Abstract
Photoactive proteins such as PYP (photoactive yellow protein) are generally accepted as model systems for studying protein signal state formation. PYP is a blue-light sensor from the bacterium Halorhodospira halophila. The formation of PYP's signaling state is initiated by trans-cis isomerization of the p-coumaric acid chromophore upon the absorption of light. The quantum yield of signaling state formation is ≈0.3. Using femtosecond visible pump/mid-IR probe spectroscopy, we investigated the structure of the very short-lived ground state intermediate (GSI) that results from an unsuccessful attempt to enter the photocycle. This intermediate and the first stable GSI on pathway into the photocycle, I0, both have a mid-IR difference spectrum that is characteristic of a cis isomer, but only the I0 intermediate has a chromophore with a broken hydrogen bond with the backbone N atom of Cys-69. We suggest, therefore, that breaking this hydrogen bond is decisive for a successful entry into the photocycle. The chromophore also engages in a hydrogen-bonding network by means of its phenolate group with residues Tyr-42 and Glu-46. We have investigated the role of this hydrogen bond by exchanging the H bond-donating residue Glu-46 with the weaker H bond-donating glutamine (i.e., Gln-46). We have observed that this mutant exhibits virtually identical kinetics and product yields as WT PYP, even though during the I0-to-I1 transition, on the 800-ps time scale, the hydrogen bond of the chromophore with Gln-46 is broken, whereas this hydrogen bond remains intact with Glu-46.
Keywords: ground state intermediate, hydrogen bond, quantum yield, picosecond, vibrational
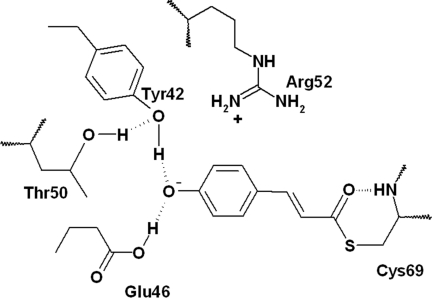
PYP (photoactive yellow protein) belongs to the Xanthopsins, a family of blue-light photoreceptors that contain 4-hydroxy-cinnamic acid as their photoactive chromophore (see refs. 1–3 for a review). PYP is a small protein and therefore an attractive model system for exploring how a chromophore and protein interact to sense light and send a biological signal. Its photocycle has been characterized by various experimental techniques, such as fluorescence (4, 5), (time-resolved) FTIR (6–8), (time-resolved) x-ray crystallography (9–12), NMR (13), Stark spectroscopy (14), and pump(-dump)-probe spectroscopy (15, 16). X-ray diffraction on PYP crystals has demonstrated that the PYP chromophore is covalently linked (see Fig. 1) to the protein backbone by means of Cys-69 (12). It is further embedded in a hydrogen-bonding network consisting of Glu-46, Tyr-42, Thr-50, and Cys-69 (12). In the ground state, the chromophore is in a deprotonated trans form, negatively charged, and possibly stabilized by the positive Arg-52 residue (12). After photoexcitation, the chromophore forms a red-shifted intermediate, referred to as I0, within a few picoseconds. This intermediate has a shifted absorption maximum from 446 to 500 nm. The second intermediate, I1 (or pR or PYPL), absorbs maximally at 480 nm and is formed in 1–3 ns (15–20). This intermediate is followed by protonation of the chromophore and a large structural change of the protein on a millisecond time scale, which is believed to be the signaling state (see refs. 1–3 for a review). A previous report of visible pump/mid-IR probe measurements on WT PYP (21) provided detailed insight in the initial structural changes taking place during chromophore isomerization on the 200-fs to 3-ns time scale, a period covering formation of the excited state (ES), I0, and I1. It was observed that a stable cis ground state formed in ≈2 ps, which Heyne et al. (22) confirmed by using the same technique in combination with normal mode calculations. In conjunction with isomerization of the chromophore, dynamic changes of the hydrogen-bonding network surrounding the chromophore were observed. For example, the carbonyl group of the chromophore breaks its hydrogen bond to the backbone of Cys-69 (21) and probably flips to the other side of the pocket, leading to the conformation observed for a cryotrapped intermediate in x-ray diffraction and during the first few nanoseconds of the I1 state in time-resolved x-ray diffraction experiments (9, 23). The hydrogen bond between the chromophore and Glu-46 is weakened during the initial events of the photocycle and subsequently strengthened (21).
Fig. 1.
Schematic drawing of the active site of WT PYP. The p-coumaric acid chromophore is covalently bound to the protein backbone by means of Cys-69; in addition, it takes part in a distal hydrogen-bonding network. In E46Q, glutamic acid (Glu) is changed to glutamine by using site-directed mutagenesis.
In this study, we investigate the functional role of the hydrogen-bonding network of the PYP chromophore by comparing the mid-IR difference spectra of the PYP photocycle intermediates in the Glu-46-Gln (referred to in shorthand notation as E46Q) mutant with those of WT PYP. Glutamine donates a weaker hydrogen bond to the chromophore because of the inherently weaker hydrogen-bonding characteristics of the amide group of the glutamine side chain (24, 25), but in the ground state, the hydrogen bond is still present (26, 27). This mutant has a considerably red-shifted absorption spectrum compared with WT PYP; it peaks at 460 nm. Its photocycle is approximately three times faster (28) (a recovery lifetime of 50 ms versus 140 ms, but this value is strongly pH-dependent).
