Abstract
An application that provides a flexible and easy to use interface to the GAMMA spectral simulation package is described that is targeted at investigations using in vivo MR spectroscopic methods. The program makes available a number of widely used spatially-localized MRS pulse sequences and NMR parameters for commonly-observed tissue metabolites, enabling spectra to be simulated for any pulse sequence parameter and viewed in an integrated display. The application is interfaced with a database for storage of all simulation parameters and results of the simulations. This application provides a convenient method for generating a priori spectral information used in parametric spectral analyses and for visual examination of the effects of difference pulse sequences and parameter settings.
Keywords: MR spectroscopy, spectral simulation, parametric spectral analysis, GAMMA, software
1. Introduction
Acquisition and analysis methods for biomedical applications of magnetic resonance spectroscopy (MRS) have changed radically in the past decade with the development of improved spatial localization techniques; improved MR instrumentation suitable for studies in animals and humans; and sophisticated spectral analysis methods that utilize the increased computational capabilities that have become available. For in vivo measurements, MR spectroscopic data is complicated by the inherent overlap and complex shapes of the signals of interest. Methods to improve detection of specific spectral contributions include spectral editing [4,18,28,33,46-48]; spectral fitting using constrained parametric model optimization and incorporating a priori metabolic information [6,7,23,24,31,35,40,45,50]; and use of multidimensional spectral acquisition methods [42-44]. In all of these areas of research, spectral simulation methods are becoming an increasingly important tool.
While simulation techniques have been used in scientific studies for decades, only the recent advent of open source tools and increases in desktop computational power have made these techniques available to individual investigators. Introduced in 1994 [36], the GAMMA C++ library is a flexible and efficient tool for describing and simulating MR experiments. Since the early 1990s, there have been reports of a number of other NMR simulation libraries [5,16,22] and application tools [1,2,15,17,27,29,32,49] which have sought to provide similar functionality in either a more optimized or user friendly fashion. The growth in available tools reflects the consensus among MR scientists that simulation methods are a necessary and desirable step in the development of increasingly sophisticated data acquisition/analysis methods for viewing complex molecular systems. Despite the increase in available tools, GAMMA has maintained a strong presence in the field due to its flexibility, performance, straightforward elegant design and standard C++ implementation. There have been numerous reports of spectral simulations that have made use of the GAMMA library [3,8-14,19,21,22,25,26,30,34,37-39,41,50,51]. However, use of the GAMMA library also requires familiarity with C++ programming and it provides only rudimentary tools for the display, storage and analysis of results. In this report we describe an application that provides a coherent development framework and convenient graphical interface to pre-compiled modules developed using GAMMA, which is termed GAVA (GAmma Visual Analysis).
The primary goal for GAVA was to shorten times for pulse sequence development and sequence parameter optimization for observation of specific metabolite structures, and to provide a method for generating and visualizing basis functions used in a parametric spectral analysis procedure [40,52]. Secondary goals included providing advanced users with source code examples of how to create MR simulations both with and without pulsed gradients, providing non-technical users with access to libraries of pre-compiled MR sequences for creating their own simulations, and creating generalized access to a database of previous simulation results, all accessed through straightforward graphical user interfaces (GUI). Finally, GAVA was designed to be extensible, cross-platform compatible, and freely available.
2. Methods
2.1. Program Structure
The GAVA program is comprised of three core program elements: 1) an IDL-based GUI front end, 2) MR simulations based on the GAMMA C++ library, and 3) a MySQL-based relational database for persistent storage of inputs and results. These elements were chosen based on ease-of-use, cross-platform interconnectivity, maturity of development and extensive documentation. A general flow diagram of the interconnectivity of the GAVA core elements is shown in Figure 1.
Figure 1.

Flow diagram for GAVA environment interconnectivity of IDL, GAMMA and MySQL.
IDL (ITT Visual Information Solutions) is a fourth generation programming language and development environment that is easy to use and includes a variety of well integrated data analysis, plotting and GUI development functionality. As an interpreted language, it lends itself to rapid code development, recompilation and debugging. IDL provides access to C++ shared libraries using a CALL_EXTERNAL mechanism that passes parameters via the argv/argc formalism. GAMMA [36] is an object-oriented library written in C++ that facilitates software development for simulation of MR experiments. It provides C++ objects that encapsulate the operations common to MR experiments. This allows abstract entities such as density matrices, operators, commutators, spin systems, and ideal and real RF pulses to be easily described and applied in program procedures. MySQL is a robust relational database [http://dev.mysql.com/downloads] that was incorporated into the GAVA project as a persistent data storage mechanism for information relative to the GAVA program, including: MR simulation inputs, MR pulse sequence storage and simulation results storage for future access. MySQL was integrated into GAVA using the C++ API (application programming interface) in both IDL and the GAMMA simulations.
