Summary
The essential Saccharomyces cerevisiae pre-messenger RNA splicing protein 24 (Prp24) has four RNA recognition motifs (RRMs) and facilitates U6 RNA base-pairing with U4 RNA during spliceosome assembly. Prp24 is a component of the free U6 small nuclear ribonucleoprotein particle (snRNP) but not the U4/U6 bi-snRNP, and so is thought to be displaced from U6 by U4/U6 base-pairing. The interaction partners of each of the four RRMs of Prp24 and how these interactions direct U4/U6 pairing are not known. Here we report the crystal structure of the first three RRMs and the solution structure of the first two RRMs of Prp24. Strikingly, RRM 2 forms extensive inter-domain contacts with RRMs 1 and 3. These contacts occupy much of the canonical RNA-binding faces (β-sheets) of RRMs 1 and 2, but leave the β-sheet of RRM 3 exposed. Previously identified substitutions in Prp24 that suppress mutations in U4 and U6 spliceosomal RNAs cluster primarily in the β-sheet of RRM 3, but also in a conserved loop of RRM 2. RNA binding assays and chemical shift mapping indicate that a large basic patch evident on the surface of RRMs 1 and 2 is part of a high affinity U6 RNA binding site. Our results suggest that Prp24 binds free U6 RNA primarily with RRMs 1 and 2, which may remodel the U6 secondary structure. The β-sheet of RRM 3 then influences U4/U6 pairing through interaction with an unidentified ligand.
Keywords: RRM, U6 RNA, splicing, RNA chaperone, Prp24
Introduction
Nuclear pre-messenger RNA splicing is an essential step of gene expression in eukaryotic organisms. Pre-mRNA splicing must be highly efficient and accurate, since failure to remove an intron or an error in splicing is likely to render an mRNA useless. Accordingly, the complex that catalyzes pre-mRNA splicing, the spliceosome, is very elaborate, comprising five RNA’s (U1, U2, U4, U5, and U6) and more than 75 proteins.1 Spliceosome function reflects this complexity, involving multiple steps: assembly, catalytic activation, two chemical reactions, and disassembly.2,3 Each of these steps requires ATP and the participation of one or more RNA-dependent ATPases. Each step is accompanied by a change in conformation of the spliceosomal RNAs and/or pre-mRNA substrate. Consequently, the spliceosome is a very interesting subject for elucidating the biochemical mechanisms for programmed rearrangements in RNA secondary, tertiary and quaternary structure.
Large changes in RNA conformation are often catalyzed by proteins called RNA chaperones.4, 5 One of the most conformationally dynamic RNAs in the spliceosome is U6 RNA. In preparation for spliceosome assembly, the U6 3′ intramolecular stem-loop unwinds and the exposed sequences pair with U4 RNA, to form the U4/U6 small nuclear ribonucleoprotein complex. After assembly of the spliceosome on an intron, spliceosome activation occurs, during which U4 RNA is unwound from U6 RNA. U6 then base pairs with U2 RNA to form the catalytically active form of the spliceosome. The essential U6 RNA-binding protein Prp24 is strongly implicated in promoting U4/U6 base-pairing,6–12 and may also be involved in dissociation of the U4/U6 complex during spliceosome activation.6,7,11,13,14,15 Thus, Prp24 is a candidate U6 RNA chaperone.
Prp24 is present in eukaryotic organisms from yeast to humans, but is quite variable in primary structure.12, 16 In the yeast S. cerevisiae, Prp24 has four RNA recognition motifs (RRMs) (Figure 1(a)), although RRM 4 is quite degenerate and was recognized only by alignment with orthologs from other fungi.12 Many other fungal species have an additional N-terminal domain of 500 to 700 residues. Plants and animals have the N-terminal extension, but have only one or two recognizable RRMs. The two RRMs of human Prp24 appear most similar to yeast RRMs 1 and 217 (Figure 1(b)), but could also correspond to RRMs 2 and 3.16
Figure 1.
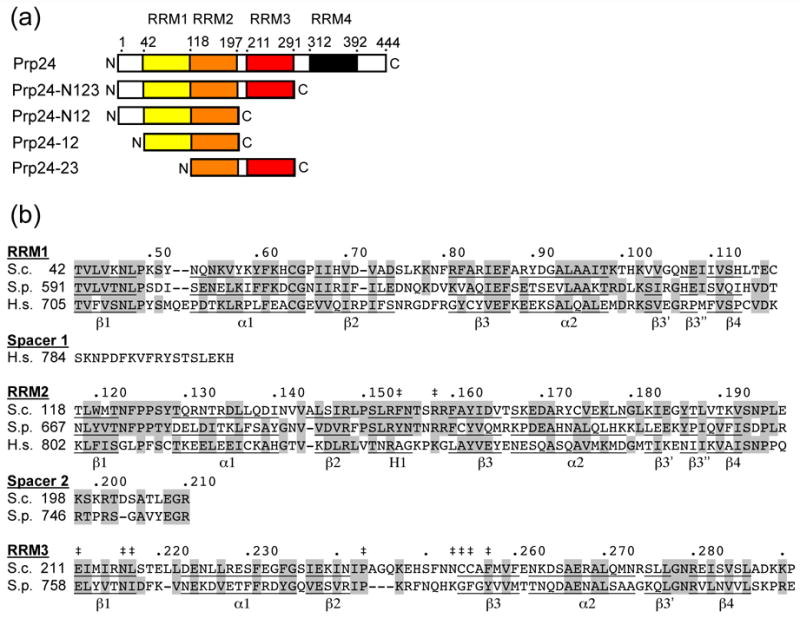
Primary structure and conservation of Prp24. (a) Domain structure of S. cerevisiae Prp24. The four RRMs are represented in different colors and the approximate boundaries of the four RRMs are indicated by amino acid residue number. Truncation constructs used in this study are shown below the full-length protein; see Materials and Methods for precise endpoints of constructs. (b) Sequence conservation of RRMs 1–3 in S. cerevisiae (S.c), Schizosaccharomyces pombe (S.p.), and human (H.s.) Prp24. S. pombe and human Prp24 have long N-terminal domains (not shown), and human Prp24 has only two RRMs. Identical and highly similar residues are highlighted grey. A dot marks every 10th residue in the S.c. sequence. Secondary structure elements are underlined and labeled (α = alpha helix, β = beta sheet, H = 310 helix). Double-daggers (‡) indicate the positions of mutations that suppress U6-A62G or U4-G14C, which include L217P, N253S, and C255R (U4-G14C)6 and F154S, R158S, E211D, N216H, P243S, C254W, and F257I (U6-A62G).11
To better understand how Prp24 influences U6 RNA conformation, we sought to determine its three dimensional structure. We solved a crystal structure of a C-terminally truncated Prp24 containing the N-terminus and RRMs 1, 2 and 3 (Prp24-N123; Figure 1(a)). NMR spectroscopy was performed on two Prp24 fragments containing either RRMs 1 and 2 (Prp24-12) or RRMs 2 and 3 (Prp24-23), and a solution structure of Prp24-12 was solved. We also performed RNA binding assays with these constructs. We find that the three RRMs all adopt a canonical fold and interact extensively with one another. The β-sheet faces of RRMs 1 and 2 are largely occluded by protein-protein interactions, while the β-sheet of RRM 3 is exposed. Previously selected mutations in Prp24 that suppress mutations in U4 and U6 RNAs cluster in the β-sheet of RRM 3 and the RRM 2/3 interface. RRMs 1 and 2, in combination with the N-terminus (Prp24-N12), are sufficient for high-affinity binding of U6 RNA, and contain a highly basic patch on their external face that may be a non-canonical RNA-binding site. Thus, rather than acting as independent “beads on a string”, RRMs 1–3 of Prp24 appear to provide a highly structured surface for the conformational remodeling of U6 RNA.