In addition, we focus on the structural events during the isomerization process that are responsible for successful entrance into the photocycle. Recently, by using visible pump-dump-probe spectroscopy, an early (ground state) intermediate other than I0 was identified (15). This intermediate was observed to form from the ES in competition with I0 but decayed to the original ground state in ≈3–4 ps. This intermediate, therefore, is originating from unsuccessful attempts of the chromophore to enter the photocycle. The quantum yield for successful entry into the photocycle is ≈0.3 (15, 21, 29). Comparing the mid-IR difference spectra of the two intermediates might yield information about which molecular factors are responsible for the relatively low yield of signaling state formation. We build on the results of the aforementioned pump-dump-probe experiments and specifically introduce this ground state intermediate (GSI) in our data analysis. The vibrational absorption-difference spectrum of this state reveals that the chromophore is structurally distorted and can most likely be considered to be a cis isomer but with the hydrogen bond between the chromophore's C O group and the Cys-69 residue of the protein still intact.
O group and the Cys-69 residue of the protein still intact.
Results
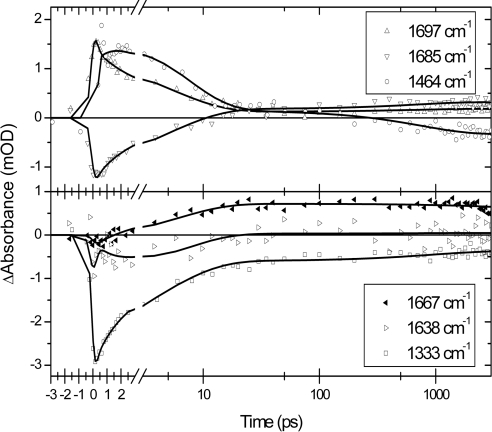
The absorption-difference data of PYP E46Q photoexcited at 475 nm consist of 160 time traces (see Fig. 2) recorded between 1,090 and 1,740 cm−1. A global analysis (i.e., an analysis of all time traces simultaneously) with a model of parallel decaying states showed that the data are well described by using four exponential decays with time constants of 1.4 (±0.1) ps, 6 (±0.3) ps, 800 (±70) ps, and >10 ns, respectively. The corresponding decay-associated difference spectra of each of these time constants in the 1,740–1,610 cm−1 region are shown in Fig. 3. Because of the presence of a small perturbed free-induction decay before t = 0, no information faster than the instrument response (≈200 fs) is extracted. The two longest time constants can be identified as the lifetimes of the states I0E46Q and I1E46Q, respectively. The 1.4-ps time constant is similar to the value(s) reported for the ES lifetime in WT PYP; a multiexponential decay with 0.6-ps and 2.8-ps lifetimes was observed by using visible pump-probe spectroscopy (15), and 2-ps and 3-ps time constants were reported by using vis/mid-IR pump-probe spectroscopy (21, 22).§ Therefore, we assign the 1.4-ps time constant to the ES lifetime of E46Q. The 6-ps component is associated with spectral changes that are distinctly different from those of the 1.4-ps ES component, and they also clearly differ from the spectral characteristics of the I0E46Q state (see Fig. 3).
Fig. 2.
Selection of time traces measured on E46Q PYP. Absorption difference (in mOD) is plotted as a function of time (in picoseconds). The time axis is linear up to 3 ps and logarithmic until 3 ns. The black lines are fits to the data based on a target analysis.
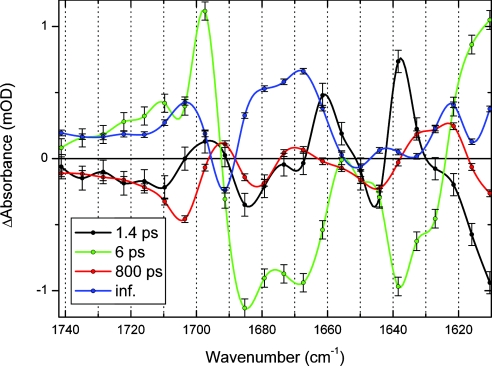
Fig. 3.
Decay-associated difference spectra (including error bars) of the E46Q data. The data are fitted with four time constants: 1.4 ps (black), 6 ps (green), 800 ps (red), and long-lived (blue).