The control and data flow logic of the GAVA environment is contained in the IDL GUI front end. The central data construct is referred to as an Experiment and is composed of one or more Simulations. Any single Simulation is composed of a set of one metabolite and one pulse sequence timing. Thus, an Experiment with five metabolites and eight variations in pulse sequence timings would contain 40 Simulations. Only one Experiment at a time can be loaded into GAVA, either from results stored in the database or from a new spectral simulation. However, multiple GAVA sessions can be run to analyze multiple data sets from the same Experiment or compare different Experiments for results stored in the database. GAVA could also be used effectively in a distributed computing environment with all instances connected to the same MySQL GAVA database.
The loaded Experiment can be acted upon by various GAVA modules, each with its own GUI, including: the ‘Experiment Browser’, ‘New Experiment’, ‘Add To Experiment’, Plot and Utility modules. Details for module features are listed below. MySQL database access for Experiment results is transparent to the user and a variety of graphical and text output functions are available in each module. There are twelve MR pulse sequence simulations provided with the standard GAVA installation as well as the functionality to develop and share stand-alone user-contributed pulse sequence libraries to create new Experiments. Non-technical users can implement the standard GAVA installation and create, store and analyze spectral simulations from the MR sequence library without any expertise in IDL, C++ or SQL programming. Users with C++ programming experience can add their own simulation functionality to the GAVA environment by modifying only the GAMMA code in the provided C++ templates, as described below.
To facilitate cross-platform compatibility and program extensibility, platform dependent code was kept to a minimum and based where possible on templates. The main GAVA C++ shared library contains only five basic MR pulse sequences and two procedures that provide database connectivity between IDL and MySQL. The MR sequence list inside GAVA is extended by importing stand-alone pulse sequence libraries. This extension mechanism is based on using a C++ pulse sequence procedure template and making changes only to the GAMMA object calls used to describe the MR simulation. The template passes in 24 standard control and I/O variables from the GAVA environment and up to 30 additional user-defined variables. These variables can be utilized at the user’s discretion to control simulation setup, metabolite initialization, ongoing timing changes (via two GAVA incremented loop variables), density matrix post-processing and the output of results back to the GAVA environment. Common uses of the user-defined variables might be to loop over effective field strengths to simulate the effects of gradients or as a list of RF pulse files to analyze the effects of various real pulses on metabolites of interest. One or more template procedures (a.k.a. pulse sequences) can be compiled into a shared library that can subsequently be imported into the GAVA environment.
2.2. Experiment Explorer Module
The Experiment Explorer module is the initial GUI that appears when GAVA is started up (Figure 2). From this module, the user can run a new Experiment or load results from Experiments stored in the database. All other GAVA modules can be launched from the Explorer, but some require that an Experiment be loaded first. Experiments are listed and differentiated in the Explorer by a short name (40 characters) and a longer comment string. The comment string is updated as each name is selected. Experiments can be sorted and listed based on the nuclei involved and the field strength for which they were run by selecting values from the relevant pull-down menus. As the user selects an Experiment name, the relevant data is loaded from the database. Only the last user-selected result is loaded into memory, and running a new Experiment will supersede the last loaded result. Also listed on the browser are the metabolites included in the selected Experiment.
Figure 2.

GAVA Experiment Browser widget, the first widget displayed upon startup, it provides access to new/saved results, utilities and visualization and analysis widgets.
Action buttons along the left of the Browser launch GAVA widgets for running new Experiments, plotting results and viewing a text listing of the loaded Experiment. Utility modules and ASCII output options are accessed from the menu bar. Utilities include a module to manage database content, and a module to set global values for simulation binning and normalization. These and the network TCP/IP module are discussed in the following section.
A network connection module allows the host address, database name, login and password to be set for connection to a GAVA MySQL database. These values can be reset at any time, allowing users access to data on either local or networked GAVA databases. Experiments loaded from one location can subsequently be saved to a different database. This is the most direct form of collaboration, although other forms of data import/export exist to accommodate various network security issues.
2.3. New/AddTo Experiment Module (Spectral Simulations)
The New Experiment module (Figure 3) launches a widget on which to enter all information necessary to run and store the results from an Experiment. Users select the pulse sequence, nucleus, field strength and one or more metabolites to be simulated from drop-down widget lists. The investigator’s name, a short Experiment name and a longer Experiment comment are typed into text widgets. Up to two loop variables can be set for a simulation. Each loop variable has a start value, a step value and a “number of steps” entry. Depending on the sequence selected, the loop widgets reconfigure their labels to provide useful information about the values that should be entered in each field, including the loop variable name and unit type. Any additional, fixed variables that need to be passed to the simulation are set up in a scrolled list in an optional third column of the widget. The Experiment is not actually performed until the Run Simulation button is pressed, and upon finishing, the results can either be sent to the Plot Widget for viewing, or submitted to the database for storage. Another more immediate output option is for GAVA to dump the results from the final transition table, consisting of frequencies, amplitudes and phases for all resonance lines for all metabolites and sequence timings, to a tab delineated text file for external processing.