Results and Discussion
Crystal structure of Prp24-N123
We solved a crystal structure of Prp24-N123 to a resolution of 2.7 Å using single wavelength anomalous diffraction (Supplementary Table I). The asymmetric unit contains two tetramers (chains A–D and E–H), the structures of which are essentially identical. There is no evidence that Prp24 functions as an oligomer, so the physiological relevance of this arrangement is uncertain. The four molecules in each tetramer have very similar structures and are related by non-crystallographic symmetry. The root mean square deviation (RMSD) between each monomer and their average structure is 0.33 Å for backbone atoms. However, loop 3 of RRM 2 was resolved only in chains B and F, so we have used chain B to represent the structure of Prp24-N123. Forty-nine of the 291 amino acid residues of chain B are unresolved, including the N-terminal domain (residues 1–40) and few short internal regions (see below).
The first three RRMs of Prp24 all exhibit canonical folds, with a four stranded β-sheet and two α-helices (Figure 2). However, the lengths of the secondary structure elements vary among the RRMs. For example, RRM 1 has a long β-strand 2 and a short loop 3, while RRMs 2 and 3 have short β-strands 2 and long loops 3 (Figures 1(b) and 2). RRM 2 loop 3 is disordered in all but one chain of the tetramer, in which it has a short helical element (Figure 2). RRM 3 loop 3 is disordered in all chains. Loop 5, between α-helix 2 and β-strand 4, has a short, two stranded β-sheet in RRMs 1 and 2, as is often seen in other RRMs.18 In RRM 3, Loop 5 has a single β-strand that extends the main β-sheet (Figure 1(b) and 2).
Figure 2.
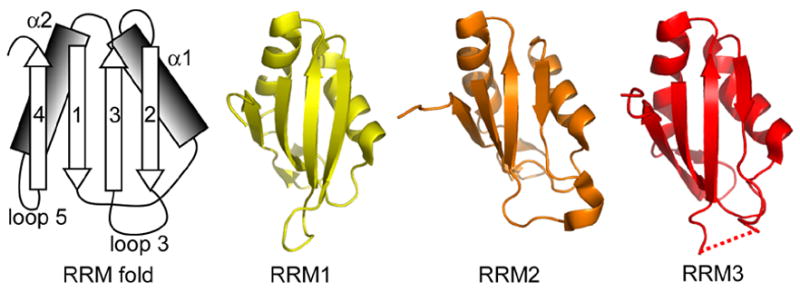
RRMs 1, 2 and 3 in Prp24-N123 crystal structure. The RRMs adopt canonical folds. The schematic at left shows the canonical β–α– β– β–α– β RRM fold, with the β-strands and α-helices numbered in order from the N-terminus. The backbone structure of RRMs 1–3 is shown at right. The dotted line indicates a disordered region in loop 3 of RRM 3.
Given the canonical folds of the RRMs, we were interested to examine their interactions. A variety of different contacts are seen in several two-RRM structures,18 some of which are RNA-dependent and some of which occur in the free protein. A three-RRM structure has not previously been reported. We find that RRM 2 interacts extensively with both RRMs 1 and 3 (Figures 3(a)–(c)). Residues in or immediately adjacent to all four β-strands of RRM 1 make close contacts with residues in or adjacent to β-strands 2 and 3 of RRM 2. Many of these contacts are hydrophobic, but there is a salt bridge between Arg84 of RRM 1 and Asp164 of RRM 2. There is about 575 Å2 of buried solvent-accessible surface area (SASA) in each RRM at the RRM 1–2 interface. This is about 10% of total SASA of each RRM and comparable to other two-RRM structures where two RRMs interact with each other without bound RNA or DNA. 18,19 This fairly small buried surface area is typical of “non-obligate” intermolecular contacts between proteins that also exist stably in a monomeric state,20 and suggests that the RRM1-RRM2 intramolecular interaction may be dynamic.
Figure 3.
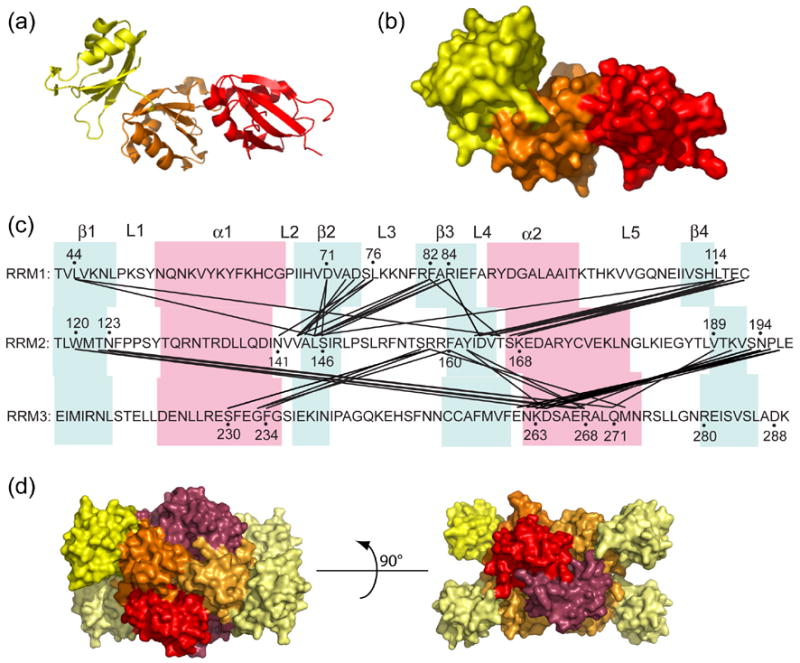
Interactions between RRMs 1, 2 and 3 of Prp24. (a) Ribbon diagram of Prp24-N123, with the RRMs colored as in Figures 1 and 2. (b) Surface structure of Prp24-N123, in the same orientation as panel A. (c) Map of inter-RRM interactions. Residues in two different RRMs that are within 4.0 Å distance from one another are connected by black lines. β-strands are highlighted in blue, and α-helices in pink. Loops between secondary structure elements are labeled L1–L5. (d) Surface structure of a Prp24-N123 tetramer (chains A–D) from the crystal structure. The three RRMs in chain B are colored as in Figure 1 and 2. Chains A, C and D are colored similar to chain B, but are light yellow, light orange and red-purple for RRMs 1–3, respectively.
Residues in or immediately adjacent to β-strands 1, 3 and 4 of RRM 2 make close contacts with residues in α-helices 1 and 2 of RRM 3. Despite the 2.7 Å resolution limit of the crystal structure, it is possible to infer the existence of hydrogen bonds based on the refined atomic positions. The side chain of Arg158 in RRM 2 makes a hydrogen bond with main chain carbonyl group of Ser230 in RRM 3. A hydrogen bond network exists between residues 193–195 and 263–264. The hydroxyl group of Tyr162 forms hydrogen bonds with side chains of Asp264 and Arg268, and the aromatic ring of Phe160 interacts with the guanidinium group of Arg268 by planar π-stacking. Overall, there is about 550 Å2 of buried SASA in each RRM at the RRM 2–3 interface, comparable in area to the RRM1–2 interface. While the β-sheets of RRMs 1 and 2 are largely occluded by inter-domain interactions, the β-sheet of RRM 3 faces away from RRM 2 and is exposed in Prp24-N123.