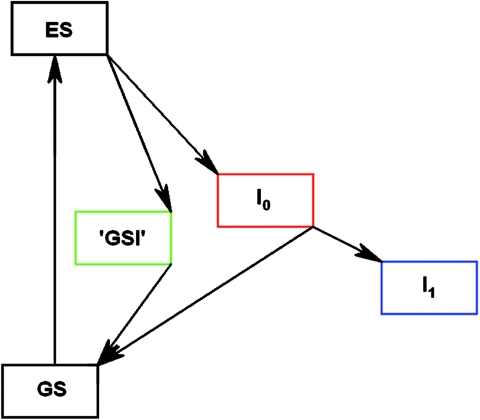
The initial dynamics of PYP have been shown to involve the parallel decay of the ES into the I0 product state and into a short-lived GSI (15). To extract the spectra of these states and to estimate their relative quantum yields, we have analyzed our data by using a target model (30) based on that reported by Larsen et al. (15). In this model, the ES can decay into the I0 ground state and, in parallel, into a short-lived GSI (see Fig. 4). From I0 the I1 state is formed, whereas GSI decays into the stable ground state. When this model is applied to the data, we find that the 1.4-ps, 6-ps, 800-ps, and 10-ns time constants correspond to the lifetimes of the ES, GSIE46Q, I0E46Q, and I1E46Q, respectively. The branching fractions, giving the relative contribution of the decay from the ES into I0E46Q and GSIE46Q, equal 0.31 ± 0.05 and 0.58 ± 0.05, respectively (see Table 1). The formation of I0E46Q seems to be the point of no return: I1E46Q is formed from I0E46Q with a yield of 90–100%. The overall quantum yield for I0E46Q formation, 0.27–0.31, is similar to that reported before for WT PYP, 0.35–0.5 (18, 21, 30). Alternate models were explored to test the possibility of a vibrational cooling process (i.e., by introducing a state in between ES and I0) and the formation of a completely separate state. Based on the resulting spectra and quantum yield, as determined for several long-living signals, we conclude that our data are best described by the presented model where two states are formed in parallel from the ES and the second species (GSI) falls directly back into the equilibrated ground state, consistent with Larsen et al. (15).
Fig. 4.
Model used for the target analysis. GS is the ground state; I0 and I1 are the first two transient, on-pathway intermediates.
Table 1.
Target analysis parameters
| State | E46Q |
WT |
||
|---|---|---|---|---|
| Yield* | Time† | Yield* | Time† | |
| ES | 1.0 | 1.4 ps (0.1) | 1.0 | 1.2 ps (0.1) |
| GSI | 0.58‡ | 6.0 ps (0.3) | 0.72 | 6.2 ps (0.3) |
| I0 | 0.31‡ | 800 ps (70) | 0.28 | 700 ps (50) |
| I1 | 0.9–1.0§ | >10 ns | 0.9–1.0§ | >10 ns |
*Error in the quantum yield is ±0.05.
†Time constant; error is shown in parentheses.
‡Remaining ESE46Q (0.11) falls back into ground state.
§Remaining I0 falls back into ground state.
Because of the detection of the GSI in the IR data, we reanalyzed the WT PYP data published earlier (21) with the added assumption of the presence of the GSI state. The obtained results are shown in Table 1. The values are reasonably close to those reported by Larsen et al. (the “homogeneous model” in ref. 15). The species-associated difference spectra of WT and E46Q that result from the target analysis are shown in Fig. 5 and will be discussed in more detail below.
Fig. 5.
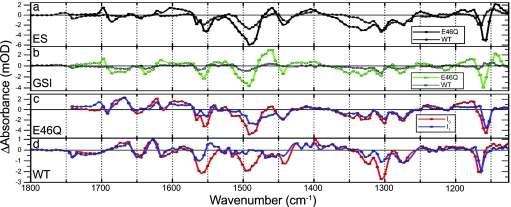
Species-associated difference spectra of WT PYP and E46Q as a result of the target analysis. A comparison of the ES spectra of WT (gray) and E46Q (black) is shown in a, and a comparison of GSIWT (gray) and GSIE46Q (green) is shown in b. The I0 (red) and I1 (blue) spectra for E46Q and WT PYP are shown in c and d, respectively. Note that c and d share the same legend. Negative features in these spectra originate from the ground state bleach, and positive ones originate from ES or product state absorption.
Discussion
The vibrational spectrum of a protein or a protein-bound chromophore contains a wealth of information about its structure, its interaction with the environment, and its electronic properties. In addition, monitoring reaction-induced IR absorption changes can reveal the response of those parts of the protein that are directly involved in the ongoing reactions. The spectra that we have resolved for the initial events of the PYP photocycle yield information about crucial questions relating to protein–chromophore interactions and function. In particular, by comparing the spectra of the I0 and the GSI states, we can characterize the factors that are important for a successful entry into the photocycle.
The spectra we obtained in this study were interpreted by using previous assignments of FTIR and Raman spectra of model chromophores in solution, isotope-labeled PYP, mutants, and normal mode analysis (7, 8, 22, 31–36). Note that the negative bands in the spectra correspond to those of PYP in its ground state, whereas the positive bands arise from band shifts or product bands that are due to the formation of the ES, GSI, I0, and I1 states, respectively.
Isomerization of the Chromophore in the I0 and I1 States.
The phenolate-ring modes dominate the signals at ≈1,160, 1,443, 1,485, and 1,550 cm−1. The instantaneous spectral changes of the phenolate-ring modes are similar to those of a protonated minus deprotonated p-coumaric acid difference spectrum (35) and imply that the phenol ring becomes less negative in the ES, because they all are also ionic markers. Electric field measurements of WT and E46Q PYP (14) showed an instantaneous change in dipole moment upon photoexcitation, which was explained by a charge translocation from the phenolic oxygen over the ethylene chain, in line with our observations. Note that this charge translocation would result in a rearrangement of the carbon bond conjugation of the chromophore, facilitating isomerization; it could give the double bond of the carbonyl group more of a single-bond character. Molecular dynamics simulations also indicated that the negative charge moves along the ethylene chain, although it was calculated to move all the way toward the nearby positively charged Arg-52 residue (37), as opposed to moving toward the thio-ester linkage of the chromophore, as was deduced from the results of Stark spectroscopy (14).
