, this branched hydroxy-naphthoquinone exhibited the most potent inhibition of the yeast bc1 complex. Synthesis of the pure S stereoisomer of #10576 enhanced this inhibition even further. Notably, this stereoisomer displayed a selectivity toward the yeast enzyme three times greater than that exhibited by the drug atovaquone. Molecular modeling of #10576 interaction with the yeast bc1 complex binding pocket shows that both stereoisomers have the branched methyl group facing in the same direction toward the Val-146 residue. However, while this decrease in degrees of freedom of the side-chain was thermodynamically advantageous for the S stereoisomer, it was detrimental for docking of the other stereoisomer. The calculated structure showed that the R stereoisomer clashed with various residues of the binding pocket that were forced to rotate in order to accommodate the more rigid conformation of the R stereoisomer. The sharp increase in IC50 and calculated binding energy between the linear C8 compound and the R stereoisomer of #10576 can be explained by the high energetic cost of this structural adjustment.
The selectivity observed with the S stereoisomer of #10576 for the yeast versus bovine bc1 complex can be rationalized by structural differences in the binding site geometry as illustrated in Fig. 9. The interaction between the inhibitor and the binding pocket is maximal when the branched side-chain is sandwiched between Leu-275 and Val-146. In the bovine enzyme, the corresponding amino acid pair Phe-274 and Val-145 is less effective, and a yeast enzyme with a L275F mutation (8) displays a 4-fold increase of IC50 toward the S stereoisomer of #10576 (data not shown). Furthermore, the Phe-278 present in the yeast enzyme is important in locking the long chains in the bent position where the inhibitor-binding pocket interactions are maximized. In the bovine enzyme, the Ala-277 failed to restrict the ligand into this conformation. Further evidence for the contribution of this residue to the difference in binding is that a yeast enzyme with a P278I mutation (9) also displays a 4-fold increase of IC50 toward the S stereoisomer of #10576 (data not shown). The cumulative effect of these differences, which affect both enthalpic and entropic binding determinants, at least partly accounts for the species selectivity.
Figure 9. Binding of the S isomer of #10576 in the ubiquinol oxidation pocket of the yeast and bovine cytochrome bc1 complexes illustrating structure differences that may contribute to differential binding efficacy.
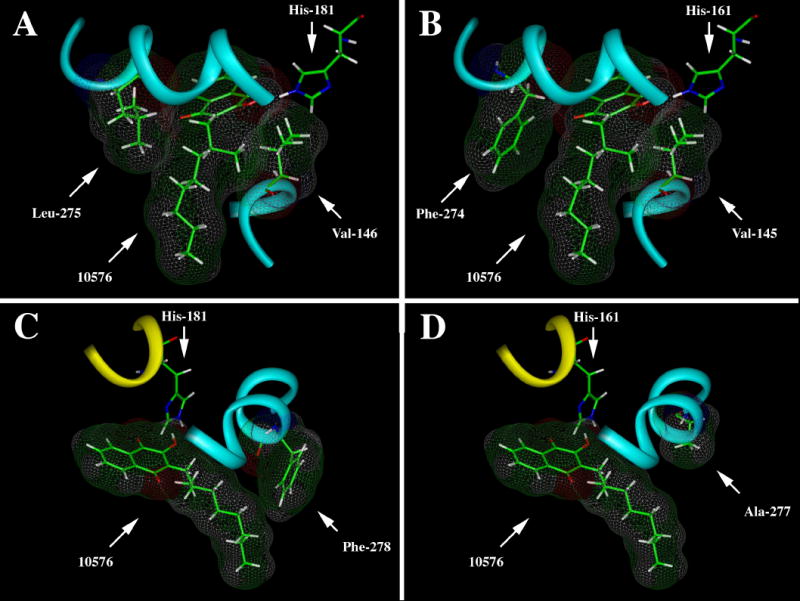
Panel A shows the van der Waals interactions between Val-146 and Leu-275 that facilitate stable binding of #10576 in the pocket. Panel B shows the corresponding Val-145 and Phe-274 in the pocket of the bovine enzyme. Note the apparent lack of interaction between Phe-274 and the
The racemic mixture of compound #10576 has been known for showing strong anti-plasmodium activity in an avian model (23), and we expect that the S stereoisomer would probably show even greater activity in this test system. Unfortunately, this compound has also been shown to be rapidly metabolized in humans by bis-hydroxylation of the terminal two carbons of the side-chain, followed by oxidative decarboxylation, which nullified its anti-plasmodium therapeutic potential (24,25). This hydroxylation reaction, presumably catalyzed by human cytochrome P450s, should be preventable by appropriate modification of the branched side-chain. Since the S stereoisomer of #10576 is more potent and exhibits better selectivity with a fungal enzyme than atovaquone, this compound should be considered as a new lead to design an improved hydroxy-naphthoquinone therapeutic.