There are six other multiple RRM structures to date, all of which have two RRMs.18,19 Only two of them (heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1) and polypyrimidine tract binding (PTB) protein) show RRM-RRM interactions in their free forms.18,19 When compared to these two structures, both RRM 1–2 and RRM 2–3 interactions of Prp24 display substantially different RRM-RRM orientations. However, it is interesting that α-helix 2 of RRM3 is involved in the contact with RRM2 as in the other two structures, where the same helices of their C-terminal RRM are used in the inter-RRM contacts. As in the hnRNPA1 and PTB structures, salt bridges, hydrogen bonds, and hydrophobic contacts primarily comprise the RRM interfaces of Prp24.
In the crystal structure, Prp24-N123 forms a tetramer in which the four chains are related by three mutually perpendicular 2-fold rotation axes. RRMs 2 and 3 participate in contacts with other chains at the tetramer interface (Figure 3(d)), but RRM 1 does not. On the surface of chain B, chain A buries about 820 Å2, chain C buries 480 Å2, and chain D buries 1150 Å2 of SASA. Loop 5 of RRM 2 in chain A is inserted into a cleft made by RRMs 2 and 3 of chain B (and vice versa), forming a “knobs into holes” packing arrangement. Chains B and C interact entirely through the β-sheet faces of RRMs 3, while chains B and D interact primarily through α-helices 2 of RRMs 2 and 3. Interestingly, RNA binding studies with Prp24-23 and Prp24-34C constructs suggested that both form homodimers when bound to RNA, consistent with a dimerization domain in RRM3.21
Solution structure of Prp24-12
We were concerned that the extensive intramolecular interactions between RRMs 1, 2 and 3 in the crystal structure might be an artifact of crystal packing, so we determined the solution structure of Prp24-12, a fragment of S. cerevisiae Prp24 encompassing the first two RRMs (Figure 1(a)), by NMR. Gel filtration chromatography (data not shown) and estimation of the rotational correlation time (τc, 13.5 +/− 0.5 ns) from 15N NMR relaxation rates (T1, T2) confirmed that Prp24-12 is a monomer in solution. Nearly complete 1H, 13C, and 15N resonance assignments were obtained.22 The Prp24-12 structure was determined independently of the crystal structure, using automated NOE assignment and structure calculation software. The ten lowest energy conformers have highly similar backbone secondary structures (Figure 4(a)). Strikingly, in solution RRMs 1 and 2 assume a fixed orientation with respect to each other (Supplementary Table II), consistent with extensive inter-domain interactions.
Figure 4.
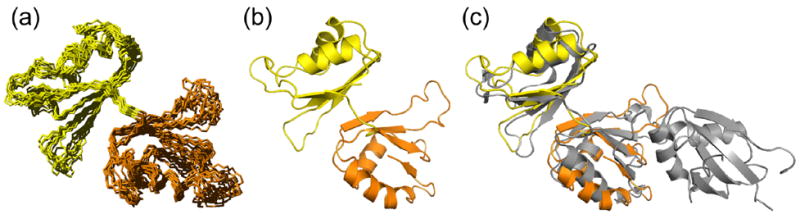
Solution structure of Prp24-12. (a) Ensemble of ten energy-minimized NMR structures of Prp24-12. The backbone RMSD of secondary structure-containing regions of RRMs 1 (yellow) and 2 (orange) is 0.97 Å ± 0.09. (b) Ribbon diagram of the lowest free energy structure of Prp24-12, with RRMs colored as in Figures 1–3. (c) Comparison of the mean NMR and crystal (depicted in grey) structures. Backbone atoms of secondary structure regions were fit to RRM domains 1 and 2 and give an RMSD of 2.0 Å ± 0.3 (Supplementary Table III).
Nine inter-domain NOEs are observed between RRM 1 (β-strands 2–3) and RRM 2 (loop 2 or β-strand 2). These contacts occur between residues Ala73/Val143, Ala73/Leu145, Ser75/Val143, Phe82/Leu145, and Arg84/Leu145, consistent with the crystal structure (Figure 3(c)). Interestingly, the inter-domain Arg84/Asp164 salt bridge identified in the x-ray structure is also observed in the NMR structure. The lowest free energy solution structure is shown in Figure 4(b), and is overlaid on the crystal structure in Figure 4(c). The solution and crystal structures are highly similar, with a secondary structure backbone RMSD of 2.0 Å for RRMs 1 and 2 (Supplementary Table III). Importantly, these results show that the extensive inter-domain contacts observed in the crystal structure are not an artifact of oligomerization or crystal packing.
Dynamics of Prp24 RRMs 1, 2 and 3
To further examine the solution structure of Prp24, we generated NMR backbone assignments for Prp24-23 (Figure 1(a)) and measured relaxation times for Prp24-12 and -23. Amide chemical shifts of RRM2 substantially differ between Prp24-12 and Prp24-23 protein constructs, supporting the idea that inter-domain contacts also occur in solution. Both x-ray and NMR data identify three highly dynamic regions in RRMs 2 and 3 of Prp24 (Figure 5, shaded in grey). In NMR relaxation experiments, loop 3 of RRMs 2 and 3 (residues 151–158 and 245–251, respectively) and the linker region between RRMs 2 and 3 (residues 196–207) exhibit pico- to nanosecond timescale motions as judged by their low 1H-15N heteronuclear NOE values and T1/T2 ratios. Similarly, high or undefined crystallographic B-factor values in the same regions of the protein indicate that the electron density of the polypeptide backbone undergoes significant dynamic disorder. Interestingly, limited trypsin proteolysis of full-length Prp24 yields a ~33 kDa fragment that includes residues 13–239, consistent with cleavage in the dynamic loop 3 region of RRM 3.21 These results indicate that RRMs 1–3 form a highly structured core of Prp24, with only a few dynamic internal segments. In the crystal structure, RRM 1 exhibits the highest average B-factor (Figure 5), probably because it is not constrained by intermolecular contacts within the tetramer, unlike RRMs 2 and 3 (Figure 3D).
Figure 5.
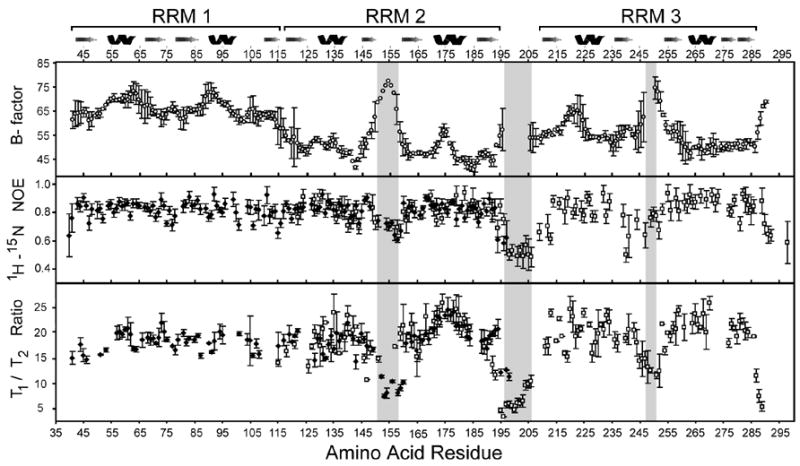
Dynamics of Prp24 RRMs 1, 2 and 3. Average backbone B-factors, 15N heteronuclear NOE values, and 15N T1/T2 relaxation time ratios are plotted versus the protein sequence (residues 41–291). Open circles are data points from the Prp24-N123 crystal structure, and filled diamonds and open squares are from NMR experiments on Prp24-12 and -23, respectively. B-factor errors incorporate the eight Prp24 chains present in the crystallographic asymmetric unit. Two Prp24-12 and Prp24-23 15N heteronuclear NOE data sets were acquired, analyzed, and incorporated into error calculations. For the T1/T2 ratio data sets, errors represent multiple 15N T1 and T2 relaxation experiments in addition to the uncertainty of the exponential fit. Disorderd regions in the crystal structure are shaded in grey (residues 151–157, 196–207 and 245–251).
