Abstract
Background
Histone modification regulates chromatin structure and influences gene expression associated with diverse biological functions including cellular differentiation, cancer, maintenance of genome architecture, and pathogen virulence. In Entamoeba, a deep-branching eukaryote, short chain fatty acids (SCFA) affect histone acetylation and parasite development. Additionally, a number of active histone modifying enzymes have been identified in the parasite genome. However, the overall extent of gene regulation tied to histone acetylation is not known.
Results
In order to identify the genome-wide effects of histone acetylation in regulating E. histolytica gene expression, we used whole-genome expression profiling of parasites treated with SCFA and Trichostatin A (TSA). Despite significant changes in histone acetylation patterns, exposure of parasites to SCFA resulted in minimal transcriptional changes (11 out of 9,435 genes transcriptionally regulated). In contrast, exposure to TSA, a more specific inhibitor of histone deacetylases, significantly affected transcription of 163 genes (122 genes upregulated and 41 genes downregulated). Genes modulated by TSA were not regulated by treatment with 5-Azacytidine, an inhibitor of DNA-methyltransferase, indicating that in E. histolytica the crosstalk between DNA methylation and histone modification is not substantial. However, the set of genes regulated by TSA overlapped substantially with genes regulated during parasite development: 73/122 genes upregulated by TSA exposure were upregulated in E. histolytica cysts (p-value = 6 × 10-53) and 15/41 genes downregulated by TSA exposure were downregulated in E. histolytica cysts (p-value = 3 × 10-7).
Conclusion
This work represents the first genome-wide analysis of histone acetylation and its effects on gene expression in E. histolytica. The data indicate that SCFAs, despite their ability to influence histone acetylation, have minimal effects on gene transcription in cultured parasites. In contrast, the effect of TSA on E. histolytica gene expression is more substantial and includes genes involved in the encystation pathway. These observations will allow further dissection of the effects of histone acetylation and the genetic pathways regulating stage conversion in this pathogenic parasite.
Background
Regulation of gene expression is a complex process controlled by sequence-specific DNA binding proteins, modulation of chromatin structure, and post-transcriptional modifications. In recent years, increased attention has been given to the role of epigenetic mechanisms, such as the modification of histone proteins, in gene regulation [1]. These modifications, including methylation, phosphorylation and acetylation, occur at specific amino acids on the N-terminal tails of histone core proteins, particularly H3 and H4, and regulate chromatin structure and gene expression [2,3]. Methylation of histones at lysine residues has typically been associated with transcriptionally silent heterochromatin [4]. In contrast, lysine acetylation is generally thought to trigger the opening of chromatin structure and transcriptional activation [5,6]. However, this is an oversimplified model and does not represent the true complexity of these processes, which can also differ between lower and higher eukaryotes [7]. Individual modifications of histones may be interdependent, with methylation of certain lysine residues blocking or enhancing the addition of acetyl groups nearby [8,9]. In addition, methylation of arginine residues may actually activate the transcription of some genes. A number of proteins have been identified which regulate these modifications, including histone acetyltransferases (HATs), histone deacetylases (HDACs), histone methyltransferases (HMT), and a recently discovered class of histone demethylases [10].
The protozoan parasite Entamoeba histolytica has two morphologically distinct life cycle forms, the infectious cyst form that transmits disease from person to person, and the trophozoite form that multiplies in the colon and eventually differentiates back into the cyst form. While in the colon, the trophozoite form causes invasive disease (colitis and liver abscess) in 50 million people per year making amebiasis a leading parasitic cause of death worldwide [11]. Despite its importance for human health, little is known about how this parasite modulates its gene expression during host invasion or conversion from one life cycle form to the other. Changes in transcript abundance in E. histolytica are associated with host invasion [12], with exposure to oxidative stress [13], and with conversion between the cyst and trophozoite forms [14], but the mechanisms regulating transcript levels are poorly understood. A number of amebic promoter elements and transcription factors have been described [15] and DNA methylation has been identified as playing a role in controlling a limited amount of amebic gene expression [16,17]. Functional histone-modifying enzymes, such as HATs of the MYST and GNAT families, and a Class I HDAC, and acetylated histones have been described in E. histolytica [18], but their activities have not yet been tied to gene expression changes.
In Entamoeba invadens, a parasite of reptiles, a role for histone modifications in the regulation of stage conversion has been proposed. Histones of in vitro cultured E. invadens trophozoites are constitutively acetylated, with the levels of acetylation increasing in the presence of Trichostatin A (TSA), but decreasing in the presence short chain fatty acids (SCFA) such as butyrate [19]. The decreased histone acetylation resulting from butyrate exposure was unexpected, as this compound induces increased histone acetylation in all other eukaryotic cells in which it has been examined [20-22]. Treatment of E. invadens trophozoites with TSA or SCFAs blocks their in vitro development to the cyst stage, suggesting a biological role for histone modification in Entamoeba development [23]. The link between cyst development and histone acetylation observed in E. invadens has not been recapitulated in E. histolytica due to lack of an in vitro system for encystation. Complicating the studies of E. histolytica is the fact that individual laboratory strains of the parasite have different baseline histone acetylation patterns [19]. For example, E. histolytica HM-1:IMSS under standard culture conditions does not have any detectable acetylated H4, whereas two other strains, E. histolytica Rahman and E. histolytica 200:NIH, have multiply-acetylated H4 populations under the same growth conditions. Additionally, both of these strains shift to a hyperacetylated H4 pattern when treated with TSA. Furthermore, when grown with SCFAs, E. histolytica Rahman and E. histolytica 200:NIH H4 histones become hypoacetylated, similar to the response of E. invadens. The unusual hypoacetylation response to butyrate of Entamoeba suggests that SCFAs regulate histone acetylation and gene expression in a unique way, one that most likely reflects parasite adaptation to growth in the presence of the large amounts of the short chain fatty acids found in the colon.
In other protozoan parasites histone modification plays important roles in life cycle progression and antigenic variation. In Toxoplasma gondii, chromatin immunoprecipitation analysis has demonstrated differential acetylation and methylation in the promoters of stage-specific genes during stage conversion [24]. In addition, treatment with drugs that affect histone acetylation or arginine methylation affected both stage-conversion and overall gene expression [24,25]. In Plasmodium falciparum histone H4 acetylation states and promoter occupation by the Sir2 transcriptional regulator have been linked to changes in the expression of var genes [26].
To gain insights into the role of histone acetylation in regulating gene expression in E. histolytica we treated E. histolytica trophozoites with SCFA or TSA and performed whole genome transcriptional profiling. The data revealed that in E. histolytica there was minimal transcriptional response to SCFAs, with ~0.1% of genes modulated ± 2-fold. In contrast, the transcriptional response to TSA was greater (~2% of genes modulated ± 2-fold), and the gene expression changes overlapped significantly with the transcriptional signature of the developmental pathway to cysts [14]. This work represents the first genome wide analysis of transcriptional changes associated with histone modifications in E. histolytica and reveals a subset of developmentally regulated genes whose expression correlates with changes in the level of histone acetylation.
Results
E. histolytica strains HM-1:IMSS, Rahman, and 200:NIH have similar but not identical expression profiles of genes encoding histone-modifying enzymes
To account for differences in levels of histone acetylation between E. histolytica strains, we analyzed previously published data from a whole genome microarray to compare the gene expression profiles of three strains of E. histolytica (HM-1:IMSS, Rahman and 200:NIH) [14]. Overall, the expression profiles of the three E. histolytica strains were highly similar although some genes whose transcript levels were significantly different between strains (± 2-fold and p-value <0.05) were identified. Overall, 127 genes had higher expression in E. histolytica HM-1:IMSS, 261 genes had higher expression in E. histolytica 200:NIH, and 71 genes had higher expression in E. histolytica Rahman compared to the other two E. histolytica strains (Additional File 1). For our purposes, we focused on the expression levels of genes involved in the regulation of histone acetylation and chromatin structure. Surprisingly, given the absence of multiply acetylated histones in E. histolytica HM-1:IMSS, several HAT genes (2.m00560 and 67.m00100) were expressed at relatively high levels in HM-1:IMSS trophozoites (Additional File 2). Some differences in expression levels of histone modification genes between the strains were identified. One HAT (100.m00145) had significantly higher expression E. histolytica 200:NIH. Two Sir2 family HDAC genes were expressed differentially between strains: 251.m00088 was expressed at significantly higher levels in E. histolytica HM-1:IMSS and 2.m00521 was more highly expressed in E. histolytica 200:NIH. The overexpression of particular Sir2 genes in yeast leads to global histone deacetylation [6]. The increased expression of a HAT gene in E. histolytica 200:NIH trophozoites and high expression of a Sir2 HDAC gene in E. histolytica HM1:IMSS is consistent with the histone acetylation patterns in these strains. However, the actual levels histone proteins and their relative enzyme activities in these parasite strains will need to be established before conclusions can be made about the causes of the differences in levels of multiply-acetylated histones in the isolates.