Supplementary Material
Acknowledgments
We would like to thank Dr. Robert Schultz at the National Cancer Institute (Drug Synthesis & Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment and Diagnosis, Bethesda, MD) for providing hydroxynaphthoquinones.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kessl JJ, Lange BB, Merbitz-Zahradnick T, Zwicker K, Hill P, Meunier B, Meshnick S, Trumpower BL. Molecular basis for atovaquone binding to the cytochrome bc1 complex. J Biol Chem. 2003;278:31312–31318. doi: 10.1074/jbc.M304042200. [DOI] [PubMed] [Google Scholar]
- 2.Gutteridge WE. Antimalarial drugs currently in development. J Roy Soc Med. 1989;82 Suppl. 17:63–66. [PMC free article] [PubMed] [Google Scholar]
- 3.Araujo FG, Huskinson J, Remington JS. Remarkable in vitro and in vivo activities of the hydroxynaphthoquinone 566C80 against tachyzoites and tissue cysts of Toxoplasma gondii. Antimicrob Agents Chemother. 1991;35:293–299. doi: 10.1128/aac.35.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughes WT, Oz HS. Successful prevention and treatment of babesiosis with atovaquone. J Infect Dis. 1995;172:1042–1046. doi: 10.1093/infdis/172.4.1042. [DOI] [PubMed] [Google Scholar]
- 5.Hughes WT, Gray VL, Gutteridge WE, Latter VS, Pudney M. Efficacy of a hydroxynaphthoquinone, 566C80, in experimental Pneumocystis carinii pneumonitis. Antimicrob Agents Chemother. 1990;34:225–228. doi: 10.1128/aac.34.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutteridge WE. 566C80, an antimalarial hydroxynaphthoquinone with broad spectrum: experimental activity against opportunistic parasitic infections of AIDS patients. J Protozool. 1991;38:141S–143S. [PubMed] [Google Scholar]
- 7.Srivastava IK, Morrisey JM, Darrouzet E, Daldal F, Vaidya AB. Resistance mutations reveal the atovaquone-binding domain of cytochrome b in malaria parasites. Mol Microbiol. 1999;33:704–711. doi: 10.1046/j.1365-2958.1999.01515.x. [DOI] [PubMed] [Google Scholar]
- 8.Kessl JJ, Hill P, Lange BB, Meshnick S, Meunier B, Trumpower BL. Molecular basis for atovaquone resistance in Pneumocystis jirovecii modeled in the cytochrome bc1 complex of Saccharomyces cerevisiae. J Biol Chem. 2004;279:2817–24. doi: 10.1074/jbc.M309984200. [DOI] [PubMed] [Google Scholar]
- 9.Kessl JJ, Ha KH, Merritt AK, Lange BB, Hill P, Meunier B, Meshnick SR, Trumpower BL. Cytochrome b mutations that modify the ubiquinol-binding Pocket of the cytochrome bc1 complex and confer anti-malarial drug resistance in Saccharomyces cerevisiae. J Biol Chem. 2005;280:17142–17148. doi: 10.1074/jbc.M500388200. [DOI] [PubMed] [Google Scholar]
- 10.Kessl JJ, Ha KH, Merritt AK, Meshnick SR, Trumpower BL. Molecular basis of Toxoplasma gondii atovaquone resistance modeled in Saccharomyces cerevisiae. Mol Biochem Parasitol. 2006;146:255–258. doi: 10.1016/j.molbiopara.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 11.Moody CJ, Norton CL. Synthesis of 1, 2- fused indoles by radical cyclisation. J Chem Soc, Perkin Trans. 1997;1(17):2639–2643. [Google Scholar]