Location of U4-G14C and U6-A62G suppressor mutations in the Prp24 structure
U4 and U6 spliceosomal RNAs form an extensive base-pairing interaction during assembly of the spliceosome.23 Mutations that disrupt this interaction, such as U4-G14C,6 or that stabilize a competing intramolecular stem in U6 RNA, such as U6-A62G,24 result in a cold-sensitive growth defect. Selections for extragenic (trans-acting) suppressors of the cold-sensitivity of both U4-G14C and U6-A62G strains yielded mutations in the PRP24 gene.6,11 Each suppressor mutation results in substitution of a single residue within loop 3 of RRM 2, β-strand 1 or 3 of RRM 3, or loop 3 of RRM 3 (Figure 1(b)). When mapped to the crystal structure, the substitutions in RRM 3 form a continuous track across the face of the β-sheet (Figure 6), consistent with disruption of an interaction interface with an unknown ligand, possibly RRM 4, U4 RNA or U6 RNA. Some of the mutations alter residues in the RRM 3 dimerization interface observed in the crystal structure. In particular, the Asn216His, Asn253Ser and Phe257Ile substitutions alter residues involved in close (<4 Å) contacts. This finding raises the possibility that Prp24 dimerization through RRM 3 occurs in vivo, for example, during U4/U6 annealing, and that suppression is due to changes in dimerization.
Figure 6.
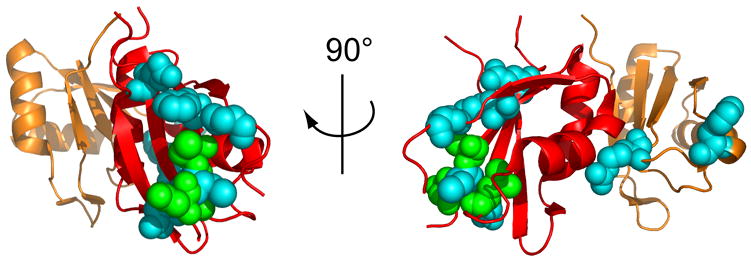
Location of U4-G14C and U6-A62G suppressor mutations in RRMs 2 and 3 of Prp24. The backbone of RRM 2 is shown in orange and RRM 3 in red. Residues at which U4-G14C (green) or U6-A62G (blue) suppressor substitutions were selected are shown in space-fill mode. Two views related by a 90° rotation about the vertical axis are shown. See Figure 1(b) for the identity of the residues shown.
Intriguingly, the substitutions in RRM 2 are far from the β-sheet face of RRM 3, but are close to the RRM 2/RRM 3 interface. In particular, the Arg158Ser substitution, which also confers a heat-sensitive growth defect,11 is expected to alter a very close contact with Ser230 in RRM 3 (Figure 3(c)). Therefore, the RRM 2 substitutions may influence RRM 3 function indirectly.
High affinity binding of Prp24-N12 to a sub-domain of U6 RNA
An earlier study implicated RRMs 1 and 2 of S. cerevisiae Prp24 as the high-affinity binding site for U6 RNA, but concluded that RRMs 1 and 2 might bind U6 RNA in a non-canonical fashion, using residues outside of β-strands 1 and 3.17 The fact that the β-sheets of RRMs 1 and 2 are largely occupied by protein-protein interactions suggests the possibility that the protein undergoes a conformational rearrangement upon RNA binding, or that U6 RNA binds other surfaces of these RRMs. If RRMs 1 and 2 are a major binding determinant for U6 RNA, we would expect Prp24-N12 to bind U6 with an affinity similar to that of Prp24-N123, while Prp24-23 should not bind U6 well. Conversely, if the β-sheet of RRM 3 is the key binding determinant for U6 RNA, the opposite results should be obtained. To discriminate between these possibilities, Prp24-N123, -N12, and -23 proteins were incubated with a 40-nucleotide, 32P-labeled RNA containing sequences identical to S. cerevisiae U6 RNA nucleotides 41–64 and 83–88 (Figure 7(a)), the primary binding site for Prp24.17 Prp24-N123 binds to this RNA with an apparent Kd of 45 ± 10 nM (Figure 7(b) and 7(d)), similar to the 24 nM apparent Kd observed for Prp24-N123 binding to full length U6 RNA.17 This result suggests that the 40-nucleotide RNA includes most of the binding determinants for Prp24-N123. Prp24-N12 binds to the 40-nucleotide RNA with only two-fold lower affinity (81 ± 10 nM; Figures 7(c) and 7(d)). In contrast, Prp24-23 exhibits little, if any, binding to the 40-nucleotide RNA (Figure 7(d), data not shown). These results are consistent with RRMs 1 and 2 providing the major binding site for U6 RNA. However, we cannot exclude the possibility that the N-terminal 41 amino acids of Prp24 also contribute to U6 RNA binding.
Figure 7.