Growth of E. histolytica in the presence of SCFA has minimal effects on parasite gene expression
SCFA have substantial effects on histone acetylation and development in Entamoeba [19,23]. To determine the effect of SCFA on amebic growth, we tested the ability of E. histolytica to grow in TYI-S-33 LG medium ± SCFA. In media with SCFA, there were no significant alterations in growth of E. histolytica HM-1:IMSS and 200:NIH parasites at 16 hours, the time points at which the microarray experiments were performed (data not shown). To determine if growth with SCFA altered parasite gene expression, we compared the transcriptional profile of E. histolytica 200:NIH trophozoites grown in TYI-S-33 LG medium ± SCFA. The E. histolytica 200:NIH strain was used for this and subsequent microarray analyses because it has an histone acetylation pattern similar to encystation-competent E. invadens and hence was the most likely to provide insights into developmental pathways regulated by histone acetylation [19]. For each strain and culture condition the number of arrays and the correlations between array data sets are outlined in Tables 1 and 2. Genes were considered differentially expressed if they had a ≥ 2-fold change and were significant with an FDR of < 0.05. Overall, few changes in gene expression were observed when we compared E. histolytica 200:NIH grown in TYI-S-33 or LG medium to those grown in LG medium with SCFA. Only 11 genes were differentially regulated by addition of SCFA, and there were no changes in genes associated with histone modifications (Table 3; Additional File 3). Thus, although growth of E. histolytica 200:NIH trophozoites with SCFAs results in hypoacetylation of H4 histones [19], these changes are apparently not associated with significant alterations in the gene expression profile of axenically grown trophozoites.
Table 1.
An overview of the microarrays generated and used in the analysis.
|
E. histolytica
200:NIH |
E. histolytica
200:NIH |
E. histolytica
200:NIH |
|
| Number of arrays | 4 | 3 | 3 |
| Minimum correlation of the arrays in condition | 0.96 | 0.96 | 0.98 |
| Culture medium | TYI-S-33 or LG | LG+SCFA | LG+TSA |
Summary of the culture conditions and number of arrays performed for each condition with E. histolytica 200:NIH.
Table 2.
Correlations of arrays used in analysis.
|
E. histolytica
200:NIH (TYI-S-33 or LG) |
E. histolytica
200:NIH (LG+SCFA) |
E. histolytica
200:NIH (LG+TSA) |
|
| 200:NIH (TYI-S-33 or LG) | X | 0.99 | 0.98 |
| 200:NIH (LG+SCFA) | X | X | 0.98 |
| 200:NIH (LG+TSA) | X | X | X |
The average normalized microarray data of a given condition was compared to another condition and the correlations are shown.
Table 3.
E. histolytica genes regulated by exposure short-chain fatty acids.
| Probe ID | GenBank ID | Description | Fold-change | FDR |
| Up-regulated | ||||
| 122.m00139_at | XM_647238 | ADP-ribosylation factor, putative | 2.11 | 1.97E-02 |
| 14.m00310_at | XM_651236 | hypothetical protein | 6.70 | 2.09E-03 |
| 205.m00100_s_at | XM_645582 | hypothetical protein | 2.57 | 2.86E-02 |
| 22.m00285_at | XM_650728 | hypothetical protein | 19.36 | 1.19E-11 |
| 295.m00030_at | XM_644416 | conserved hypothetical protein | 20.75 | 3.12E-14 |
| 418.m00028_at | XM_643537 | 70 kDa heat shock protein, putative | 6.03 | 1.97E-02 |
| 522.m00017_at | XM_643097 | hypothetical protein | 3.41 | 6.42E-03 |
| 522.m00018_x_at | XM_643100 | hypothetical protein | 2.78 | 1.19E-03 |
| 585.m00015_s_at | XM_642978 | conserved hypothetical protein | 3.16 | 1.82E-03 |
| 72.m00179_at | XM_648717 | hypothetical protein | 6.25 | 5.69E-04 |
| Down-regulated | ||||
| 5.m00482_at | XM_651923 | protein kinase, putative | -2.82 | 2.86E-02 |
The mean trophozoite expression value for E. histolytica 200:NIH strain under standard culture conditions, the fold-change in SCFA treated parasites, FDR, GenBank ID number, and gene annotations are shown. Ten genes are upregulated and one is downregulated. 205.m00100_s_at also represents 105.m00129.
Growth of E. histolytica 200:NIH in the presence of TSA changes the amebic transcriptional profile
To determine the effect of TSA on amebic growth, E. histolytica strains HM-1:IMSS and 200:NIH were grown in TYI-S-33 LG medium ± TSA. E. histolytica HM-1:IMSS trophozoites died within 2–3 days in medium with 150 nM TSA, whereas E. histolytica 200:NIH survived and grew but at a much slower rate than in TYI-S-33 LG medium alone (Figure 1). As treatment with TSA has been shown to cause histone H4 hyperacetylation [19], the effects of TSA on parasite gene expression were determined by comparing the transcriptional profile of E. histolytica 200:NIH trophozoites grown in TYI-S-33 LG medium ± TSA. Four microarrays were hybridized with RNA from E. histolytica 200:NIH parasites grown in TYI-S-33 or LG medium and compared to three microarrays hybridized with RNA from E. histolytica 200:NIH parasites grown in LG medium plus TSA for 16–72 hours. In contrast to the minimal transcriptional changes seen in the SCFA treatment, TSA exposure resulted in significant changes in gene expression. Overall, 163 genes, ~2% of the genes tested, showed altered transcript abundance, with 122 genes upregulated and 41 genes downregulated by TSA exposure (Tables 4 and 5; Additional File 4). Of the 122 genes whose expression increased with TSA treatment, 46 (38%) had normalized expression < 0.2 in 200:NIH trophozoites grown in TYI-S-33 or LG, indicating that they may be silenced under normal in vitro growth conditions.
Figure 1.
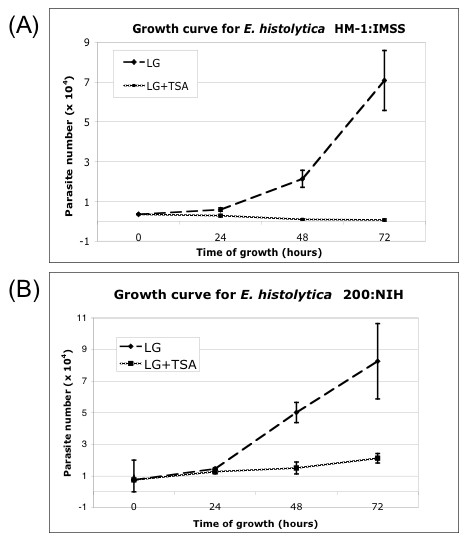
Growth rates of E. histolytica HM-1:IMSS and 200:NIH in medium with Trichostatin A. Log-phase trophozoites (7,500 cells) were seeded into 14 ml tubes containing fresh media (LG or LG+TSA). Aliquots were counted every 24 hours. (A) E. histolytica HM1:IMSS parasites stopped proliferating immediately upon transfer to LG+TSA containing media. (B) E. histolytica 200:NIH can grow in LG+TSA, although at a reduced rate compared to growth in LG medium. Experiments were performed a minimum of two times and standard deviation is shown.
Table 4.