Figure 3.

GAVA New Experiment widget, allows users to define and run spectral simulations.
An Experiment can consist of up to three dimensions depending on the metabolites selected and settings for the two loop variables. Loop variable usage depends on the pulse sequence timings or other parameter variations inherent to GAMMA simulation procedures. For example, an ideal pulse FID simulation has no sequence timings and creates only a 1D result (for the metabolites selected) while an ideal STEAM simulation gives a 3D result based on the metabolites selected and the TE and TM pulse sequence timings that are linked to the loop variables. However, loop variables are not limited to use as sequence timings. The raw loop variables are passed in the standard list of parameters to each simulation and can be used to iterate over any variable set up by the programmer (e.g. pulse train length, RF file lists, etc.).
The basic GAVA installation includes five predefined pulse sequences packaged in a single library and seven stand-alone libraries. The five basic sequences make use of ideal RF pulses (defined in GAMMA) in their simulations, including: one-pulse, spin-echo, STEAM, PRESS and J-PRESS sequences. The seven stand-alone libraries demonstrate a variety of extensions to the standard PRESS pulse sequence. Two are examples of how to add a Carr-Purcell pulse train, and loop over either sequence timings or the number of pulses in the train. One generates basis functions for TE-averaged PRESS acquisitions [20], and four investigate corrections for PRESS acquisitions due to chemical shift artifacts caused by real RF localization pulses by including the effects of spatial variations of the B1 profiles and a full simulation using complete user-defined time-domain RF pulse shapes [25]. The source code for all seven libraries serves as examples for how to create and integrate additional pulse sequences into the GAVA environment.
2.4. Plot Widget Module
The Plot module is used to display and analyze the currently loaded Experiment. The majority of spectral simulations are run to either maximize or minimize the signal coherence of one or more metabolites and/or resonance groups based on variations in pulse sequence timings. The tools used most often to determine signal areas are plots of individual metabolites or integral plots over specified PPM spectral ranges. The four main windows for the Plot widget (Figure 4) are the Plot Controls widget and three display windows used to visualize 1) 1D and stack plots, 2) regional integral plots and 3) integral contour plots which also include a grayscale image display option. All three displays can be independently turned on or off, resized and controlled by settings located on the Control Widget.
Figure 4.

GAVA Plot widget display with the 1D/Stack Plot, Integral Plot and Contour Plot windows open. The 1D/Stack Plot display is in 1D mode, showing results for both glutamate and myo-inositol for a single simulation timing.
The 1D and Stack Plot window displays one or more metabolite spectra plotted for specific Index1 and Index2 values, which are linked to the Experiment loop variables, or it can show a stack plot of spectra for one metabolite along either the Index1 or Index2 range (Figure 5). The horizontal axis is always displayed in field-independent PPM values, and can be zoomed in and out along any sub-section using the mouse. A left and right cursor can also be set using the mouse. The digital resolution, and hence display quality and analysis accuracy, of the displayed spectra and plots is controlled by the user-defined linewidth, sweep width and spectral points widget entries. Both Gaussian and Lorentzian line shapes are supported and real, imaginary, magnitude and overlaid complex plots can be displayed.
Figure 5.

GAVA Plot widget display in Stack Plot mode, showing results for glutamate at 20 simulated TE timings in a STEAM simulation at 1.5 Tesla.
The displays in both the Integral Plot and Contour Plot base their range of integral calculations on the cursor locations set in the 1D Plot display and the first metabolite selected from the display list. Changes to the cursor locations trigger automatic updates in both displays. Integral Plot controls allow the user to select for integration along either Index1 or Index2 dimension. Contour Plot controls include the number of steps that are used to delineate the iso-lines in the plot and also the ability to flip between contour plots and grayscale images that reflect the integral values. All display window contents can be output to a variety of graphical file formats including: CGM, TIFF, JPEG and encapsulated postscript. Alternatively, displays can be sent directly to an attached printer or output to a tab-delineated text file of plot values.