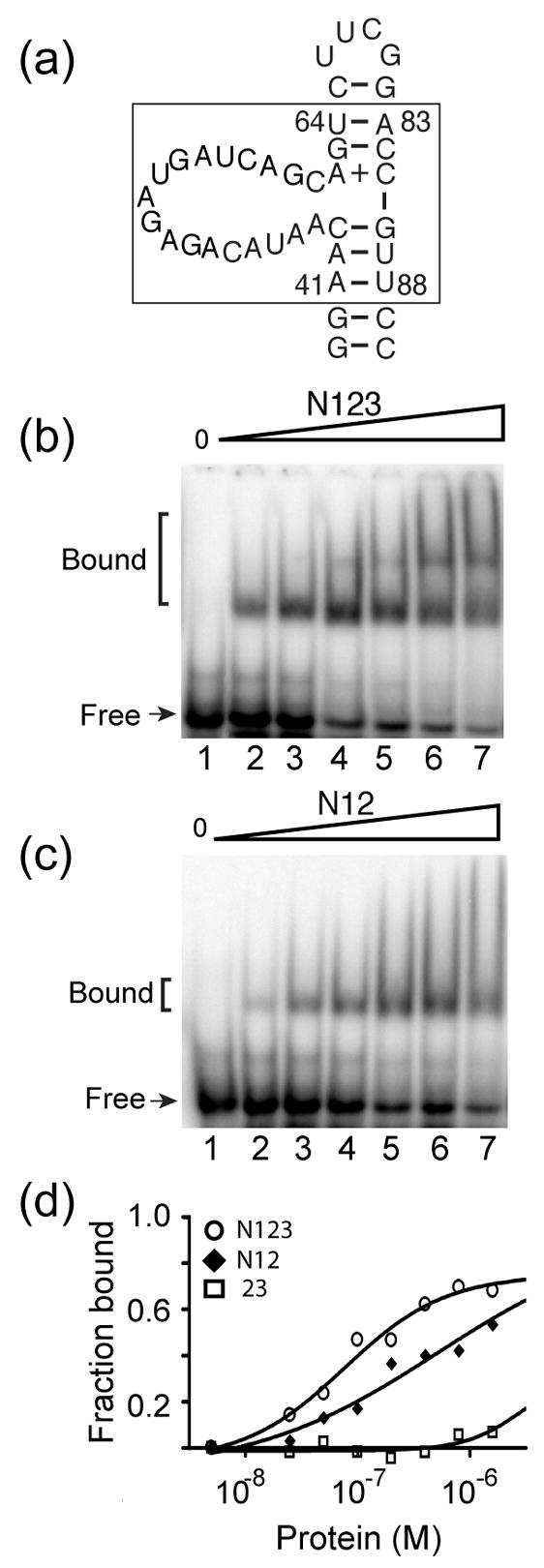
Binding of truncated Prp24 proteins to a domain of U6 RNA. (a) Primary and possible secondary structure of a minimal U6 RNA construct (40-nucleotide RNA) used in gel mobility shift assays (S. cerevisiae nucleotides 41–64 and 83–88 are boxed). 15N-labeled (b) Prp24-N123 and (c) Prp24-N12 proteins at concentrations of 0, 25, 50, 100, 200, 400, and 800 nM (lanes 1–7, respectively) were incubated with 32P-labeled 40-nucleotide RNA and resolved on a 6% native polyacrylamide gel. (d) Fraction of RNA bound plotted against the total concentration of Prp24-N123 (open circles), N12 (filled diamonds), and 23 (open squares). Data were fit to a one-site hyperbolic binding function (Y= (Bmax*X)/{Kd +X}), where Y is the fraction bound and X is the concentration of Prp24. The apparent Kd (in nM) is 45 ± 10 for Prp24-N123 and 81 ± 10 for Prp24-N12. An accurate apparent Kd value could not be extrapolated for Prp24-23.
Mapping the RNA-binding region on the surface of RRMs 1 and 2
The amino acid residues of Prp24-N12 that contact RNA were mapped by NMR, using the minimal 40-nucleotide construct (Figure 7(a)), as well as a 21-nucleotide RNA corresponding to residues 40–60 of yeast U6. The two RNAs gave highly similar results. Addition of sub-stoichiometric amounts of either RNA to 2H,15N-labeled Prp24-N12 protein resulted in line broadening of selective resonances in 15N HSQC-TROSY spectra (Figure 8(a) and data not shown), revealing specific residues that undergo millisecond exchange kinetics due to RNA binding. The resonances that are specifically affected by the RNA interaction map primarily to the α-helical face and loops 3 and 5 of RRM 1, and loop 3 and the β-sheet of RRM 2 (Figure 8(b)). Several of these residues are positively charged, including Lys59, Lys62, Lys77, Lys99, Arg131, Arg134, Arg148, and Lys191. Poisson-Boltzmann calculation of the electrostatic surface of Prp24-N123 reveals a large electropositive surface that spans RRM 1 and RRM 2 but not RRM 3 (Figure 8(c)), which has an unusually acidic pI of about 5.0.17 Many of the residues that exhibit altered chemical shift upon addition of RNA map to this electropositive surface, suggesting that binding is largely electrostatic.
Figure 8.
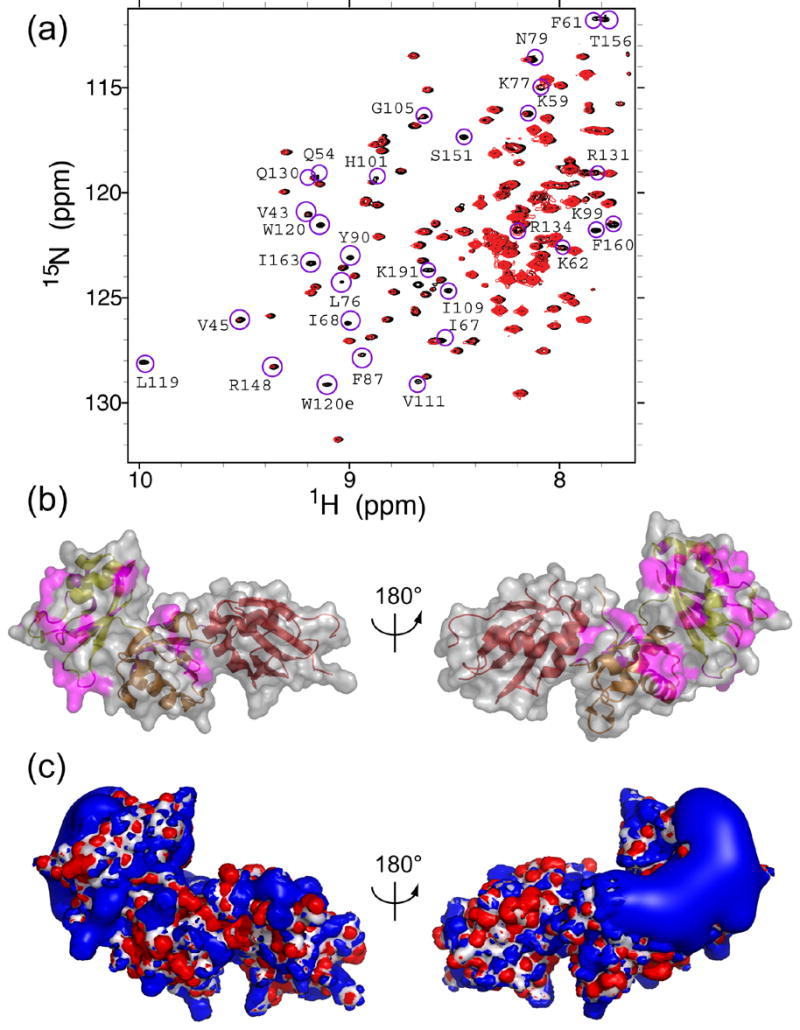
Mapping of U6 RNA binding interactions on the surface of Prp24. (a) Region of the 1H-15N HSQC-TROSY spectra showing amide peaks of 0.2 mM 2H,15N-labeled Prp24-N12 protein in the absence (black) and presence (red) of 50 μM 21-nucleotide RNA (yeast U6 nucleotides 40–60). Assigned resonances that are selectively line broadened upon the addition of RNA are circled in magenta and labeled. (b) Location of the line broadened amino acids (shown in magenta) in the crystal structure. The structure at left is in the same orientation as is shown in Figures 3(a) and 3B, and is rotated 180° around the vertical axis relative to the structure at right. The ribbon is colored as in Figure 1. (c) Electrostatic potential isosurface of the Prp24-N123 crystal structure calculated by the Poisson-Boltzmann equation. The isosurface is contoured at ± 4 kT and is superposed on the molecular surface (white). A prominent electropositive region (blue) exists on one side of the protein, possibly corresponding to an RNA binding site. Electronegative regions (red) are small and distributed more uniformly around the protein. Structures are in the same orientation as in Figure 8(b).