- 12.Mori K. Revision of the absolute configuration of A-factor. Tetrahedron. 1983;39:3107–3109. [Google Scholar]
- 13.Shirai S, Seki M, Mori K. Pheromone Synthesis, CXCIX. Eur J Org Chem. 1999:3139–3145. [Google Scholar]
- 14.Gallardo H, Merlo AA. Ethyl lactate as a convenient precursor for synthesis of chiral liquid crystals. Synthetic Comm. 1993;23:2159–2169. [Google Scholar]
- 15.Fouquet C, Schlosser M. Improved carbon-carbon linking by controlled copper catalysis. Angew Chem. 1974;13:83–84. [Google Scholar]
- 16.Ljungdahl PO, Pennoyer JD, Robertson D, Trumpower BL. Purification of highly active cytochrome bc1 complexes from phylogenetically diverse species by a single chromatographic procedure. Biochim Biophys Acta. 1987;891:227–242. doi: 10.1016/0005-2728(87)90218-0. [DOI] [PubMed] [Google Scholar]
- 17.Snyder CH, Trumpower BL. Ubiquinone at center N is responsible for triphasic reduction of cytochrome b in the cytochrome bc1 complex. J Biol Chem. 1999;274:31209–31216. doi: 10.1074/jbc.274.44.31209. [DOI] [PubMed] [Google Scholar]
- 18.Hunte C, Koepke J, Lange C, Rossmanith T, Mitchel H. Structure at 2.3 A resolution of the cytochrome bc1 complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure. 2000;8:669–684. doi: 10.1016/s0969-2126(00)00152-0. [DOI] [PubMed] [Google Scholar]
- 19.Palsdottir H, Gomez Lojero C, Trumpower BL, Hunte C. Structure of the yeast cytochrome bc1 complex with a hydroxyquinone anion Qo site inhibitor bound. J Biol Chem. 2003;278:31303–31311. doi: 10.1074/jbc.M302195200. [DOI] [PubMed] [Google Scholar]
- 20.Sacau EP, Estevez-Braun A, Ravelo AG, Ferro EA, Tokuda H, Mukainaka T, Nishino H. Inhibitory effects of lapachol derivatives on epstein-barr virus activation. Bioorg Med Chem. 2003;11:483–488. doi: 10.1016/s0968-0896(02)00542-4. [DOI] [PubMed] [Google Scholar]
- 21.Lima NM, Correia CS, Leon LL, Machado GM, Madeira MF, Santana AE, Goulart MO. Antileishmanial activity of lapachol analogues. Mem Inst Oswaldo Cruz. 2004;99:757–761. doi: 10.1590/s0074-02762004000700017. [DOI] [PubMed] [Google Scholar]
- 22.Sek L, Boyd BJ, Charman WN, Porter CJ. Examination of the impact of a range of Pluronic surfactants on the in-vitro solubilization behavior and oral bioavailability of lipidic formulations of atovaquone. J Pharm Pharmacol. 2006;58:809–820. doi: 10.1211/jpp.58.6.0011. [DOI] [PubMed] [Google Scholar]
- 23.Fieser LF, Richardson AP. Naphthoquinone antimalarials. J Am Chem Soc. 1948a;70:3151–3165. doi: 10.1021/ja01190a001. [DOI] [PubMed] [Google Scholar]
- 24.Fieser LF, Chang FC, Dauben WG, Heidelberger C, Heymann H, Seligman AM. Naphthoquinone antimalarials. Metabolic oxidation products. J Pharmacol Exp Ther. 1948b;94:85–96. [PubMed] [Google Scholar]
- 25.Hudson AT, Dickins M, Ginger CD, Gutteridge WE, Holdich T, Hutchinson DBA, Pudney M, Randall AW, Latter VS. 566C80: a potent broad spectrum anti-infective agent with activity against malaria and opportunistic infections in AIDS patients. Drugs Exptl Clin Res. 1991;17:427–435. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