Signals originating from C C modes appear mainly in the 1,280–1,330 cm−1 region; the negative features at 1,300/05 and 1,327/33 cm−1 are C
C modes appear mainly in the 1,280–1,330 cm−1 region; the negative features at 1,300/05 and 1,327/33 cm−1 are C C trans-cis markers (8, 32), indicating the disappearance of the trans ground state of the chromophore. The upward band at 1,289 cm−1 is a marker for the cis product state. Structurally, it has been assigned to the
C trans-cis markers (8, 32), indicating the disappearance of the trans ground state of the chromophore. The upward band at 1,289 cm−1 is a marker for the cis product state. Structurally, it has been assigned to the  C
C C
C C(
C( S
S )
) O skeleton stretch (31, 32).
O skeleton stretch (31, 32).
The C C vibrations (also trans-cis markers) appear mainly at ≈1,600–1,635 cm−1, and those of the C
C vibrations (also trans-cis markers) appear mainly at ≈1,600–1,635 cm−1, and those of the C O of the chromophore appear in the 1,640- to 1,670-cm−1 region. This spectral region deserves special attention and is discussed in more detail below. In general, however, the WT I1 spectrum shows a very close resemblance to the steady-state FTIR (difference) spectrum of cryotrapped I1WT (or PYPL) (8) and with the time-resolved 50-ns step-scan FTIR spectrum reported by Brudler et al. (6), as noted in ref. 21. In time-resolved x-ray Laue diffraction experiments, I1WT has been shown to be a cis isomer (38). From the disappearance of the C
O of the chromophore appear in the 1,640- to 1,670-cm−1 region. This spectral region deserves special attention and is discussed in more detail below. In general, however, the WT I1 spectrum shows a very close resemblance to the steady-state FTIR (difference) spectrum of cryotrapped I1WT (or PYPL) (8) and with the time-resolved 50-ns step-scan FTIR spectrum reported by Brudler et al. (6), as noted in ref. 21. In time-resolved x-ray Laue diffraction experiments, I1WT has been shown to be a cis isomer (38). From the disappearance of the C C and C
C and C C trans modes and the appearance of the C
C trans modes and the appearance of the C C mode at 1,289 cm−1, we concluded that also in I0WT the chromophore had isomerized (21). This conclusion was confirmed by the observation of an additional cis marker mode at 1,000 cm−1 in the I0WT state (22). Changes in the C
C mode at 1,289 cm−1, we concluded that also in I0WT the chromophore had isomerized (21). This conclusion was confirmed by the observation of an additional cis marker mode at 1,000 cm−1 in the I0WT state (22). Changes in the C C(
C( S
S )
) O bond region occurring in ≈800 ps (see features at ≈1,300 cm−1) indicate that a relaxation of the cis configuration marks the I0WT-to-I1WT transition. Overall, the spectra of I0 and I1 of E46Q and WT PYP are very similar in these regions. Therefore, we conclude that, in E46Q, the chromophore follows the same initial events as in WT: The cis isomer ground state I0 is formed in 1.4 ps (slightly faster in WT), followed by a structural relaxation in 800 ps with the I0-to-I1 transition.
O bond region occurring in ≈800 ps (see features at ≈1,300 cm−1) indicate that a relaxation of the cis configuration marks the I0WT-to-I1WT transition. Overall, the spectra of I0 and I1 of E46Q and WT PYP are very similar in these regions. Therefore, we conclude that, in E46Q, the chromophore follows the same initial events as in WT: The cis isomer ground state I0 is formed in 1.4 ps (slightly faster in WT), followed by a structural relaxation in 800 ps with the I0-to-I1 transition.
Hydrogen-Bond Breaking in E46Q.
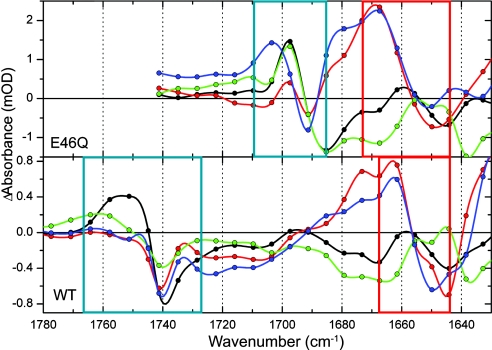
The E46Q spectra differ from WT at ≈1,160 cm−1 and in the 1,700- to 1,740-cm−1 region. The negative band at 1,165 cm−1 in WT PYP has shifted to 1,160 cm−1 in the mutant. This band has been assigned to phenolate-ring mode Y9a (32–34) and is apparently sensitive to the altered hydrogen-bond strength because of the replacement of Glu by Gln. The other main difference is in the C O region of the (mutated) amino acid at position 46: For WT, we observe band shifts in the 1,735- to 1,760-cm−1 region due to the C
O region of the (mutated) amino acid at position 46: For WT, we observe band shifts in the 1,735- to 1,760-cm−1 region due to the C O mode of Glu-46, which are absent in the mutant. On the other hand, new signals appear in E46Q between 1,685 and 1,705 cm−1 that we assign to the C
O mode of Glu-46, which are absent in the mutant. On the other hand, new signals appear in E46Q between 1,685 and 1,705 cm−1 that we assign to the C O mode of Gln-46 (see blue boxes in Fig. 6). This C
O mode of Gln-46 (see blue boxes in Fig. 6). This C O mode shifts from 1,685 cm−1 in the ground state to 1,697 cm−1 in the excited, GSIE46Q, and I0E46Q states. In I1E46Q, it further shifts to 1,704 cm−1, which is the same frequency as that reported for this state by Brudler et al. (6) using FTIR spectroscopy with nanosecond time resolution. With time-resolved x-ray Laue diffraction spectroscopy, it was shown that the hydrogen bond between Gln-46 and the chromophore is lost in I1 (38). We thus demonstrate here that disruption of this hydrogen bond occurs during the I0E46Q-to-I1E46Q transition on the 800-ps time scale. In WT PYP, the hydrogen bond between Glu-46 and the chromophore is still intact on this time scale and is even slightly strengthened (see Fig. 6 and ref. 21).