Additional residues implicated in RNA-binding by chemical shift analysis map to the β-sheet regions of RRMs 1 and 2. These residues include Val 43, Val45, Ile67, Ile68, Leu76, Phe87, Ile110, Val111, Leu119, Trp120, Phe160 and Ile163. Since some of these residues are buried in the absence of RNA, the data suggest that binding of RNA alters the intramolecular contacts between RRMs 1 and 2. Such an induced change in Prp24 conformation may allow canonical, sequence-specific contacts between the RNA and the β-sheets of RRMs 1 and 2.
Prp24 binds to U6 RNA sequence-specifically and facilitates pairing of U6 RNA bases with U4 RNA bases. The annealing or “matchmaking” activity of Prp24 may require unwinding intramolecular U6 RNA stems and exposing the bases to solvent. Indeed, increasing the stability of the 3′ intramolecular stem decreases U6 RNA affinity for purified Prp24.17 Furthermore, Prp24-bound U6 RNA is more reactive with single strand-specific base modifying reagents than is unbound U6 RNA.25 The structure of another matchmaking protein bound to RNA was recently solved.26 MRP1/MRP2 protein promotes pairing of guide RNA with pre-mRNA by unwinding an intramolecular helix and anchoring the ribose-phosphate backbone of the guide RNA to a highly basic region of the protein, leaving the nucleotide bases exposed for pairing with pre-mRNA. We suggest that the electropositive surface of RRMs 1 and 2 is an RNA annealing domain, while the cleft between RRMs 1 and 2, including the β-sheet face of RRM 2, is a sequence-specific RNA-binding site.
Conclusions and prospects
The structure of the N-terminal two-thirds of splicing factor Prp24 reveals extensive interactions between the three RRMs that comprise most of its length. We conclude that the interactions betweens RRMs 1 and 2 are not an artifact of crystal packing, since they are also present in solution in the NMR structure. We show that RRMs 1 and 2 comprise the primary binding determinant for U6 RNA, yet their canonical RNA-binding faces are largely occupied by protein-protein interactions. Thus, RNA binding either requires a large conformational change in Prp24, or U6 RNA binds to Prp24 in a non-conventional manner. Chemical shift analysis of RNA binding suggests that both may occur, since the presence of RNA alters resonances of residues on the surface of RRMs 1 and 2, as well as residues buried in the interface between the RRMs. Given the matchmaker activity of Prp24, we propose that its association with U6 RNA remodels the U6 RNA structure, in part by stabilization of unwound U6 RNA on the large basic patch spanning RRMs 1 and 2. Confirmation of this hypothesis will require solving the structure of the RNA-protein complex.
The Prp24-N123 structure does not yet explain the mechanism of suppression of the cold-sensitive growth defect of the U4-G14C and U6-A62G mutations by substitutions in RRMs 2 and 3. The suppressor mutations cluster primarily in the β-sheet of RRM3, which forms intermolecular contacts in the crystal. Given that only truncated forms of Prp24 have been observed to form oligomers in solution, its is possible that full-length Prp24 forms oligomers in a regulated fashion in vivo, although it is unclear how disruption of this process would suppress mutations that destablize the U4/U6 complex. Alternatively, it may be the interaction with some other ligand that is disrupted by the suppressor mutations. Candidate ligands are the C-terminal one-third of Prp24, other splicing proteins, or U4 or U6 RNAs. Further studies will be required to identify the ligand of RRM 3, and the effect of the suppressor mutations on interaction with this ligand.
Materials and Methods
X-ray crystallography and anaylsis
The Prp24-N123 construct, which encodes residues 1–291 as well as an N-terminal Ser residue that remains after TEV cleavage, was cloned and selenomethionyl protein was purified following the standard Center for Eukaryotic Structural Genomics (CESG) pipeline protocol for cloning,27 protein expression,28 protein purification29 and overall information management30. Crystals of the selenomethionyl protein were grown by the hanging-drop method from 10 mg/ml protein solution in buffer (50 mM NaCl, 3 mM NaN3, 0.3 mM TCEP, 5 mM Tris pH 8.0) mixed with an equal amount of reservoir solution containing 14% MEPEG 5000, 0.2 M tetramethylammonium chloride, 0.1 M succinate pH 4.0 at 4°C. The crystals were soaked in reservoir solutions supplemented with increasing amounts of ethylene glycol up to 30% and were flash-frozen in a stream of cryogenic nitrogen gas.
Diffraction data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source (APS), Argonne National Laboratory at a wavelength of 0.97911 Å at 100 K. The diffraction images were processed with HKL2000.31 The selenium substructure of the crystal was determined by using SHELXD32 and HySS from PHENIX,33,34 and the selenium positions were used for single wavelength anomalous diffraction phasing in AUTOSHARP.35 Initial phase information to 4.0 Å was further improved and extended to 2.7 Å by density modification and phase extension using NCS averaging as implemented in RESOLVE.36 The initial model was built by ARP/wARP,37 and the structure was completed using alternate cycles of manual building in COOT38 and refinement in REFMAC5.39 NCS restraints between the corresponding monomers of the two tetramers (A and E, B and F, C and G, D and H) in the asymmetric unit were applied during the refinement. TLS refinement of 24 groups corresponding to the individual RRMs was also used.
The stereochemical quality of the final model was assessed using MolProbity.40 The electrostatic surface calculation of the structure was performed with APBS41 using 2 and 80 for the dielectric constants of the protein and solvent respectively. The charging model was produced by PyMOL (http://www.pymol.org) as was the visualization. Buried solvent-accessible surface area calculations were done with the program WHATIF using a probe radius of 1.4 Å. 42 The final coordinates were deposited in the RCSB Protein Data Bank43 with accession number 2GHP.
NMR structure determination
Recombinant proteins in all NMR studies contained a carboxyl-terminal 6x His tag and were prepared as previously described.17, 22 Prp24-12 and -23 (residues 39–197 and 115–291, respectively) were overexpressed in BL21 (DE3) cells in M9 minimal media (15NH4Cl and 13C-glucose), containing 250 μg/ml ampicillin. Both Prp24 constructs purified as monomeric proteins and were 90–95% pure as judged by SDS-PAGE, gel filtration chromatography, and their estimated rotational correlation times. NMR protein samples were dialyzed three times against 2 L of NMR buffer (50 mM KPO4 pH 6.5, 100 mM KCl, and 2 mM DTT) and subsequently concentrated to 0.3–1.0 mM using a Centricon filter (Millipore).
NMR experiments for the Prp24-12 and Prp24-23 proteins were carried out at 25°C using Varian Inova 800 and 600 MHz spectrometers equipped with cryogenic triple resonance probes. Sequence specific backbone assignments were achieved using 2D 1H-15N HSQC, 2D 1H-13C HSQC (aliphatic and aromatic optimized), 3D HNCO, 3D HNCA, 3D HNCACB, 3D CBCA(CO)NH, and 3D HBHA(CO)NH experiments. For Prp24-12, additional 1H and 13C side-chain assignments were assigned and confirmed using 3D H(C)CH-TOCSY, 3D H(C)CH-COSY experiments. All spectra were processed and initially analyzed with NMRPipe44 and Sparky (http://www.cgl.ucsf.edu/home/sparky) software packages, respectively. For Prp24-12, the CARA software program45 was used to determine the side chain 1H, 13C, 15N resonance assignments (BioMagResBank database accession number 7070).22
For Prp24-12, 3D 1H-15N NOESY-HSQC and 3D 1H-13C NOESY-HSQC (aliphatic and aromatic optimized) spectra (60 ms mixing time) were acquired and analyzed based upon the sequence specific resonance assignments and input into the automated NOESY peak picking and NOE assignment method ATNOS/CANDID 1.1.46 The ATNOS/CANDID approach included only the chemical shift list obtained from the sequence specific assignments and the three 3D NOESY spectra. A total of 1670 NOE meaningful upper limit constraints were obtained from the ATNOS/CANDID calculation. In addition to this NOE data, 212 torsion angle constraints (105 for φ and 107 for ψ) from the program TALOS47 were used for the structure calculations in CYANA 2.1.48 Based on the 20 lowest target function conformers, 64 hydrogen bonds were identified by CYANA, manually confirmed in the NMR data, and used in the final structure calculation. The 20 conformers with the lowest target function values were energy-minimized with respect to the AMBER force field49 in a water shell of 8 Å minimal thickness, using the program OPAL.50 The final coordinates were deposited in the RCSB Protein Data Bank43 with accession number 2GO9.