2.5. Utilities Module
The Utilities module provides a full set of widgets for managing metabolite parameters, Experiments and pulse sequences. Metabolite chemical shift and J-coupling descriptions can be deleted, added, viewed and edited through a widget interface or imported from a file in GAMMA ‘sysfile’ format. Experiments can be deleted or accessed for a textual description of the simulation settings used to create it. Pulse sequences can be deleted or new ones added through a widget interface that collects the information necessary to call the shared library/function and to properly label the widget entries in the New Experiment module. An Import/Export scheme exists where all the relevant settings are written to an SQL script file to facilitate pulse-sequence library collaborations.
The Global SimParams widget is also part of the Utilities module, and plays an important role in running new Experiments. Spectral simulation results need to be binned to merge the values in the final transition table into a manageable number of spectral lines and normalized to ensure that results are comparable between pulse sequence timings and the ranges of metabolites being simulated. This is accomplished using a single reference proton at a spectral location distant from all other resonance groups. The GAVA environment passes in parameters for locating the reference peak and binning lines via the standard variables passed to the pulse sequence C++ template. However it is up to the user to include the reference proton in the metabolite description, and ensure that the Global SimParams reflect those settings. This setup provides the flexibility to process a wide variety of nuclei, RF pulses, J-coupling and chemical shifts in any simulation library.
3. Results
With the release of the GAVA environment, we have achieved our initial development goal of providing a GUI based control and display program that interfaces the functionality of the GAMMA library with the powerful query and storage functions of the MySQL database. Research applications for this tool have included: 1) Simulating multiple metabolites for a single TE and pulse sequence for use as basis functions in spectral analysis tools [39,50]. 2) Simulating one metabolite for multiple, progressive sequence timings to determine optimal pulse sequence settings for a clinical MR spectroscopic data acquisition. 3) Simulation of high field NMR metabolite basis groups for dynamically labeled multi-nuclear metabolomics probes. 4) Evaluation of chemical shift artifacts for clinical MR spectroscopic data acquisitions using non-ideal RF pulses [25]. All of these applications were explored using only the built-in pulse sequence simulations. To date, a variety of specific add-on pulse sequences simulations exist and more are under development in response to a growing number of research questions addressable using spectral simulation.
Two examples of spectral development and analysis are shown in Figures 6 and 7. The results shown in both figures were created using multiple CGM outputs of the 1D/Stack, Integral and Contour windows for the various metabolites. Results from a 1H PRESS sequence simulation at 3T that depict the effects of TE averaging for four overlapping metabolites are shown in Figure 6. The four stack plots display metabolite peak variations for 30 different TE values due to J-coupling. Region integral plots on the right show how the signals from the various multiplet resonance groups disperse or refocus at increasing TE. The TE averaged spectra at bottom left, show the effects of summing the 30 spectra. Metabolite peaks for GABA, glutamine, glutamate and myo-inositol resolve into simpler spectral patterns with less overlap.
Figure 6.
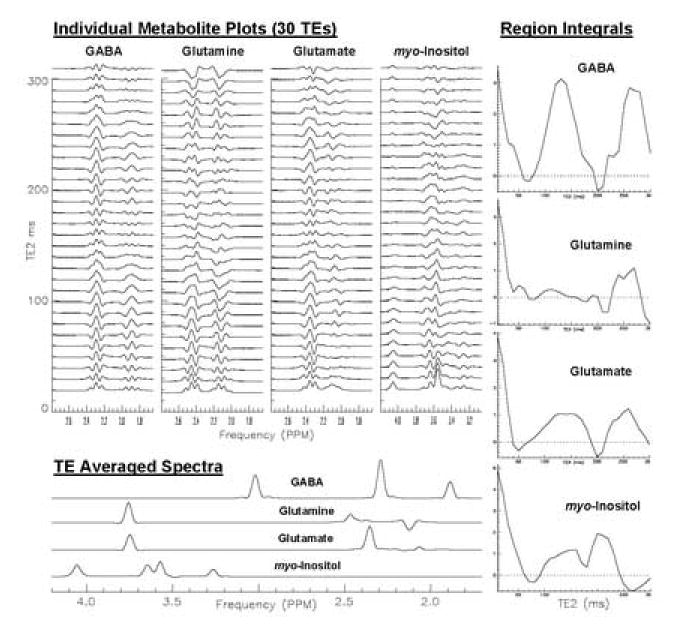
Results from a PRESS sequence simulation that shows the effects of TE averaging of 3T PRESS spectra for four overlapping metabolites. The four stack plots (top left) show metabolite peak variations for 30 different TE values due to J-coupling. Region integral plots (right) show how the signals from the various multiplet resonance groups refocus at each TE. The TE averaged spectra (bottom left), show the effects of summing the 30 spectra for each metabolite. Metabolite peaks for GABA, glutamine, glutamate and myo-inositol resolve into simpler spectral patterns with less overlap when TE averaging is applied.
Figure 7.