Subset of E. histolytica genes upregulated by exposure to Trichostatin A.
| Probe ID | GenBank ID | Description | Fold-change | FDR |
| 489.m00024_at | XM_643195 | hypothetical protein | 39.96 | 7.60E-16 |
| 14.m00310_at | XM_651236 | hypothetical protein | 39.67 | 2.32E-20 |
| 295.m00030_at | XM_644416 | conserved hypothetical protein | 38.77 | 2.29E-20 |
| 522.m00017_at | XM_643097 | hypothetical protein | 20.45 | 4.53E-15 |
| 418.m00028_at | XM_643537 | 70 kDa heat shock protein, putative | 17.45 | 6.71E-10 |
| 36.m00204_s_at | XM_650008 | hypothetical protein | 14.84 | 1.22E-08 |
| 451.m00037_s_at | XM_643383 | hypothetical protein | 14.68 | 3.96E-04 |
| 556.m00022_x_at | XM_643031 | hypothetical protein | 14.37 | 5.75E-07 |
| 12.m00306_at | XM_651386 | hypothetical protein | 13.25 | 1.64E-05 |
| 376.m00054_s_at | XM_643784 | hypothetical protein | 13.02 | 1.62E-10 |
| 564.m00020_x_at | XM_643013 | hypothetical protein | 12.93 | 8.19E-10 |
| 82.m00164_s_at | XM_648353 | hypothetical protein | 12.88 | 8.00E-12 |
| 28.m00298_s_at | XM_650406 | hypothetical protein | 11.79 | 5.97E-09 |
| 110.m00118_at | XM_647568 | Rho family GTPase | 11.60 | 2.26E-14 |
| 50.m00195_s_at | XM_649449 | hypothetical protein | 11.36 | 3.02E-05 |
| 135.m00094_at | XM_646923 | hypothetical protein | 10.98 | 2.38E-08 |
| 493.m00030_x_at | XM_643175 | hypothetical protein | 10.25 | 1.40E-24 |
| 337.m00049_at | XM_644075 | hypothetical protein | 9.93 | 5.43E-07 |
| 164.m00105_x_at | XM_646368 | hypothetical protein | 9.92 | 4.32E-05 |
| 847.m00011_x_at | XM_642805 | hypothetical protein | 9.63 | 9.76E-05 |
| 205.m00100_s_at | XM_645582 | hypothetical protein | 9.48 | 3.01E-08 |
| 373.m00052_at | XM_643804 | hypothetical protein | 8.71 | 1.14E-06 |
| 395.m00030_x_at | XM_643673 | protein kinase, putative | 8.68 | 6.64E-04 |
| 749.m00013_s_at | XM_642848 | hypothetical protein | 8.61 | 1.85E-03 |
| 227.m00077_at | XM_645230 | hypothetical protein | 8.40 | 1.70E-08 |
| 728.m00012_x_at | XM_642855 | hypothetical protein | 8.37 | 5.83E-06 |
| 460.m00025_x_at | XM_643328 | hypothetical protein | 8.00 | 3.05E-02 |
| 167.m00116_x_at | XM_646306 | hypothetical protein | 7.98 | 3.77E-03 |
| 621.m00019_at | XM_642928 | hypothetical protein | 7.84 | 8.57E-06 |
| 584.m00019_at | XM_642979 | heat shock protein 70, putative | 7.82 | 5.54E-06 |
| 477.m00021_at | XM_643239 | hypothetical protein | 7.60 | 1.27E-03 |
| 123.m00123_x_at | XM_647209 | conserved hypothetical protein | 7.48 | 3.84E-05 |
| 76.m00146_at | XM_648554 | hypothetical protein | 7.34 | 1.75E-04 |
| 89.m00125_s_at | XM_648139 | hypothetical protein | 7.32 | 2.51E-05 |
| 220.m00068_x_at | XM_645332 | hypothetical protein | 7.14 | 2.48E-03 |
| 263.m00053_at | XM_644795 | hypothetical protein | 6.95 | 2.56E-04 |
| 804.m00006_x_at | XM_642821 | hypothetical protein | 6.94 | 2.44E-04 |
| 72.m00179_at | XM_648717 | hypothetical protein | 6.85 | 1.17E-04 |
| 211.m00072_at | XM_645468 | zinc finger protein, putative | 6.80 | 5.04E-03 |
| 102.m00082_at | XM_647784 | hypothetical protein | 6.74 | 6.19E-03 |
| 411.m00025_x_at | XM_643570 | hypothetical protein | 6.68 | 8.26E-03 |
| 162.m00085_at | XM_646412 | predicted protein | 6.61 | 3.81E-06 |
| 157.m00087_x_at | XM_646502 | conserved hypothetical protein | 6.50 | 8.44E-04 |
| 12.m00326_at | XM_651342 | hypothetical protein | 6.42 | 4.10E-04 |
| 52.m00169_x_at | XM_649399 | hypothetical protein | 6.39 | 2.21E-03 |
| 22.m00288_at | XM_650731 | hypothetical protein | 6.28 | 6.49E-04 |
| 71.m00129_at | XM_648728 | hypothetical protein | 6.16 | 5.70E-05 |
| 6.m00428_at | XM_651814 | hypothetical protein | 6.10 | 1.46E-04 |
| 496.m00027_x_at | XM_643167 | hypothetical protein | 5.98 | 9.24E-06 |
| 135.m00113_at | XM_646942 | hypothetical protein | 5.91 | 4.26E-03 |
The mean trophozoite expression value for E. histolytica 200:NIH strain under standard culture conditions, the fold-change in TSA treated parasites, FDR, GenBank ID number, and gene annotations are shown. Genes in bold have been confirmed by RT-PCR. Genes regulated during trophozoite to cyst development are shown in italics. Additional loci represented by crosshybridizing _s_at probe sets are as follows: 36.m00204_s_at: 88.m00180; 451.m00037_s_at: 467.m00031 and 50.m00196; 376.m00054_s_at: 116.m00123; 82.m00164_s_at: 82.m00157; 28.m00298_s_at: 481.m00033 and 90.m00158; 205.m00100_s_at: 105.m00129; 749.m00013_s_at: 142.m00151; 89.m00125_s_at: 357.m00038.
Table 5.