15N NMR relaxation of RRMs 1, 2 and 3
Multiple 15N NMR relaxation experiments of the Prp24-12 and 23 proteins were acquired at 600 MHz and 800 MHz.51 Longitudinal (T1) experiments were collected with relaxation delays of 10, 100, 300, 500, 700, 900, and 1300 ms while transverse (T2) experiments were measured with relaxation delays of 10, 30, 50, 70, 90, 110, 130, 150, and 170 ms. Spectra were processed using NMRPipe and relaxation curves were fitted using the relaxation heights extension in SPARKY (http://www.cgl.ucsf.edu/home/sparky). Uncertainties for relaxation times (T1 and T2) were estimated from multiple relaxation datasets (acquired 3 times for Prp24-12 and 2 times for Prp24-23) as well as from the error in the mono-exponential decay fit of the measured signal intensity. Two 15N heteronuclear NOE datasets were acquired for each construct with the unenhanced and enhanced (3 s 1H excitation period) spectra interleaved. 15N heteronuclear NOE values represent the average ratio of peak heights between the enhanced and unenhanced spectra from the two data sets. Errors were measured from the ratio of the peak height to the spectrum noise.
Protein-RNA binding assays
The Prp24-N123 and -N12 proteins used for RNA binding assays contain residues 1–291 and 1–197, respectively, and a carboxyl-terminal 6x His tag. The Prp24-23 protein is identical to that used for NMR studies. Prp24-U6 binding was performed and dissociation constants calculated as previously described,17 with the following modifications. RNA was incubated with 15N-labeled protein in the absence of carrier tRNA or BSA. The 40-nucleotide RNA (Figure 8(a)) was transcribed in vitro using His6-tagged T7 RNA polymerase, synthetic DNA oligonulceotides (IDT, Inc.), 1 mM CTP, GTP, and UTP, 0.1 mM ATP, and 1 mCi/mL [α-32P]ATP. RNA was purified by denaturing 15% polyacrylamide gel electrophoresis, identified by UV absorbance, and excised from the gel. RNA was quantitated by scintillation counting and subsequently brought to pH 7.0 by the addition of 1 M NaOH.
1H-15N HSQC-TROSY spectra of the 0.2 mM 2H,15N-labeled Prp24-N12 protein were acquired on a Varian Unity Inova 900 MHz spectrometer equipped with a cold probe. Separate RNA titrations were performed using the 40-nucleotide (Figure 7(a)) and the 21-nucleotide U6 RNA constructs. RNA was directly added to the NMR samples at room temperature and subsequently incubated for 30 minutes at 4ºC at each titration stage. For the 21-nucleotide RNA, spectra were collected at 50, 100, 150, 200, 300, 500, and 900 μM concentrations. Line broadening was enhanced in other regions of the spectra at concentrations above 150 μM and a distinct set of HN resonances were observed at 900 μM, corresponding to an approximate 5:1 (RNA:protein) stoichiometry.
Supplementary Material
Acknowledgments
We thank Gregg Sahli at APS, members of the CESG team, Yun-Xing Wang (NCI-Frederick), NMRFAM (www.nmrfam.wisc.edu), the Markley lab (especially Marco Tonelli and Greg Zornetzer), Joel McManus, and Nicole Aulik for helpful discussions and technical assistance, and Jim Keck and Joel McManus for comments on the manuscript. Use of the APS SER-CAT beamline was supported by the DOE Office of Basic Energy Sciences, under Contract W-31-109-Eng-38, and by the member institutions (www.ser-cat.org/members.html). This study made use of the National Magnetic Resonance Facility at Madison, which is supported by National Institutes of Health grants P41RR02301 (Biomedical Research Technology Program, National Center for Research Resources) and P41GM66326 (National Institute of General Medical Sciences). Equipment in the facility was purchased with funds from the University of Wisconsin, the National Institutes of Health (P41GM66326, P41RR02301, RR02781, RR08438), the National Science Foundation (DMB-8415048, OIA-9977486, BIR-9214394), and the U.S. Department of Agriculture. This investigation was supported by NIH grants GM74901 and GM64598 (CESG), GM65166 (to S.E.B.), and GM54018 (to D.A.B.). N.J.R. and S.S.K. received support from NIGMS pre-doctoral training grants GM08293 and GM08349, respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Will CL, Lührmann R. Spliceosome structure and function. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 3. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 369–400. [Google Scholar]
- 2.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–26. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 3.Brow DA. Allosteric cascade of spliceosome activation. Annu Rev Genet. 2002;36:333–60. doi: 10.1146/annurev.genet.36.043002.091635. [DOI] [PubMed] [Google Scholar]
- 4.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–4. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 5.Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat Rev Mol Cell Biol. 2004;5:908–19. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 6.Shannon KW, Guthrie C. Suppressors of a U4 snRNA mutation define a novel U6 snRNP protein with RNA-binding motifs. Genes Dev. 1991;5:773–85. doi: 10.1101/gad.5.5.773. [DOI] [PubMed] [Google Scholar]
- 7.Ghetti A, Company M, Abelson J. Specificity of Prp24 binding to RNA: a role for Prp24 in the dynamic interaction of U4 and U6 snRNAs. RNA. 1995;1:132–45. [PMC free article] [PubMed] [Google Scholar]
- 8.Jandrositz A, Guthrie C. Evidence for a Prp24 binding site in U6 snRNA and in a putative intermediate in the annealing of U6 and U4 snRNAs. Embo J. 1995;14:820–32. doi: 10.1002/j.1460-2075.1995.tb07060.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raghunathan PL, Guthrie C. A spliceosomal recycling factor that reanneals U4 and U6 small nuclear ribonucleoprotein particles. Science. 1998;279:857–60. doi: 10.1126/science.279.5352.857. [DOI] [PubMed] [Google Scholar]
- 10.Lygerou Z, Christophides G, Seraphin B. A novel genetic screen for snRNP assembly factors in yeast identifies a conserved protein, Sad1p, also required for pre-mRNA splicing. Mol Cell Biol. 1999;19:2008–20. doi: 10.1128/mcb.19.3.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vidaver RM, Fortner DM, Loos-Austin LS, Brow DA. Multiple functions of Saccharomyces cerevisiae splicing protein Prp24 in U6 RNA structural rearrangements. Genetics. 1999;153:1205–18. doi: 10.1093/genetics/153.3.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rader SD, Guthrie C. A conserved Lsm-interaction motif in Prp24 required for efficient U4/U6 di-snRNP formation. RNA. 2002;8:1378–92. doi: 10.1017/s1355838202020010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strauss EJ, Guthrie C. A cold-sensitive mRNA splicing mutant is a member of the RNA helicase gene family. Genes Dev. 1991;5:629–41. doi: 10.1101/gad.5.4.629. [DOI] [PubMed] [Google Scholar]