Results for lactate from a PRESS sequence simulation at 1.5T using real RF pulses with Gaussian weighted Sinc pulse envelopes. This shows the effects of chemical shift across the spatial dimension for the doublet and quartet peaks at 1.3 and 4.17 ppm. Resonance groups at different chemical shifts are rotated by varying flip angles based on the bandwidth of the RF pulses applied. Parameters: TE=288ms, 30x30 spatial locations simulated across a FOV=30mm for RF pulses and applied gradients with an effective bandwidth of 20mm. Stack plots show peak variations in SpatDim1 for SpatDim2 fixed on center. The top and bottom contour/grayscale images show the variation in regional areas of the quartet and doublet peak areas, respectively, for all spatial locations in the PRESS voxel.
In Figure 7 are shown the results for lactate from a PRESS sequence simulation at 1.5T using real RF pulses with Gaussian weighted Sinc envelopes with a FWHM width of 1399 Hz and a pulse length of 1.7ms. RF pulses were applied at the resonance frequency (4.7 ppm). This shows the effects of the different chemical shift values for the doublet and quartet resonance groups, 1.3 and 4.17 ppm respectively, across the two spatial dimensions in the simulation. Resonance groups at different chemical shifts are rotated by varying flip angles based on the bandwidth and shape of the RF pulses applied. Simulation parameters were: TE=288ms, 30x30 spatial locations simulated across a FOV=30mm for RF pulses and applied gradients with effective bandwidths of 20mm. The stack plots show peak variations in the SpatDim1 (spatial dimension 1) direction for the SpatDim2 (spatial dimension 2) location fixed at the center of the FOV. The top and bottom contour/grayscale images show the variation in regional areas of the quartet and doublet peak areas, respectively, for all spatial locations in the PRESS voxel. Note that spatially, at the edges of the RF pulse bandwidth where the effective flip angles of the refocusing pulses are much lower than 180°, metabolite areas show a decreasing spread of areas, indicated by the darkening grayscale and closely spaced contour lines, respectively. Also, note that the chemical shift effects for lactate are more noticeably displayed by the blackened regions in the grayscale images than by the contour lines as the metabolite signals not affected by both refocusing pulses end up out of phase with other spatial regions.
4. Discussion
The GAVA program is available on request from the authors at no cost. The end-user download includes pre-compiled GAVA, GAMMA and MySQL binaries for Windows and source code and Makefiles only for Linux (x86), and is distributed for use with the IDL Virtual Machine. Extensions to GAVA developed by outside users can be redistributed freely, and a number of pulse sequence add-on libraries have already been added to the standard distribution. For users that wish to contribute to the development of the GAVA environment, a development download is also available. It contains the GAVA source code for the main library, source code for selected pulse sequence add-on libraries and the GAMMA and MySQL installations used in the current GAVA release. A full IDL development license is necessary to extend the GAVA GUI code, and a C++ compiler is required to create standalone pulse sequence libraries. Suggestions for compiling the GAVA main library on platforms other than Windows and Linux (x86) are available in the development appendix of the user manual.
A short term and simple extension of the functionality of the current application will be the simulation and graphical representation of two dimension NMR experiments. The current infrastructure allows for one or both of the looping variables to account for the sequence timings. In addition, the graphical representation of the results is straightforward in terms of stack plots and 3D contour plots. All that is necessary is the addition of a means for the user to identify the pulse sequence as a 2D sequence which could then be used to optionally apply a 2D Fourier transform to simulation results.
Finally, the performance of much of the functionality provided by GAVA is inherently amenable to speedup via straightforward parallelization. A number of standard multi-processor and distributed solutions could be applied to the GAVA architecture to take advantage of the greatly increased performance such systems offer.