E. histolytica genes downregulated by exposure to Trichostatin A.
| Probe ID | GenBank ID | Description | Fold-change | FDR |
| 111.m00118_at | XM_647540 | leishmaniolysin-related peptidase, putative | -44.44 | 0.00E+00 |
| 233.m00105_at | XM_645153 | hypothetical protein | -29.24 | 0.00E+00 |
| 7.m00429_at | XM_651789 | Beige BEACH domain protein, putative | -21.88 | 8.51E-10 |
| 223.m00071_at | XM_645286 | hypothetical protein | -10.88 | 3.04E-04 |
| 31.m00224_at | XM_650243 | hypothetical protein | -9.43 | 2.76E-03 |
| 1.m00712_at | XM_652408 | hypothetical protein | -8.62 | 1.64E-05 |
| 223.m00075_at | XM_645290 | lipid phosphatase, putative | -6.71 | 1.34E-03 |
| 105.m00133_at | XM_647680 | NADP-dependent alcohol dehydrogenase | -5.81 | 8.19E-05 |
| 223.m00077_x_at | XM_645292 | hypothetical protein | -5.65 | 8.47E-03 |
| 585.m00015_s_at | XM_642978 | conserved hypothetical protein | -5.52 | 1.11E-02 |
| 223.m00069_at | XM_645284 | hypothetical protein | -5.49 | 9.05E-03 |
| 223.m00068_at | XM_645283 | scavenger mRNA decapping enzyme, putative | -5.24 | 2.27E-03 |
| 17.m00307_s_at | XM_651008 | fatty acid elongase, putative | -5.08 | 1.34E-02 |
| 234.m00042_at | XM_645139 | hypothetical protein | -5.00 | 2.34E-02 |
| 66.m00150_at | XM_648887 | high mobility group protein, putative | -4.78 | 3.68E-02 |
| 131.m00139_at | XM_647034 | M-phase inducer phosphatase, putative | -4.72 | 2.36E-02 |
| 249.m00083_at | XM_644976 | hypothetical protein | -4.44 | 3.58E-03 |
| 7.m00436_at | XM_651745 | actobindin homolog, putative | -4.31 | 7.67E-03 |
| 223.m00079_at | XM_645294 | cysteinyl-tRNA synthetase, putative | -4.20 | 1.54E-02 |
| 425.m00057_s_at | XM_643497 | conserved hypothetical protein | -4.10 | 2.14E-02 |
| 223.m00070_at | XM_645285 | protein kinase, putative | -4.08 | 2.28E-02 |
| 4.m00636_at | NA | pseudogene, galactose-specific adhesin light subunit | -4.00 | 4.34E-02 |
| 217.m00081_s_at | XM_645388 | hypothetical protein | -3.91 | 1.64E-02 |
| 92.m00148_s_at | XM_648072 | conserved hypothetical protein | -3.88 | 4.66E-02 |
| 223.m00078_at | XM_645293 | ribonuclease, putative | -3.85 | 3.89E-02 |
| 180.m00114_at | XM_646068 | hypothetical protein | -3.73 | 1.75E-03 |
| 223.m00067_at | XM_645282 | integral membrane protein, putative | -3.73 | 3.58E-03 |
| 223.m00074_at | XM_645289 | hypothetical protein | -3.72 | 1.80E-02 |
| 223.m00076_at | XM_645291 | hypothetical protein | -3.42 | 9.24E-03 |
| 10.m00362_at | XM_651510 | cysteine proteinase, putative | -3.41 | 3.20E-09 |
| 54.m00183_at | XM_649345 | hypothetical protein | -3.33 | 4.69E-02 |
| 242.m00078_s_at | XM_645064 | cysteine protease 1 | -2.89 | 3.47E-03 |
| 77.m00173_at | XM_648536 | hypothetical protein | -2.76 | 2.82E-04 |
| 17.m00351_at | XM_651053 | galactose-inhibitable lectin 35 kda subunit precursor | -2.75 | 2.84E-02 |
| 9.m00419_at | XM_651593 | Fe-hydrogenase, putative | -2.72 | 1.73E-03 |
| 52.m00148_at | XM_649403 | lysozyme, putative | -2.61 | 4.36E-02 |
| 154.m00120_at | XM_646541 | glucosidase II alpha subunit, putative | -2.57 | 3.53E-02 |
| 9.m00416_at | XM_651590 | hypothetical protein | -2.49 | 4.28E-02 |
| 47.m00182_at | XM_649559 | fatty acid elongase, putative | -2.26 | 3.57E-02 |
| 22.m00269_s_at | XM_650745 | hypothetical protein | -2.19 | 2.76E-02 |
| 77.m00178_s_at | XM_648541 | surface antigen ariel1-related | -2.08 | 3.15E-02 |
The mean trophozoite expression value for E. histolytica 200:NIH strain under standard culture conditions, the fold-change in TSA treated parasites, FDR, GenBank ID number, and gene annotations are shown. Genes in bold have been confirmed by RT-PCR. Genes regulated during trophozoite to cyst development are shown shaded in italics. Additional loci represented by crosshybridizing _s_at probe sets are as follows: 17.m00307_s_at: 17.m00304; 425.m00057_s_at: 10.m00394; 217.m00081_s_at: 20.m00277; 92.m00148_s_at: 250.m00108 and 284.m00087; 242.m00078_s_at: 79.m00156; 77.m00178_s_at: 19.m00343 and 533.m00017.
Semi-quantitative RT-PCR confirmation of array results
To confirm the expression patterns observed by microarray analysis, we performed semi-quantitative RT-PCR on 5 genes upregulated by exposure to TSA (135.m00113, 14.m00310, 337.m00049, 340.m00050 and 146.m00117), and 4 genes downregulated by exposure to TSA (1.m00712, 223.m00071, 223.m00075, and 77.m00173) (Figure 2). The gene for ssRNA, 247.m00075, 13.m00291 and 7.m00480 were used as loading controls. Serial dilutions of cDNA were performed for each sample. In all cases, RT-PCR results confirmed the array data.
Figure 2.
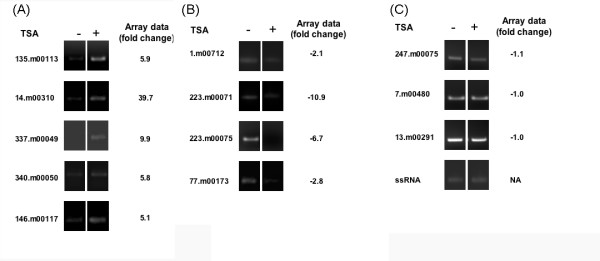
RT-PCR confirms microarray data for genes regulated by TSA. Genes found to be differentially expressed based on the array data were tested by semi-quantitative RT-PCR. RNA from log phase E. histolytica 200:NIH trophozoites exposed to 150 nM TSA for 16 hours was used to generate cDNA for the analysis. (A) Genes identified by the array analysis as upregulated in TSA treated parasites (135.m00113, 14.m00410, 337.m00049, 340.m00050 and 146.m00117) are shown. (B) Genes identified as downregulated in TSA parasites (1.m00712, 223.m00071, 223.m00075, and 77.m00173) are shown. (C) Genes identified as being unchanged in TSA treated parasites (247.m00075 and 7.m00480, 13.m00291) and small subunit ribosomal RNA (X61116) were used as a loading control. For all genes, the trends indicated by the array data were recapitulated by the RT-PCR analysis. For all samples, a control reaction without reverse transcriptase control was performed, and was negative.
A substantial number genes regulated by TSA are also developmentally regulated in E. histolytica
We compared the lists of genes regulated by TSA to a number of transcriptional profiles previously generated with E. histolytica parasites. There was no significant overlap of genes modulated by TSA with parasite genes modulated in a mouse model of colitis [27] or after exposure to 5-Azacytidine [16]. However, the profile of genes regulated by TSA was found to overlap substantially with the profiles of genes differentially expressed in the two developmental stages (trophozoites and cysts) of E. histolytica [14] (Figure 3 and Tables 4 and 5). There was significant overlap between genes upregulated by TSA treatment and cyst-specific genes, with 73 of the 122 genes upregulated by TSA also upregulated in cysts (p-value = 6 × 10-53). Genes downregulated by TSA treatment overlapped significantly with trophozoite-specific genes, with 15 of the 41 genes downregulated by TSA also downregulated in cysts (p-value = 3 × 10-7). There was no significant overlap in the opposite direction (4 genes downregulated by TSA were upregulated in cysts and 4 genes upregulated by TSA were downregulated in cysts). Genes that were upregulated in both TSA-treated trophozoites and in cysts include some of the most highly induced genes under both conditions. An example is the hypothetical protein (489.m00024), which shows a ~40-fold increase in expression in TSA treated parasites and >500-fold increase in cysts [14]. Also included in this group are several genes encoding heat shock proteins (418.m00028 and 136.m00105) and putative signaling molecules (acid sphingomyelinase, 18.m00321 and a protein kinase, 395.m00030).
Figure 3.
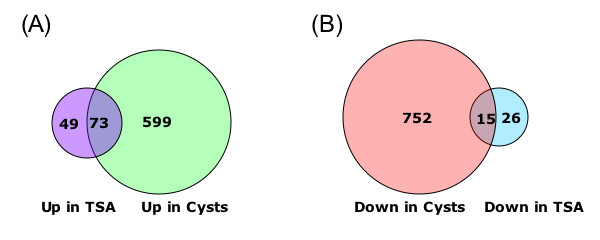
Venn diagram of genes regulated by TSA and during stage conversion. Overlap of genes regulated by TSA with developmentally regulated genes is shown. Of 122 genes upregulated by TSA treatment, 73 also show increased expression in cysts. Of the 41 genes downregulated by TSA treatment, 15 have increased expression in trophozoites. Both of these overlaps are statistically significant (p = 6 × 10-53 and p = 3 × 10-7 respectively).
Genes regulated in E. histolytica 200:NIH by exposure to Trichostatin A
Heat shock proteins
A number of heat shock proteins, including Hsp70 isoforms (64.m00148, 584.m00019, 65.m00150 and 418.m00028) were induced by TSA treatment. Whether these genes are regulated by histone acetylation, or whether their induction is due to a stress response of the parasites to growth in TSA is unclear at this point. A gene expression response to heat shock was previously reported to be linked to encystation in E. invadens [28], thus high expression of these genes indeed appears to be characteristic of the transcriptional profile of stage conversion.