O mode shifts from 1,685 cm−1 in the ground state to 1,697 cm−1 in the excited, GSIE46Q, and I0E46Q states. In I1E46Q, it further shifts to 1,704 cm−1, which is the same frequency as that reported for this state by Brudler et al. (6) using FTIR spectroscopy with nanosecond time resolution. With time-resolved x-ray Laue diffraction spectroscopy, it was shown that the hydrogen bond between Gln-46 and the chromophore is lost in I1 (38). We thus demonstrate here that disruption of this hydrogen bond occurs during the I0E46Q-to-I1E46Q transition on the 800-ps time scale. In WT PYP, the hydrogen bond between Glu-46 and the chromophore is still intact on this time scale and is even slightly strengthened (see Fig. 6 and ref. 21).
Fig. 6.
Comparison of the species-associated difference spectra of E46Q and WT in the Glu/Gln C O (blue box) and chromophore C
O (blue box) and chromophore C O (red box) regions.
O (red box) regions.
The initial photocycle events, such as the rate and yield of I0 and I1 formation, are very similar for WT and E46Q despite the weaker hydrogen bond in E46Q and its breakage on the 800-ps time scale. We conclude, therefore, that the strength of the hydrogen bond between the chromophore and residue 46 does not seem to play a crucial role in the initial part of the photocycle. At longer time scales, however, this mutation causes a faster ground-state recovery rate (28, 39). Different ground-state recovery pathways for WT and E46Q were also reported to show up in dehydrated films (40).
Successful or Unsuccessful Photocycle Entry Attempts.
Larsen et al. (15) have shown that the state that we resolve here as GSI is one through which a major part of the ES decays to the ground state. The species-associated difference spectra of this state differs from both the I0 and I1 spectra, most notably in the 1,600- to 1,700-cm−1 region (see Fig. 5).
In this region, there is a positive band at 1,667 cm−1 in the I0E46Q and I1E46Q spectra (see red box in Fig. 6), which has been assigned to the chromophore's C O group, free from interaction with the N backbone atom of Cys-69 (6, 36). Breaking the hydrogen bond, which occurs because of the rotation of the C
O group, free from interaction with the N backbone atom of Cys-69 (6, 36). Breaking the hydrogen bond, which occurs because of the rotation of the C O group around the chromophore's long axis as observed in nanosecond time-resolved x-ray studies (38) and cryotrapped intermediates (41), causes the frequency of this mode to upshift, leaving a bleached band at 1,650 cm−1 in the I0E46Q and I1E46Q spectra. Remarkably, the positive features are notably absent in ES and GSI, indicating that the hydrogen bond between the C
O group around the chromophore's long axis as observed in nanosecond time-resolved x-ray studies (38) and cryotrapped intermediates (41), causes the frequency of this mode to upshift, leaving a bleached band at 1,650 cm−1 in the I0E46Q and I1E46Q spectra. Remarkably, the positive features are notably absent in ES and GSI, indicating that the hydrogen bond between the C O of the chromophore and Cys-69 is intact in ES and GSI.
O of the chromophore and Cys-69 is intact in ES and GSI.
We note that although there is no upshifted product band, the ES spectrum does contain a bleached band at 1,644 cm−1, which could be due to a more single-bond character of the carbonyl group in the ES because of electron migration upon excitation, as discussed above. Such migration would shift the mode into the ≈1,200-cm−1 region, where, unfortunately, it is hard to recognize. In addition, there is a negative band in the GSI spectrum at 1,638 cm−1. This band has been proposed to originate primarily from the central C C bond (32). The bleaching of this band could therefore be interpreted as GSI's assumption of a backbone conformation that is slightly different from I0. We note that in the I1 state as well as in the cryotrapped PYPL spectrum, both the 1,650- and the 1,638-cm−1 band are fully resolved (8).
C bond (32). The bleaching of this band could therefore be interpreted as GSI's assumption of a backbone conformation that is slightly different from I0. We note that in the I1 state as well as in the cryotrapped PYPL spectrum, both the 1,650- and the 1,638-cm−1 band are fully resolved (8).