- 14.Vaidya VC, Seshadri V, Vijayraghavan U. An extragenic suppressor of prp24-1 defines genetic interaction between PRP24 and PRP21 gene products of Saccharomyces cerevisiae. Mol Gen Genet. 1996;250:267–76. doi: 10.1007/BF02174384. [DOI] [PubMed] [Google Scholar]
- 15.Kuhn AN, Brow DA. Suppressors of a cold-sensitive mutation in yeast U4 RNA define five domains in the splicing factor Prp8 that influence spliceosome activation. Genetics. 2000;155:1667–82. doi: 10.1093/genetics/155.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell M, Schreiner S, Damianov A, Reddy R, Bindereif A. p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. Embo J. 2002;21:2724–35. doi: 10.1093/emboj/21.11.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwan SS, Brow DA. The N- and C-terminal RNA recognition motifs of splicing factor Prp24 have distinct functions in U6 RNA binding. RNA. 2005;11:808–20. doi: 10.1261/rna.2010905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maris C, Dominguez C, Allain FH. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. Febs J. 2005;272:2118–31. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 19.Vitali F, Henning A, Oberstrass FC, Hargous Y, Auweter SD, Erat M, Allain FH. Structure of the two most C-terminal RNA recognition motifs of PTB using segmental isotope labeling. Embo J. 2006;25:150–62. doi: 10.1038/sj.emboj.7600911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nooren IM, Thornton JM. Diversity of protein-protein interactions. Embo J. 2003;22:3486–92. doi: 10.1093/emboj/cdg359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kwan SS. Thesis (PhD) University of Wisconsin-Madison; 2005. Protein-mediated U6 RNA conformational changes in the yeast spliceosome. [Google Scholar]
- 22.Reiter NJ, Lee D, Wang YX, Tonelli M, Cornilescu C, Bahrami A, Butcher SE. Resonance assignments for the two N-terminal RNA recognition motifs (RRMs) of the S. cerevisiae Pre-mRNA Processing protein Prp24. J Biomol NMR. 2006;2006;36(Suppl 5):58. doi: 10.1007/s10858-006-9039-4. [DOI] [PubMed] [Google Scholar]
- 23.Brow DA, Guthrie C. Spliceosomal RNA U6 is remarkably conserved from yeast to mammals. Nature. 1988;334:213–8. doi: 10.1038/334213a0. [DOI] [PubMed] [Google Scholar]
- 24.Fortner DM, Troy RG, Brow DA. A stem/loop in U6 RNA defines a conformational switch required for pre-mRNA splicing. Genes Dev. 1994;8:221–33. doi: 10.1101/gad.8.2.221. [DOI] [PubMed] [Google Scholar]
- 25.Karaduman R, Fabrizio P, Hartmuth K, Urlaub H, Luhrmann R. RNA structure and RNA-protein interactions in purified yeast U6 snRNPs. J Mol Biol. 2006;356:1248–62. doi: 10.1016/j.jmb.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 26.Schumacher MA, Karamooz E, Zikova A, Trantirek L, Lukes J. Crystal structures of T. brucei MRP1/MRP2 guide-RNA binding complex reveal RNA matchmaking mechanism. Cell. 2006;126:701–11. doi: 10.1016/j.cell.2006.06.047. [DOI] [PubMed] [Google Scholar]
- 27.Thao S, Zhao Q, Kimball T, Steffen E, Blommel PG, Riters M, Newman CS, Fox BG, Wrobel RL. Results from high-throughput DNA cloning of Arabidopsis thaliana target genes using site-specific recombination. J Struct Funct Genomics. 2004;5:267–76. doi: 10.1007/s10969-004-7148-4. [DOI] [PubMed] [Google Scholar]
- 28.Sreenath HK, Bingman CA, Buchan BW, Seder KD, Burns BT, Geetha HV, Jeon WB, Vojtik FC, Aceti DJ, Frederick RO, Phillips GN, Jr, Fox BG. Protocols for production of selenomethionine-labeled proteins in 2-L polyethylene terephthalate bottles using auto-induction medium. Protein Expr Purif. 2005;40:256–67. doi: 10.1016/j.pep.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 29.Jeon WB, Aceti DJ, Bingman CA, Vojtik FC, Olson AC, Ellefson JM, McCombs JE, Sreenath HK, Blommel PG, Seder KD, Burns BT, Geetha HV, Harms AC, Sabat G, Sussman MR, Fox BG, Phillips GN., Jr High-throughput purification and quality assurance of Arabidopsis thaliana proteins for eukaryotic structural genomics. J Struct Funct Genomics. 2005;6:143–7. doi: 10.1007/s10969-005-1908-7. [DOI] [PubMed] [Google Scholar]
- 30.Zolnai Z, Lee PT, Li J, Chapman MR, Newman CS, Phillips GN, Jr, Rayment I, Ulrich EL, Volkman BF, Markley JL. Project management system for structural and functional proteomics: Sesame. J Struct Funct Genomics. 2003;4:11–23. doi: 10.1023/a:1024684404761. [DOI] [PubMed] [Google Scholar]
- 31.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–26. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 32.Uson I, Sheldrick GM. Advances in direct methods for protein crystallography. Curr Opin Struct Biol. 1999;9:643–8. doi: 10.1016/s0959-440x(99)00020-2. [DOI] [PubMed] [Google Scholar]
- 33.Grosse-Kunstleve RW, Adams PD. Substructure search procedures for macromolecular structures. Acta Crystallogr D Biol Crystallogr. 2003;59:1966–73. doi: 10.1107/s0907444903018043. [DOI] [PubMed] [Google Scholar]
- 34.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 35.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D Biol Crystallogr. 2003;59:2023–30. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 36.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr D Biol Crystallogr. 2000;56:965–72. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nat Struct Biol. 1999;6:458–63. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 40.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi, psi and Cbeta deviation. Proteins. 2003;50:437–50. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 41.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–41. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vriend G. WHAT IF: a molevular modeling and drug design program. J Mol Graph. 1990;8:52–6. 29. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 43.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 45.Masse JE, Keller R. AutoLink: automated sequential resonance assignment of biopolymers from NMR data by relative-hypothesis-prioritization-based simulated logic. J Magn Reson. 2005;174:133–51. doi: 10.1016/j.jmr.2005.01.017. [DOI] [PubMed] [Google Scholar]
- 46.Herrmann T, Guntert P, Wuthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319:209–27. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 47.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 48.Guntert P, Mumenthaler C, Wuthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273:283–98. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 49.Ponder JW, Case DA. Force fields for protein simulations. Adv Protein Chem. 2003;66:27–85. doi: 10.1016/s0065-3233(03)66002-x. [DOI] [PubMed] [Google Scholar]
- 50.Luginbuhl P, Guntert P, Billeter M, Wuthrich K. The new program OPAL for molecular dynamics simulations and energy refinements of biological macromolecules. J Biomol NMR. 1996;8:136–46. doi: 10.1007/BF00211160. [DOI] [PubMed] [Google Scholar]
- 51.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