Signaling molecules
Genes regulated by treatment with TSA include several that are likely to have functions in signal transduction. These include protein kinases (14.m00339 and 395.m00030) and a Rho family GTPase (110.m00118), all with increased expression in TSA-treated parasites. A protein kinase (223.m00070) and a protein phosphatase (131.m00139) are both downregulated during TSA treatment. The regulation of these putative signaling molecules by TSA may suggest a role for histone acetylation in modulating signal transduction and responses to environmental factors in E. histolytica. Also upregulated by TSA are several genes, which could play a role in transcriptional regulation such as a Myb family protein (175.m00117 and zinc finger domain containing proteins (211.m00072 and 68.m00203).
Virulence
Several genes with roles in E. histolytica virulence were downregulated by TSA treatment. This includes two genes encoding cysteine proteases: CP1 (242.m00078) and a putative CP (10.m00362), lysozyme (52.m00148) and a gene encoding the 35 kDa subunit of the amebic Gal/GalNAc lectin (17.m00351). Several of these genes have previously been identified as being trophozoite-specific, thus their down regulation is a further indication of the transcriptional activation of the encystation pathway in TSA-treated parasites [14].
Genomic regions controlled by histone acetylation
We investigated whether there were genomic regions containing multiple genes that were regulated by TSA. Such regions may be indicative of regions where gene expression is regulated by chromatin structure. We identified a cluster of three genes on scaffold 123 (123.m00113, 123.m00122, and 123.m00123) that were all upregulated by TSA. Additionally, a large cluster of genes strongly down regulated by TSA was observed on scaffold 223 (223.m00067, 223.m00068, 223.m00069, 223.m00070, 223.m00071, 223.m00074, 223.m00075, 223.m00076, 223.m00077, 223.m00078 and 223.m00079). The 223 chromosomal region had also been identified as being enriched for trophozoite-specific genes [14]. Whether expression from these genomic regions is repressed by histone acetylation, or whether the effect is indirect, needs to be determined experimentally.
Discussion
Gene expression can be transcriptionally controlled by epigenetic mechanisms including DNA methylation and histone modification. In order to define the genome-wide extent of regulation of gene expression by histone modification in Entamoeba histolytica, we performed expression profiling of E. histolytica trophozoites with short chain fatty acids and Trichostatin A (both histone deacetylase inhibitors). Our results identified that in contrast to effects seen in other eukaryotic systems, and despite inducing changes in histone acetylation, SCFA induce minimal transcriptional changes in E. histolytica trophozoites. However, the parasites do modulate gene expression significantly in response to TSA. The TSA induced transcriptional signature was distinct from changes induced by inhibition of DNA methylation but strongly overlapped with the gene expression profile of encystation in E. histolytica.
E. histolytica trophozoites normally grow and differentiate in the presence of SCFA while they reside in the lumen of the colon. SCFA are known to regulate gene expression in colonic epithelial cells which are normally exposed to SCFA [29-32]. When Entamoeba parasite isolates are initially collected from infected individuals, the trophozoites are cultured with the accompanying bacteria, which produce SCFA. Subsequently, E. histolytica isolates are selected for an ability to grow in medium that does not contain bacteria or SCFA. As only a small number of genes changed expression levels in response to SCFA, either the axenic parasites have lost nearly all of their transcriptional response to SCFA, or these compounds do not normally exert a large influence on gene expression at the transcriptional level in parasites in vivo. SCFAs do inhibit encystment, however, and based on the described results here, this may be occurring via more subtle changes in transcript levels (that did not meet the fold-change criteria applied to the data) or more likely through post-transcriptional mechanisms.
In contrast to SCFA, treatment of E. histolytica 200:NIH trophozoites with TSA demonstrated changes in gene transcript levels. This indicates that when class I/II HDAC enzymes are specifically targeted in Entamoeba and increased amounts of histone hyperacetylation occur [19], transcriptional changes follow. Like other eukaryotic cells, then, the expression of a small fraction of the genome of Entamoeba parasites appears to be sensitive to hyperacetylation of core histones. Transcriptional profiling was previously performed on E. histolytica parasites treated with 5-azacytidine (5-AzaC), an inhibitor of DNA methyltransferase, showing that ~2.1% of genes were differentially regulated by 5-AzaC exposure [16]. There was no significant overlap between the genes found here to be regulated by TSA and those regulated by 5-AzaC. Thus, epigenetic types of regulation, including both DNA methylation and histone acetylation, do play roles in gene expression mechanisms in E. histolytica, but the set of genes regulated by these processes is limited and non-overlapping. This is similar to the situation in Arabidopsis thaliana, in which genes regulated by 5-AzaC and TSA do not overlap, although a synergistic effect of treatment with both compounds has been observed [33]. In contrast, in human carcinoma cells TSA treatment results in DNA demethylation [34], one indication of the increasing levels of complexity of the mechanisms that establish histone codes in higher eukaryotes [7].
The greater significance of the gene expression changes induced by TSA was their overlap with the transcription profile of parasites undergoing differentiation. Initially these data may seem at odds with previously published data in which addition of TSA to encysting cultures of E. invadens was found to block encystment [23]. However, there are several possible explanations for this result. First, here we added TSA to vegetative E. histolytica trophozoite stage cells, whereas previous studies tested the effects of TSA on encysting E. invadens, and TSA effects on trophozoites and encysting parasites may be distinct. Second, the conclusions that TSA inhibits encystation in E. invadens were based on its ability to prevent production of a chitin-containing cyst, the end product of the differentiation pathway. The transcriptome data, in contrast, is a more revealing assessment of induction of the differentiation pathway. In fact, no genes known to encode proteins involved in cyst wall synthesis, such as chitin synthase or the glycoprotein Jacob, were regulated by TSA. Histone acetylation may therefore play an early role in cell fate determination and not regulate genes involved in the terminal stages of differentiation. Another possibility is that E. histolytica and E. invadens have opposing responses to TSA. However, this seems unlikely given the recent observation that conditions that support encystation in E. histolytica also permit spontaneous encystation in E. invadens [14], and both species respond to TSA with similar hyperacetylation responses [19].
Another model to consider is based on results from Toxoplasma gondii, in which the expression of both cyst and tachyzoite-specific genes is regulated by histone acetylation states [24]. This finding implied the existence of HATs and HDACs whose activities are developmentally regulated. If a similar situation were also the case in Entamoeba, trophozoite-specific HDAC activity would normally block activation of cyst-specific genes in trophozoites, and this block would be lifted upon TSA treatment, leading to the observed expression of a fraction of the cyst-specific genes. However, TSA treatment after induction of the encystation program would repress cyst-specific HDACs and inhibit the repression of trophozoite-specific genes, hence arresting the encystation program (Figure 4). Overall, the overlap between TSA-induced and cyst-specific genes and the overlap between TSA-repressed and trophozoite-specific genes is strong evidence that histone acetylation states are part of the mechanisms that regulate developmental pathways in E. histolytica.
Figure 4.
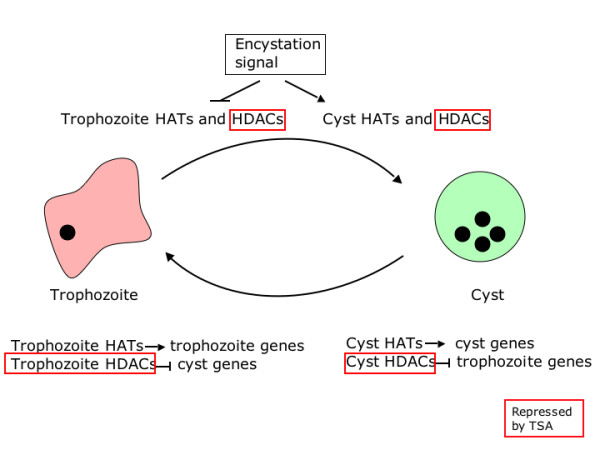
A proposed model for the role of histone acetylation in Entamoeba stage conversion. Under axenic growth conditions, trophozoite-specific HATs and HDACs induce the expression of trophozoite genes while suppressing the expression of cyst genes. When TSA is added to these cultures, the repression of HDAC activity allows expression of cyst genes. In contrast, during encystation cyst-specific HATs and HDACs become active, turning off trophozoite-specific genes and inducing cyst genes. TSA treatment of cells induced to encyst may repress the activity of cyst-specific HDACs, allowing continued expression of trophozoite genes and blocking completion of the encystation pathway. Steps sensitive to TSA treatment are highlighted in red.