The spectral changes in the remaining part of the GSI spectrum, in particular in the 1,400- to 1,200-cm−1 region, which is dominated by C C(
C( S
S )
) O modes, and the appearance of a positive cis marker at 1,289 cm−1, show that the trans backbone of the chromophore in the GSI state has been distorted and could even be considered to be in a cis configuration, although, as noted above, the hydrogen bond of the C
O modes, and the appearance of a positive cis marker at 1,289 cm−1, show that the trans backbone of the chromophore in the GSI state has been distorted and could even be considered to be in a cis configuration, although, as noted above, the hydrogen bond of the C O of the chromophore to Cys-69 is still intact. Probably, the inability of the chromophore to break the hydrogen bond on a sufficiently fast time scale leads to its decay to the original ground state by means of a mixture of vibrational cooling and reisomerization to the trans configuration, rather than to its entry into the photocycle by a relaxation of the cis isomer. The important role of the strength of the chromophore's hydrogen bond with the backbone N atom of Cys-69 in determining the success of the isomerization is compatible with the higher quantum yield of isomerization in the P68A mutant, where this hydrogen bond is weaker (42).
O of the chromophore to Cys-69 is still intact. Probably, the inability of the chromophore to break the hydrogen bond on a sufficiently fast time scale leads to its decay to the original ground state by means of a mixture of vibrational cooling and reisomerization to the trans configuration, rather than to its entry into the photocycle by a relaxation of the cis isomer. The important role of the strength of the chromophore's hydrogen bond with the backbone N atom of Cys-69 in determining the success of the isomerization is compatible with the higher quantum yield of isomerization in the P68A mutant, where this hydrogen bond is weaker (42).
Isomerization Mechanism.
The observation of a short-lived GSI with the chromophore in a distorted trans-cis configuration and its hydrogen bond of the C O group to the backbone intact is in agreement with the molecular dynamics simulations performed by Groenhof et al. (43). They showed that the breaking of the hydrogen bond is uncoupled from the isomerization process: In their simulations, isomerization around the C
O group to the backbone intact is in agreement with the molecular dynamics simulations performed by Groenhof et al. (43). They showed that the breaking of the hydrogen bond is uncoupled from the isomerization process: In their simulations, isomerization around the C C double bond takes places partly in the ES and continues rapidly with the reformation of the ground state. This step is followed by the breaking of the hydrogen bond (i.e., in the cis ground state) in a few picoseconds (43), in agreement with our observation of a cis isomer with its hydrogen bond intact. However, in these simulations, an additional intermediate is predicted between ES and I0, which has not been observed in our measurements. Possibly, our data do not contain a sufficient signal-to-noise ratio to confirm this, or the concentration of an I0-like intermediate with the C
C double bond takes places partly in the ES and continues rapidly with the reformation of the ground state. This step is followed by the breaking of the hydrogen bond (i.e., in the cis ground state) in a few picoseconds (43), in agreement with our observation of a cis isomer with its hydrogen bond intact. However, in these simulations, an additional intermediate is predicted between ES and I0, which has not been observed in our measurements. Possibly, our data do not contain a sufficient signal-to-noise ratio to confirm this, or the concentration of an I0-like intermediate with the C O hydrogen-bonded could be too low in the experiment because of a faster decay rate than formation rate. The large number of unsuccessful isomerization attempts observed in the molecular dynamics simulations could be consistent with the existence of a GSI. An initial inspection has revealed that these unsuccessful isomerizations originate from a “twisted-state” formation (G. Groenhof, personal communication). The involvement of twisted charge-transfer states in the photocycle of PYP, either on or off pathway, deserves further attention. The unsuccessful attempts to form the GSI favor the hula-twist mechanism (see ref. 44), because this mechanism is consistent with an intact, hydrogen-bonded C
O hydrogen-bonded could be too low in the experiment because of a faster decay rate than formation rate. The large number of unsuccessful isomerization attempts observed in the molecular dynamics simulations could be consistent with the existence of a GSI. An initial inspection has revealed that these unsuccessful isomerizations originate from a “twisted-state” formation (G. Groenhof, personal communication). The involvement of twisted charge-transfer states in the photocycle of PYP, either on or off pathway, deserves further attention. The unsuccessful attempts to form the GSI favor the hula-twist mechanism (see ref. 44), because this mechanism is consistent with an intact, hydrogen-bonded C O. For the entry into the photocycle, however, the multiple-bond flip mechanism apparently prevails, because we do not observe an I0 intermediate with the H bond intact. However, as mentioned above, experimental difficulties could have hampered the observation of such a state, and, consequently, this question is still open. Note that in Groenhof's simulations (43), a successful entry into the photocycle occurs by means of the hula-twist mechanism.
O. For the entry into the photocycle, however, the multiple-bond flip mechanism apparently prevails, because we do not observe an I0 intermediate with the H bond intact. However, as mentioned above, experimental difficulties could have hampered the observation of such a state, and, consequently, this question is still open. Note that in Groenhof's simulations (43), a successful entry into the photocycle occurs by means of the hula-twist mechanism.
Overview of the Initial Events of the PYP Photocycle.