Acknowledgments
The authors would like to thank Dr. Govindaraju Varanavasi for his extensive beta testing of the development code. This work was supported by PHS grants R01EB00207 and R01EB000822.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Allman T, Bain A, Garbow J. SIMPLTN, a Program for the Simulation of Pulse NMR Spectra. J Magn Reson. 1996;A123(1):26–31. doi: 10.1006/jmra.1996.0210. [DOI] [PubMed] [Google Scholar]
- 2.Bak M, Rasmusson JT, Nielsen NC. SIMPSON: a general simulation program for solid-state NMR spectroscopy. J Magn Reson. 2000;147(2):296–330. doi: 10.1006/jmre.2000.2179. [DOI] [PubMed] [Google Scholar]
- 3.Baldus M, Petkova AT, Herzfeld J, Griffin RG. Cross-polarization in the tilted frame - Assignment and spectral simplification in heteronuclear spin systems. Mol Phys. 1998;95:1197–1207. [Google Scholar]
- 4.Bax A, De Jong PG, Mehlkopf AF, Smidt J. Separation of the different orders of NMR multiple-quantum transitions by the use of pulsed field gradients. Chem Phys Lett. 1980;69:567–570. [Google Scholar]
- 5.Blanton WB. BlochLib: a fast NMR C++ tool kit. J Magn Reson. 2003;162(2):269–283. doi: 10.1016/s1090-7807(03)00035-1. [DOI] [PubMed] [Google Scholar]
- 6.Choi C, Coupland NJ, Hanstock CC, Ogilvie CJ, Higgins AC, Gheorghiu D, Allen PS. Brain gamma-aminobutyric acid measurement by proton double-quantum filtering with selective J rewinding. Magn Reson Med. 2005;54:272–279. doi: 10.1002/mrm.20563. [DOI] [PubMed] [Google Scholar]
- 7.De Graaf AA, Bovee WMMJ. Improved quantification of in vivo 1H NMR spectra by optimization of signal acquisition and processing and by incorporation of prior knowledge into the spectral fitting. Magn Reson Med. 1990;15:305–319. doi: 10.1002/mrm.1910150212. [DOI] [PubMed] [Google Scholar]
- 8.Devaux PF, Hoatson GL, Favre E, Fellmann P, Farren B, McKay AL, Bloom M. Interaction of cytochrome c with mixed dimyristoylphosphatidylcholine-dimiristoylphosphatidylserine bilayers: a deuterium nuclearXmagnetic resonance study. Biochemistry. 1986;25:3804–3812. doi: 10.1021/bi00361a011. [DOI] [PubMed] [Google Scholar]
- 9.Dusold S, Milius W, Sebald A. Iterative lineshape fitting of MAS NMR spectra: A tool to investigate homonuclearJ-coupling in isolated spin pairs. J Magn Reson. 1998;135(2):500–513. doi: 10.1006/jmre.1998.1603. [DOI] [PubMed] [Google Scholar]
- 10.Ebel A, Soher BJ, Maudsley AA. Assessment of 3D 1H NMR echo-planar spectroscopic imaging using automated spectral analysis. Magn Reson Med. 2001;46:1072–1078. doi: 10.1002/mrm.1301. [DOI] [PubMed] [Google Scholar]
- 11.Eykyn TR, Ghose R, Bodenhausen G. Offset profiles of selective pulses in isotopically labeled macromolecules. J Magn Reson. 1999;136:211–213. doi: 10.1006/jmre.1998.1623. [DOI] [PubMed] [Google Scholar]
- 12.Filip C, Hafner S, Schnell I, Demco DE, Spiess HW. Solid-state nuclear-magneticresonance spectra of dipolar-coupled multi-spin systems under fast magic-angle-spinning. J Chem Phys. 1999;110:423–440. [Google Scholar]
- 13.Govindaraju V, Gauger G, Manley G, Ebel A, Meeker M, Maudsley AA. Volumetric proton spectroscopic imaging of mild traumatic brain injury. AJNR. 2004;25:730–737. [PMC free article] [PubMed] [Google Scholar]
- 14.Govindaraju V, Meyerhoff DJ, Maudsley AA, Vermathen M, Weiner MW. Effects of brain membranes on 1H nuclear magnetic resonance signal intensity of ethanol in vitro. Alcohol and Alcoholism. 1997;32:671–681. doi: 10.1093/oxfordjournals.alcalc.a008317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graveron-Demilly D, Diop A, Briguet A, Fenet B. Product-operator algebra for strongly coupled spin systems. Journal of Magnetic Resonance. 1993;101:233–239. [Google Scholar]
- 16.Guntert P, Schaefer N, Otting G, Wuthrich K. POMA: A complete mathematica implementation of the NMR product-operator formalism. J Magn Reson. 1993;(A101):103–105. [Google Scholar]
- 17.Helgstrand M, Allard P. QSim, a program for NMR simulations. J Biolmol NMR. 2004;30:71–80. doi: 10.1023/B:JNMR.0000042962.00088.f4. [DOI] [PubMed] [Google Scholar]