Conclusion
We have used whole-genome expression profiling to demonstrate that E. histolytica 200:NIH trophozoites have dichotomous responses to SCFA and TSA, both histone deacetylase inhibitors. Despite affecting changes in histone acetylation, and in contrast to data from other eukaryotic systems, short chain fatty acids induce minimal transcriptional changes in E. histolytica trophozoites. In contrast, TSA has both a significant effect on histone acetylation and induces transcriptional changes. Importantly the transcriptional pathway modulated by TSA overlaps significantly with the gene expression changes seen with developmental conversion from trophozoites to cysts. This work identifies for the first time a molecular signature of TSA effects on E. histolytica parasites and lays the groundwork for further dissection of the roles of histone acetylation on amebic development.
Methods
Entamoeba strains and culture methods
The strains used in this study were E. histolytica HM-1:IMSS and E. histolytica 200:NIH both of which were grown axenically in TYI-S-33 medium under standard culture conditions at 36.5°C [35]. Additionally, both strains were grown in TYI-S-33 medium in the absence of glucose (LG medium), in TYI-S-33 medium plus SCFA (70 mM sodium acetate, 20 mM sodium propionate and 10 mM sodium butyrate) (SCFA medium) [23], and in TYI-S-33 medium plus TSA (150 nM or 300 nM) (TSA medium). Genotypes of the E. histolytica strains (HM-1:IMSS and 200:NIH) were confirmed by PCR and RFLP based on previously published methods [36,37].
RNA isolation and microarray hybridization
Total RNA was isolated using Trizol reagent (Invitrogen) using the manufacturer's protocol and purified using a Qiagen RNeasy kit before being used for microarray analysis [14]. Samples were processed for microarray hybridization by the Stanford University Protein and Nucleic Acids facility [38] using standard protocols. For each sample the RNA quality was checked using an Agilent BioAnalyzer QC and 4 μg subjected to the standard labeling and hybridization method [39]. E. histolytica 200:NIH parasites were grown in SCFA for 16 hours, harvested and RNA extracted for microarray experiments. E. histolytica 200:NIH parasites were grown in TSA (150 nM or 300 nM) for 16 or 72 hours (150 nM), harvested and RNA extracted for microarray experiments.
Labeled samples were hybridized to a custom generated Affymetrix platform full genome microarray (E_his-1a520285F), which has been previously described [27]. This array has 7,712 unique probe sets, which represent 9,435 open reading frames. Due to the highly repetitive nature of the E. histolytica genome, some of the probe sets are predicted to cross-hybridize with other sequences. Probe sets that represent a single gene and do not cross hybridize are labeled as (_at). Probe sets in which at least one probe may cross-hybridize with another gene(s) are labeled as (_x_at). In situations where all the probes for a given gene cross-hybridize with another gene(s), the probe sets is labeled as (_s_at) and additional genes that cross-hybridize with this probe set are listed in Additional File 3B. This array also contains probes for intergenic non-coding regions, however, these probe sets were excluded from all analysis. After hybridization, arrays were scanned and probe intensities calculation using Affymetrix GCOS software [40].
Microarray data normalization and analysis
Normalized expression values for each probe set were obtained from raw probe intensities in R 2.2.0 downloaded from the BioConductor project [41], using robust multi-array averaging with correction for oligo sequence (gcRMA) [42]. To identify differentially expressed genes, we used local pooled error testing [43] along with Benjamini-Hochberg multiple test correction [44]. In addition, fold-change was calculated in Genespring GX [45]. A minimum of three arrays from each condition were used for analysis of SCFA or TSA effects. Correlation coefficients were calculated in Genespring using standard correlation. Probe sets were considered differentially expressed between two conditions if they had at least a 2-fold change and were significant with a false discovery rate (FDR) of < 0.05, and were identified as "present" in at least one array. Datasets of transcriptional profiles from E. histolytica HM-1:IMSS, E. histolytica Rahman, parasites from an in vivo model of colitis, encystation, and 5-AzaC treatment were obtained from previously published data [14,16,27].
Semi-quantitative reverse transcriptase polymerase chain reaction (RT-PCR)
E. histolytica 200:NIH trophozoites grown in TYI-S-33 LG medium were transferred in mid log phase (3 × 105 per ml) into TYI-S-33 LG or TYI-S-33 LG/150nM TSA and incubated for 16 hrs. Total RNA was isolated with RNAzol, and 2ug RNA was treated with DNAase I for 5 minutes at 37°C. cDNA was synthesized with oligo-dT and Superscript III reverse transcriptase (Invitrogen) at 50°C for 2 hr. Ten-fold dilutions of cDNA were used as template for 30 cycles of PCR amplification with gene-specific primers. PCR products were fractionated on 1.5% agarose gels, stained with ethidium bromide, and photographed with a GE/Amersham ImageQuant ECL recorder. Primers used in the study are:
135.m00113 Sense (5'-CCGAATCTGCATTTCCAACT-3') and
135.m00113 Antisense (5'-CAATCCCTCCTCCAAGTGAA-3');
135.m00113 Sense (5'-TCTACTTGGAGGAGGGATTC-3') and
135.m00113 Antisense (5'-AATGAATTTGCATTGCATGG-3');
14.m00310 Sense (5'-GCCAGTTTCATTCCATGGTT-3') and
14.m00310 Antisense (5'-TCAGGACCACCAACATTTGA-3');
337.m00049 Sense (5'-TCAATGAATTGGTCGTTTGC-3') and
337.m00049 Antisense (5'-TCGTTTTGGTGTGAAATGTTG-3');
146.m00117 Sense (5'-CCCCATCCAAAATTGAACAG-3') and
146.m00117 Antisense (5'-GGATGGGGATTAGAAACCAAA-3');
223.m00071 Sense (5'-CCTAAACTTCAGCAAGTTCATTCA-3') and
223.m00071 Antisense (5'-GAAAGAAGTTGAGCCCAAAGCA-3');
1.m00712 Sense (5'-AACAATTGGTCAATGCTTCTCA-3') and
1.m00712 Antisense (5'-TCCCAAATGAACGAATAGGC-3');
223.m00075 Sense (5'-TGCAAAAATTAATAACCTTCTTCG-3') and
223.m00075 Antisense (5'-TCCACCAACAAAACCTGAAA-3');
77.m00173 Sense (5'-CAACATCTATTGGAAAAAGACCA-3') and
77.m00173 Antisense (5'-TGGAGATAACTCCTTCTCCATCA-3');
340.m00050 Sense (5'-CATCGAATATGATATTACATCAAATG-3') and
340.m00050 Antisense (5'-TTTATTGGAATTGGGTCAATAGCATTC-3');
247.m00075 Sense (5'-TGCAAAGTCCATTTCCAACA-3') and
247.m00075 Antisense (5'-TTTCAGGAGAAAAAGTGGCTTC-3');
7.m00480 Sense (5'-TGATTGCAAAAGATTCAGAAACA-3') and
7.m00480 Antisense (5'-ACTTGACCCAAAGTCATCACG-3');
13.m00291 Sense (5'-TGCTCAATGGCATCAATGTT-3') and
13.m00291 Antisense (5-'GCTTCCATTTGGGACGTAGA-3');
ssRNA Sense: (5'-ACGAACGAGACTGAAACCTAT-3') and
ssRNA Antisense: (5'-TGTTACGACTTCTCCTTCCTC-3').
Abbreviations
TSA: Trichostatin A; HDAC: histone deacetylases; RT-PCR: reverse transcriptase polymerase chain reaction; SCFA: short chain fatty acids; 5-AzaC: 5-Azacytidine; FDR: false discovery rate.
Authors' contributions
GME, US and DJE designed the experiments, GME and DJE performed the experiments, and GME, US, and DJE wrote the manuscript. All authors have read the final version of the manuscript and agree with its contents.