The envisioned sequence of events that occurs in PYP upon excitation, based on our current results and the molecular dynamics simulations by Groenhof et al. (43), is the following. In the ES, the negative charge on the phenolic oxygen migrates toward the thio-ester linkage, giving the double bond of the hydrogen-bonded carbonyl group more of a single-bond character. The hydrogen bond to the backbone remains initially intact. Isomerization proceeds partially in the ES and continues in the ground state, after which the chromophore follows one of two possible pathways: (i) it breaks its hydrogen bond and forms the I0 intermediate or (ii) the hydrogen bond remains intact, and the chromophore reisomerizes to the ground state. The isomerization process and the breaking of the hydrogen bond are therefore independent processes. We propose that the strength of the hydrogen bond with Cys-69 determines whether the chromophore will enter the photocycle. Upon light absorption, the hydrogen bond is weak enough to be broken in only ≈30% of the protein molecules. In the remaining fraction, the chromophore relaxes to its stable ground state in ≈6 ps. The strength of the hydrogen bond could determine the isomerization mechanism (using the multiple-bond flip to form I0 or the hula-twist mechanism to form the GSI), but it is not unambiguously shown by these results. Changing the H bond-donating residue Glu-46 near the phenolic oxygen with the weaker H bond-donating Gln-residue hardly alters the kinetics and product yields, even though, during the I0-to-I1 transition, the hydrogen bond of the chromophore with Gln-46 is broken but remains intact with Glu-46. The hydrogen bond with Glu-46, therefore, does not seem to play a crucial role in the initial part of the photocycle.
Materials and Methods
Sample was prepared in H2O buffer as described in ref. 45. It consisted of a highly concentrated protein solution pressed between two CaF2 windows, separated by a 20-μm Teflon spacer, at OD446 ≈ 1.0.
The experimental setup (see ref. 21 for a more detailed description) consists of an integrated Ti:sapphire oscillator/regenerative amplifier (Hurricane; SpectraPhysics, Mountain View, CA) operating at 1 kHz and producing 0.8-mJ pulses of 80 fs. The output of this laser is used to pump a commercial optical parametric generator and amplifier with difference frequency generation (TOPAS; Light Conversion, Vilnius, Lithuania), which results in a tunable output (2.5–10 μm) with a spectral width of ≈200 cm−1. A home-built HgCdT camera system placed behind a spectrograph is read out every shot at a repetition rate of 1 kHz and a sampling resolution of ≈6 cm−1. Another part of the Hurricane output is used to pump a home-built noncollinear optical parametric amplifier to generate pulses at 475 nm with a duration of ≈60 fs (uncompressed) and 90-nJ excitation energy. Pump-probe spectra were measured between −10 ps and 3 ns with an instrument response function of ≈200 fs (cross-correlation in GaAs). The pump beam polarization is set with a Berek rotator (Model 5540; New Focus, San Jose, CA) to the magic angle with respect to the probe beam. A phase-locked chopper at 500 Hz ensures that with every other shot the sample is excited and an absorbance difference spectrum can be calculated. To ensure a fresh spot for each laser shot, the sample is moved with a home-built Lissajous scanner. The setup is contained in a nitrogen-purged box to reduce distortions of the IR beam by water vapor absorption.
Data were collected in the spectral window from 1,090 to 1,740 cm−1 in five partially overlapping windows. To check reproducibility, each data set, typically consisting of 60 scans taking ≈70 min in total, was measured several times. The data were processed and analyzed by using global and target analysis software (30). A correction was made for the presence of a pre-time-zero offset due to thermal lens effects (46, 47). Because no reference probe pulse was used, the noise in the measured spectra consisted mainly of so-called baseline noise (i.e., a flat, structureless offset in the spectra), which is easily recognized from a singular vector decomposition of the residual matrix (30, 48). The signal-to-noise ratio of the data was enhanced by subtracting the outer product of the first two singular vector pairs of the residual matrix (being structureless in the time domain and smooth in the wavelength domain) from the data, leading to a factor of 2 reduction in the noise for both WT and E46Q. The previously recorded WT data (21) were reanalyzed with these two corrections. A good quality data set had a typical noise level of <100 μOD.
Acknowledgments
L.J.G.W.v.W. thanks J. Key for carefully reading the manuscript. This work was supported by The Netherlands Organization for Scientific Research through the Dutch Foundation for Earth and Life Sciences (Investment Grant 812.08.001 and Molecule to Cell Grant 805.47.123 to K.J.H. and Fellowship 834.01.002 to M.L.G.).
Abbreviations
- PYP
photoactive yellow protein
- ES
excited state
- GSI
ground state intermediate
Footnotes
The authors declare no conflict of interest.
This paper was submitted directly (Track II) to the PNAS office. G.F. is a guest editor invited by the Editorial Board.
The inclusion of a fast, 0.7-ps (fixed) time constant with an amplitude of 10% relative to that of the 1.4-ps time constant led to a small improvement of the fit. However, because we could not resolve it independently from our data and because it possessed a spectrum identical to that of the 1.4-ps component, we ignored its presence. Apparently, the vis-vis pump-probe experiments are more sensitive to this fast initial relaxation of the ES than the mid-IR experiments, which indicates that this relaxation is most likely solvational rather than structural.