- 18.Hennig J, Thiel T, Speck O. Improved sensitivity to overlapping multiplet signals in in vivo proton spectroscopy using a multiecho volume selective (CPRESS) experiment. Magn Reson Med. 1997;37:816–820. doi: 10.1002/mrm.1910370603. [DOI] [PubMed] [Google Scholar]
- 19.Hughes E, Brouwer EB, Harris RK. 19F solid-state NMR magic-angle-turning experiments using multiple-pulse homonuclear decoupling. J Magn Reson. 1999;138:256–267. doi: 10.1006/jmre.1999.1740. [DOI] [PubMed] [Google Scholar]
- 20.Hurd R, Sailasuta N, Srinivasan R, Vigneron DB, Pelletier D, Nelson SJ. Measurement of brain glutamate using TE-averaged PRESS at 3T. Magn Reson Med. 2004;51(3):435–440. doi: 10.1002/mrm.20007. [DOI] [PubMed] [Google Scholar]
- 21.Kaikkonen A, Ylinen EE, Punkkinen M. Multiple-quantum coherences of molecular groups - 19F in Cf3Cooag. Appl Magn Reson. 1998;15:509–518. [Google Scholar]
- 22.Kanters RPF, Char BW, Addison AW. A computer-algebra application for the description of NMR experiments using the product-operator formalism. J Magn Reson. 1993;A 101:23–29. [Google Scholar]
- 23.Kim H, Thompson RB, Hanstock CC, Allen PS. Variability of metabolite yield using STEAM or PRESS sequences in vivo at 3.0 T, illustrated with myo-inositol. Magn Reson Med. 2005;53(4):760–769. doi: 10.1002/mrm.20434. [DOI] [PubMed] [Google Scholar]
- 24.Kim H, Wild JM, Allen PS. Strategy for the spectral filtering of myo-inositol and other strongly coupled spins. Magn Reson Med. 2004;51(2):263–272. doi: 10.1002/mrm.10697. [DOI] [PubMed] [Google Scholar]
- 25.Maudsley A, Govindaraju V, Young K, Aygula Z, Pattany PM, Soher BJ, Matson G. Numerical simulation of PRESS localized MR spectroscopy. J Magn Reson. 2005;173(1):54–63. doi: 10.1016/j.jmr.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 26.McClung RED. Coherence transfer pathways and phase cycles - The decoding of a pulse sequence. Concepts Magn Reson. 1999;11:1–28. [Google Scholar]
- 27.Meresi GH, Cuperlovic M, Palke WE, Gerig JT. Pulsed Field Gradients in Simulations of One- and Two-Dimensional NMR Spectra. J Magn Reson. 1999;137(1):186–195. doi: 10.1006/jmre.1998.1665. [DOI] [PubMed] [Google Scholar]
- 28.Mescher M, Merkle H, Kirsch J, Garwood M, Gruetter R. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. 1998;11:266–272. doi: 10.1002/(sici)1099-1492(199810)11:6<266::aid-nbm530>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 29.Naressi A, Couturier C, Devos JM, Janssen M, Mangeat C, de Beer R, Graveron-Demilly D. Java-based graphical user interface for the MRUI quantitation package. Magma. 2001;12(23):141–152. doi: 10.1007/BF02668096. [DOI] [PubMed] [Google Scholar]
- 30.Pelupessy P, Chiarparin E, Bodenhausen G. Excitation of selected proton signals in NMR of isotopically labeled macromolecules. J Magn Reson. 1999;138:178–181. doi: 10.1006/jmre.1999.1715. [DOI] [PubMed] [Google Scholar]
- 31.Provencher SW. Estimation of metabolite concentrations from localized in vivo proton NMR spectra. Magn Reson Med. 1993;30(6):672–679. doi: 10.1002/mrm.1910300604. [DOI] [PubMed] [Google Scholar]
- 32.Ratiney H, Sdika M, Coenradie Y, Cavassila S, van Ormondt D, Graveron-Demilly D. Time-domain semi-parametric estimation based on a metabolite basis set. NMR Biomed. 2005;18(1):1–13. doi: 10.1002/nbm.895. [DOI] [PubMed] [Google Scholar]
- 33.Rothman DL, Arias-Mendoza F, Shulman GI, Shulman RG. A pulse sequence for simplifying hydrogen NMR spectra of biological tissues. J Magn Reson. 1984;60:430–436. [Google Scholar]
- 34.Sierra GA, Schuler MA, Schweiger A. Right-angle spinning for sensitivity improvement in pulse EPR experiments. Chem Phys Lett. 1999;303:475–481. [Google Scholar]
- 35.Slotboom J, Boesch C, Kreis R. Versatile frequency domain fitting using time domain models and prior knowledge. Magn Reson Med. 1998;39:899–911. doi: 10.1002/mrm.1910390607. [DOI] [PubMed] [Google Scholar]