Supplementary Material
Genes differentially expressed in E. histolytica HM-1:IMSS, E. histolytica Rahman, and E. histolytica 200:NIH strains. (A) Genes differentially expressed in E. histolytica HM1:IMSS compared to E. histolytica Rahman and 200:NIH. The TIGR gene number, annotation, and fold-change are shown. Normalized expression data for E. histolytica HM-1:IMSS and Rahman strains grown in TYI-S-33 media were obtained from previously published literature [14] and subjected to local pooled error testing along with Benjamini-Hochberg multiple test correction. Genes that have higher expression in E. histolytica HM-1:IMSS than in 200:NIH or Rahman are listed under "up". Genes that have lower expression in E. histolytica HM-1:IMSS than in 200:NIH or Rahman are listed under "down". (B) Genes differentially expressed in E. histolytica Rahman compared to E. histolytica 200:NIH and HM-1:IMSS. The TIGR gene number, annotation, and fold-change are shown. Genes that have higher expression in E. histolytica Rahman than in HM-1:IMSS or 200:NIH are listed under "up". Genes that have lower expression in E. histolytica Rahman than in HM-1:IMSS or 200:NIH are listed under "down". (C) Genes differentially expressed in E. histolytica 200:NIH compared to E. histolytica Rahman and HM-1:IMSS. The TIGR gene number, annotation, and fold-change are shown. Genes that have higher expression in E. histolytica 200:NIH than in HM-1:IMSS or Rahman are listed under "up". Genes that have lower expression in E. histolytica 200:NIH than in HM-1:IMSS or Rahman are listed under "down".
Expression profiles of histone and chromatin modifying genes in E. histolytica strains HM-1:IMSS, 200:NIH, and Rahman. The mean trophozoite expression value for E. histolytica HM-1:IMSS, 200:NIH, and Rahman strains under standard culture conditions, GenBank ID number, and gene annotations are shown. Additional loci represented by crosshybridizing _s_at probe sets are as follows – 49.m00192_s_at: 334.m00048, 361.m00053 and 472.m00060; 100.m00145_s_at: 48.m00184; 4.m00618_s_at: 280.m00075; 9.m00405_s_at: 117.m00168 and 67.m00101; 39.m00254_s_at: 35.m00257; 444.m00042_s_at: 25.m00257. Genes that are statistically different in one strain compared to the other two are in bold.
Data for all arrays based on MIAME format. (A) Normalized array data for all genes. The Probe ID, TIGR gene number, GenBank accession, Annotation, fold-change, and FDR are shown. The array data for E. histolytica 200:NIH grown in TYI-S-33 media and E. histolytica 200:NIH exposed to low glucose (LG), Trichostatin A (TSA), and short chain fatty acids (SCFA) are listed. Normalized expression data for E. histolytica HM-1:IMSS and Rahman strains grown in TYI-S-33 media were obtained from previously published literature [14]. (B) Representation table for all _s_at probes.
Genes differentially regulated in E. histolytica 200:NIH by exposure to TSA media. (A) Genes with increased expression in TSA media. (B) Genes with decreased expression in TSA media. The Probe ID, TIGR gene number, GenBank accession, Annotation, fold-change, and FDR are shown. All array data are available at the GEO database website at NCBI (Accession number GSE8047).
Acknowledgments
Acknowledgements
We gratefully acknowledge all members of the Singh and Eichinger labs for helpful comments and suggestions, especially Jason Hackney for input on statistical analyses. GME was supported by NIH grant AI-068899. US was supported in part by NIH grants AI-053724 and AI-068899. DJE was supported by NIH grant AI-044893.
Contributor Information
Gretchen M Ehrenkaufer, Email: gehrenk@stanford.edu.
Daniel J Eichinger, Email: eichid01@med.nyu.edu.
Upinder Singh, Email: usingh@stanford.edu.
References
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Luo RX, Dean DC. Chromatin remodeling and transcriptional regulation. J Natl Cancer Inst. 1999;91:1288–1294. doi: 10.1093/jnci/91.15.1288. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Garcia BA, Hake SB, Diaz RL, Kauer M, Morris SA, Recht J, Shabanowitz J, Mishra N, Strahl BD, Allis CD, Hunt DF. Organismal differences in post-translational modifications in histones H3 and H4. J Biol Chem. 2007;282:7641–7655. doi: 10.1074/jbc.M607900200. [DOI] [PubMed] [Google Scholar]
- Earley K, Lawrence RJ, Pontes O, Reuther R, Enciso AJ, Silva M, Neves N, Gross M, Viegas W, Pikaard CS. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes Dev. 2006;20:1283–1293. doi: 10.1101/gad.1417706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daujat S, Bauer UM, Shah V, Turner B, Berger S, Kouzarides T. Crosstalk between CARM1 methylation and CBP acetylation on histone H3. Curr Biol. 2002;12:2090–2097. doi: 10.1016/S0960-9822(02)01387-8. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- WHO Entamoeba taxonomy. Bulletin of the World Health Organization. 1997;75:291–294. [PMC free article] [PubMed] [Google Scholar]
- Weber C, Guigon G, Bouchier C, Frangeul L, Moreira S, Sismeiro O, Gouyette C, Mirelman D, Coppee JY, Guillen N. Stress by heat shock induces massive down regulation of genes and allows differential allelic expression of the Gal/GalNAc lectin in Entamoeba histolytica. Eukaryot Cell. 2006;5:871–875. doi: 10.1128/EC.5.5.871-875.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbar MA, Chatterjee NS, Sen P, Debnath A, Pal A, Bera T, Das P. Genes induced by a high-oxygen environment in Entamoeba histolytica. Mol Biochem Parasitol. 2004;133:187–196. doi: 10.1016/j.molbiopara.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Ehrenkaufer GM, Haque R, Hackney JA, Eichinger DJ, Singh U. Identification of developmentally regulated genes in Entamoeba histolytica: insights into mechanisms of stage conversion in a protozoan parasite. Cell Microbiol. 2007 doi: 10.1111/j.1462-5822.2006.00882.x. [DOI] [PubMed] [Google Scholar]
- Singh U, Rogers JB, Mann BJ, Petri WA., Jr. Transcription initiation is controlled by three core promoter elements in the hgl5 gene of the protozoan parasite Entamoeba histolytica. Proc Natl Acad Sci U S A. 1997;94:8812–8817. doi: 10.1073/pnas.94.16.8812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali IK, Ehrenkaufer GM, Hackney JA, Singh U. Growth of the protozoan parasite Entamoeba histolytica in 5-azacytidine has limited effects on parasite gene expression. BMC Genomics. 2007;8:7. doi: 10.1186/1471-2164-8-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S, Fisher O, Lohia A, Ankri S. Entamoeba histolytica DNA methyltransferase (Ehmeth) is a nuclear matrix protein that binds EhMRS2, a DNA that includes a scaffold/matrix attachment region (S/MAR) Mol Biochem Parasitol. 2005;139:91–97. doi: 10.1016/j.molbiopara.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan G, Gilchrist CA, Musa H, Torok MS, Grant PA, Mann BJ, Petri WA., Jr. Histone acetyltransferases and deacetylase in Entamoeba histolytica. Mol Biochem Parasitol. 2004;138:205–216. doi: 10.1016/j.molbiopara.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Byers J, Eichinger D. Acetylation of the Entamoeba histone H4 N-terminal domain is influenced by short chain fatty acids that enter trophozoites in a pH-dependent manner. Submitted to IJP. 2007. [DOI] [PMC free article] [PubMed]
- Kiefer J, Beyer-Sehlmeyer G, Pool-Zobel BL. Mixtures of SCFA, composed according to physiologically available concentrations in the gut lumen, modulate histone acetylation in human HT29 colon cancer cells. Br J Nutr. 2006;96:803–810. doi: 10.1017/bjn20061948. [DOI] [PubMed] [Google Scholar]
- Blottiere HM, Buecher B, Galmiche JP, Cherbut C. Molecular analysis of the effect of short-chain fatty acids on intestinal cell proliferation. Proc Nutr Soc. 2003;62:101–106. doi: 10.1079/PNS2002215. [DOI] [PubMed] [Google Scholar]
- Hinnebusch BF, Meng S, Wu JT, Archer SY, Hodin RA. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J Nutr. 2002;132:1012–1017. doi: 10.1093/jn/132.5.1012. [DOI] [PubMed] [Google Scholar]