- 36.Smith SA, Levante TO, Meier BH, Ernst RR. Computer simulations in magnetic resonance. An object-oriented programming approach. J Magn Reson. 1994;A106:75–105. [Google Scholar]
- 37.Smith SA, Murali N. Relaxation effects in a system of a spin-1/2 nucleus coupled to a quadrupolar spin subjected to RF irradiation - Evaluation of broad-band decoupling schemes. J Magn Reson. 1999;136:211–213. doi: 10.1006/jmre.1998.1582. [DOI] [PubMed] [Google Scholar]
- 38.Soher BJ, Maudsley AA. Evaluation of variable line-shape models and prior information in automated 1H spectroscopic imaging analysis. Magn Reson Med. 2004;52(6):1246–1254. doi: 10.1002/mrm.20295. [DOI] [PubMed] [Google Scholar]
- 39.Soher BJ, Vermathen P, Schuff N, Wiedermann D, Meyerhoff D, Weiner M, Maudsley AA. Short TE in vivo 1H MR spectroscopic imaging at 1.5 T: Automated spectral analysis and metabolite image formation. Magn Reson Imaging. 2000;18:1159–1165. doi: 10.1016/s0730-725x(00)00212-5. [DOI] [PubMed] [Google Scholar]
- 40.Soher BJ, Young K, Govindaraju V, Maudsley AA. Automated spectral analysis III: Application to in vivo proton MR spectroscopy and spectroscopic imaging. Magn Reson Med. 1998;40:822–831. doi: 10.1002/mrm.1910400607. [DOI] [PubMed] [Google Scholar]
- 41.Suhy J, Miller RG, Rule R, Schuff N, Licht J, Dronsky V, Gelinas D, Maudsley AA, Weiner MW. Early detection and longitudinal changes in amyotrophic lateral sclerosis by 1H MRSI. Neurology. 2002;58(5):773–779. doi: 10.1212/wnl.58.5.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas MA, Binesh N, Yue K, DeBruhl N. Volume-localized two-dimensional correlated magnetic resonance spectroscopy of human breast cancer. Magn Reson Imaging. 2001;14:181–186. doi: 10.1002/jmri.1170. [DOI] [PubMed] [Google Scholar]
- 43.Thomas MA, Ryner LN, Mehta MP, Turski PA, Sorenson JA. Localized 2D J-resolved 1H MR spectroscopy of human brain tumors in vivo. Magn Reson Imaging. 1996;6:453–459. doi: 10.1002/jmri.1880060307. [DOI] [PubMed] [Google Scholar]
- 44.Thomas MA, Yue K, Binesh N, Davanzo P, Kumar A, Siegel B, Frye M, Curran J, Lufkin R, Martin P, Guze B. Localized two-dimensional shift correlated MR spectroscopy of human brain. Magn Reson Med. 2001;46:58–67. doi: 10.1002/mrm.1160. [DOI] [PubMed] [Google Scholar]
- 45.Thompson RB, Allen PS. Response of metabolites with coupled spins to the STEAM sequence. Magn Reson Med. 2001;45:955–965. doi: 10.1002/mrm.1128. [DOI] [PubMed] [Google Scholar]
- 46.Trabesinger AH, Meier D, Boesiger P. In vivo 1H NMR spectroscopy of individual human brain metabolites at moderate field strengths. Magn Reson Imaging. 2003;21:1295–1302. doi: 10.1016/j.mri.2003.08.029. [DOI] [PubMed] [Google Scholar]
- 47.Trabesinger AH, Meier D, Dydak U, Lamerichs R, Boesiger P. Optimizing PRESS localized citrate detection at 3 Tesla. Magn Reson Med. 2005;54:51–58. doi: 10.1002/mrm.20544. [DOI] [PubMed] [Google Scholar]
- 48.van Dijk JE, Mehlkopf AF, Bovee WMMJ. Comparison of double and zero quantum NMR editing techniques for in vivo use. NMR Biomed. 1992;5:75–86. doi: 10.1002/nbm.1940050206. [DOI] [PubMed] [Google Scholar]
- 49.Veshtort M, Griffin RG. SPINEVOLUTION: a powerful tool for the simulation of solid and liquid state NMR experiments. J Magn Reson. 2006;178:248–282. doi: 10.1016/j.jmr.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 50.Young K, Govindaraju V, Soher BJ, Maudsley AA. Automated spectral analysis I: Formation of a priori information by spectral simulation. Magn Reson Med. 1998;40:812–815. doi: 10.1002/mrm.1910400605. [DOI] [PubMed] [Google Scholar]
- 51.Young K, Matson GB, Govindaraju V, Maudsley AA. Spectral simulations incorporating gradient coherence selection. J Magn Reson. 1999;140:146–152. doi: 10.1006/jmre.1999.1809. [DOI] [PubMed] [Google Scholar]
- 52.Young K, Soher BJ, Maudsley AA. Automated spectral analysis II: Application of wavelet shrinkage for characterization of non-parameterized signals. Magn Reson Med. 1998;40:816–821. doi: 10.1002/mrm.1910400606. [DOI] [PubMed] [Google Scholar]