- Byers J, Faigle W, Eichinger D. Colonic short-chain fatty acids inhibit encystation of Entamoeba invadens. Cell Microbiol. 2005;7:269–279. doi: 10.1111/j.1462-5822.2004.00457.x. [DOI] [PubMed] [Google Scholar]
- Saksouk N, Bhatti MM, Kieffer S, Smith AT, Musset K, Garin J, Sullivan WJ, Jr., Cesbron-Delauw MF, Hakimi MA. Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Mol Cell Biol. 2005;25:10301–10314. doi: 10.1128/MCB.25.23.10301-10314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle JP, Saeij JP, Cleary MD, Boothroyd JC. Analysis of gene expression during development: lessons from the Apicomplexa. Microbes Infect. 2006;8:1623–1630. doi: 10.1016/j.micinf.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Freitas-Junior LH, Hernandez-Rivas R, Ralph SA, Montiel-Condado D, Ruvalcaba-Salazar OK, Rojas-Meza AP, Mancio-Silva L, Leal-Silvestre RJ, Gontijo AM, Shorte S, Scherf A. Telomeric heterochromatin propagation and histone acetylation control mutually exclusive expression of antigenic variation genes in malaria parasites. Cell. 2005;121:25–36. doi: 10.1016/j.cell.2005.01.037. [DOI] [PubMed] [Google Scholar]
- Gilchrist CA, Houpt E, Trapaidze N, Fei Z, Crasta O, Asgharpour A, Evans C, Martino-Catt S, Baba DJ, Stroup S, Hamano S, Ehrenkaufer G, Okada M, Singh U, Nozaki T, Mann BJ, Petri WA., Jr. Impact of intestinal colonization and invasion on the Entamoeba histolytica transcriptome. Mol Biochem Parasitol. 2006;147:163–176. doi: 10.1016/j.molbiopara.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Field J, Van Dellen K, Ghosh SK, Samuelson J. Responses of Entamoeba invadens to heat shock and encystation are related. J Eukaryot Microbiol. 2000;47:511–514. doi: 10.1111/j.1550-7408.2000.tb00083.x. [DOI] [PubMed] [Google Scholar]
- Sakata T, von Engelhardt W. Stimulatory effect of short chain fatty acids on the epithelial cell proliferation in rat large intestine. Comp Biochem Physiol A. 1983;74:459–462. doi: 10.1016/0300-9629(83)90631-X. [DOI] [PubMed] [Google Scholar]
- Fusunyan RD, Quinn JJ, Fujimoto M, MacDermott RP, Sanderson IR. Butyrate switches the pattern of chemokine secretion by intestinal epithelial cells through histone acetylation. Mol Med. 1999;5:631–640. [PMC free article] [PubMed] [Google Scholar]
- Drozdowski LA, Dixon WT, McBurney MI, Thomson AB. Short-chain fatty acids and total parenteral nutrition affect intestinal gene expression. JPEN J Parenter Enteral Nutr. 2002;26:145–150. doi: 10.1177/0148607102026003145. [DOI] [PubMed] [Google Scholar]
- Sanderson IR. Short chain fatty acid regulation of signaling genes expressed by the intestinal epithelium. J Nutr. 2004;134:2450S–2454S. doi: 10.1093/jn/134.9.2450S. [DOI] [PubMed] [Google Scholar]
- Chang S, Pikaard CS. Transcript profiling in Arabidopsis reveals complex responses to global inhibition of DNA methylation and histone deacetylation. J Biol Chem. 2005;280:796–804. doi: 10.1074/jbc.M409053200. [DOI] [PubMed] [Google Scholar]
- Ou JN, Torrisani J, Unterberger A, Provencal N, Shikimi K, Karimi M, Ekstrom TJ, Szyf M. Histone deacetylase inhibitor Trichostatin A induces global and gene-specific DNA demethylation in human cancer cell lines. Biochem Pharmacol. 2007 doi: 10.1016/j.bcp.2006.12.032. [DOI] [PubMed] [Google Scholar]
- Diamond LS, Harlow DR, Cunnick CC. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans R Soc Trop Med Hyg. 1978;72:431–432. doi: 10.1016/0035-9203(78)90144-X. [DOI] [PubMed] [Google Scholar]
- Ali IK, Zaki M, Clark CG. Use of PCR amplification of tRNA gene-linked short tandem repeats for genotyping Entamoeba histolytica. J Clin Microbiol. 2005;43:5842–5847. doi: 10.1128/JCM.43.12.5842-5847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghighi A, Kobayashi S, Takeuchi T, Thammapalerd N, Nozaki T. Geographic diversity among genotypes of Entamoeba histolytica field isolates. J Clin Microbiol. 2003;41:3748–3756. doi: 10.1128/JCM.41.8.3748-3756.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CMGM Stanford Pan facility http://cmgm.stanford.edu/pan/
- Affymetrix P. Affymetrix protocols http://www.affymetrix.com/support/technical/manual/expression_manual.affx
- GCOS GCOS manual http://www.affymetrix.com/products/software/specific/gcos.affx
- Bioconductor Bioconductor http://www.bioconductor.org
- Wu Z, Irizarry R, Gentleman R, Murillo F, F S. A model based background adjustment for oligonucleotide expression arrays. Technical report; John Hopkins University, Dept of Biostatistics Working Papers. 2004.
- Jain N, Thatte J, Braciale T, Ley K, O'Connell M, Lee JK. Local-pooled-error test for identifying differentially expressed genes with a small number of replicated microarrays. Bioinformatics. 2003;19:1945–1951. doi: 10.1093/bioinformatics/btg264. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Genespring Genespring http://www.chem.agilent.com/scripts/pds.asp?lpage=27881
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genes differentially expressed in E. histolytica HM-1:IMSS, E. histolytica Rahman, and E. histolytica 200:NIH strains. (A) Genes differentially expressed in E. histolytica HM1:IMSS compared to E. histolytica Rahman and 200:NIH. The TIGR gene number, annotation, and fold-change are shown. Normalized expression data for E. histolytica HM-1:IMSS and Rahman strains grown in TYI-S-33 media were obtained from previously published literature [14] and subjected to local pooled error testing along with Benjamini-Hochberg multiple test correction. Genes that have higher expression in E. histolytica HM-1:IMSS than in 200:NIH or Rahman are listed under "up". Genes that have lower expression in E. histolytica HM-1:IMSS than in 200:NIH or Rahman are listed under "down". (B) Genes differentially expressed in E. histolytica Rahman compared to E. histolytica 200:NIH and HM-1:IMSS. The TIGR gene number, annotation, and fold-change are shown. Genes that have higher expression in E. histolytica Rahman than in HM-1:IMSS or 200:NIH are listed under "up". Genes that have lower expression in E. histolytica Rahman than in HM-1:IMSS or 200:NIH are listed under "down". (C) Genes differentially expressed in E. histolytica 200:NIH compared to E. histolytica Rahman and HM-1:IMSS. The TIGR gene number, annotation, and fold-change are shown. Genes that have higher expression in E. histolytica 200:NIH than in HM-1:IMSS or Rahman are listed under "up". Genes that have lower expression in E. histolytica 200:NIH than in HM-1:IMSS or Rahman are listed under "down".
Expression profiles of histone and chromatin modifying genes in E. histolytica strains HM-1:IMSS, 200:NIH, and Rahman. The mean trophozoite expression value for E. histolytica HM-1:IMSS, 200:NIH, and Rahman strains under standard culture conditions, GenBank ID number, and gene annotations are shown. Additional loci represented by crosshybridizing _s_at probe sets are as follows – 49.m00192_s_at: 334.m00048, 361.m00053 and 472.m00060; 100.m00145_s_at: 48.m00184; 4.m00618_s_at: 280.m00075; 9.m00405_s_at: 117.m00168 and 67.m00101; 39.m00254_s_at: 35.m00257; 444.m00042_s_at: 25.m00257. Genes that are statistically different in one strain compared to the other two are in bold.
Data for all arrays based on MIAME format. (A) Normalized array data for all genes. The Probe ID, TIGR gene number, GenBank accession, Annotation, fold-change, and FDR are shown. The array data for E. histolytica 200:NIH grown in TYI-S-33 media and E. histolytica 200:NIH exposed to low glucose (LG), Trichostatin A (TSA), and short chain fatty acids (SCFA) are listed. Normalized expression data for E. histolytica HM-1:IMSS and Rahman strains grown in TYI-S-33 media were obtained from previously published literature [14]. (B) Representation table for all _s_at probes.
Genes differentially regulated in E. histolytica 200:NIH by exposure to TSA media. (A) Genes with increased expression in TSA media. (B) Genes with decreased expression in TSA media. The Probe ID, TIGR gene number, GenBank accession, Annotation, fold-change, and FDR are shown. All array data are available at the GEO database website at NCBI (Accession number GSE8047).