References
- 1.Hellingwerf KJ, Hendriks J, Gensch T. J Phys Chem A. 2003;107:1082–1094. [Google Scholar]
- 2.Cusanovich MA, Meyer TE. Biochemistry. 2003;42:4759–4770. doi: 10.1021/bi020690e. [DOI] [PubMed] [Google Scholar]
- 3.Larsen DS, van Grondelle R. Chem Phys Chem. 2005;6:828–837. doi: 10.1002/cphc.200400351. [DOI] [PubMed] [Google Scholar]
- 4.Chosrowjan H, Mataga N, Nakashima N, Yasushi I, Tokunaga F. Chem Phys Lett. 1997;270:267–272. [Google Scholar]
- 5.Mataga N, Chosrowjan H, Shibata Y, Imamoto Y, Tokunaga F. J Phys Chem B. 2000;104:5191–5199. [Google Scholar]
- 6.Brudler R, Rammelsberg R, Woo TT, Getzoff ED, Gerwert K. Nat Struct Biol. 2001;8:265–270. doi: 10.1038/85021. [DOI] [PubMed] [Google Scholar]
- 7.Xie A, Hoff WD, Kroon AR, Hellingwerf KJ. Biochemistry. 1996;35:14671–14678. doi: 10.1021/bi9623035. [DOI] [PubMed] [Google Scholar]
- 8.Imamoto Y, Shirahige Y, Tokunaga F, Kinoshita T, Yoshihara K, Kataoka M. Biochemistry. 2001;40:8997–9004. doi: 10.1021/bi010021l. [DOI] [PubMed] [Google Scholar]
- 9.Genick UK, Soltis SM, Kuhn P, Canestrelli IL, Getzoff ED. Nature. 1998;392:206–209. doi: 10.1038/32462. [DOI] [PubMed] [Google Scholar]
- 10.Perman B, Srajer V, Ren Z, Teng T, Pradervand C, Ursby T, Bourgeois D, Schotte F, Wulff M, Kort R, et al. Science. 1998;279:1946–1950. doi: 10.1126/science.279.5358.1946. [DOI] [PubMed] [Google Scholar]
- 11.Ihee H, Rajagopal S, Srajer V, Pahl R, Anderson S, Schmidt M, Schotte F, Anfinrud PA, Wulff M, Moffat K. Proc Natl Acad Sci USA. 2005;102:7145–7150. doi: 10.1073/pnas.0409035102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Borgstahl GE, Williams DR, Getzoff ED. Biochemistry. 1995;34:6278–6287. doi: 10.1021/bi00019a004. [DOI] [PubMed] [Google Scholar]
- 13.Dux P, Rubinstenn G, Vuister GW, Boelens R, Mulder FA, Hard K, Hoff WD, Kroon AR, Crielaard W, Hellingwerf KJ, Kaptein R. Biochemistry. 1998;37:12689–12699. doi: 10.1021/bi9806652. [DOI] [PubMed] [Google Scholar]
- 14.Premvardhan LL, van der Horst MA, Hellingwerf KJ, van Grondelle R. Biophys J. 2003;84:3226–3239. doi: 10.1016/S0006-3495(03)70047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larsen DS, van Stokkum IHM, Vengris M, van der Horst MA, de Weerd FL, Hellingwerf KJ, van Grondelle R. Biophys J. 2004;87:1858–1872. doi: 10.1529/biophysj.104.043794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ujj L, Devanathan S, Meyer TE, Cusanovich MA, Tollin G, Atkinson GH. Biophys J. 1998;75:406–412. doi: 10.1016/S0006-3495(98)77525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baltuška A, van Stokkum IHM, Kroon A, Monshouwer R, Hellingwerf KJ, van Grondelle R. Chem Phys Lett. 1997;270:263–266. [Google Scholar]
- 18.Devanathan S, Pacheco A, Ujj L, Cusanovich M, Tollin G, Lin S, Woodbury N. Biophys J. 1999;77:1017–1023. doi: 10.1016/S0006-3495(99)76952-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imamoto Y, Kataoka M, Tokunaga F, Asahi T, Masuhara H. Biochemistry. 2001;40:6047–6052. doi: 10.1021/bi002437p. [DOI] [PubMed] [Google Scholar]
- 20.Gensch T, Gradinaru CC, van Stokkum IHM, Hendricks J, Hellingwerf KJ, van Grondelle R. Chem Phys Lett. 2002;356:347–356. [Google Scholar]
- 21.Groot ML, van Wilderen LJGW, Larsen DS, van der Horst MA, van Stokkum IHM, Hellingwerf KJ, van Grondelle R. Biochemistry. 2003;42:10054–10059. doi: 10.1021/bi034878p. [DOI] [PubMed] [Google Scholar]
- 22.Heyne K, Mohammed OF, Usman A, Dreyer J, Nibbering ETJ, Cusanovich MA. J Am Chem Soc. 2005;127:18100–18106. doi: 10.1021/ja051210k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imamoto Y, Kataoka M, Tokunaga F. Biochemistry. 1996;35:14047–14053. doi: 10.1021/bi961342d. [DOI] [PubMed] [Google Scholar]
- 24.Taylor R, Kennard O. Acc Chem Res. 1984;17:320–326. [Google Scholar]
- 25.Chosrowjan H, Mataga N, Shibata Y, Imamoto Y, Tokunaga F. J Phys Chem B. 1998;102:7695–7698. [Google Scholar]
- 26.Sugishima M, Tanimoto N, Soda K, Hamada N, Tokunaga F, Fukuyama K. Acta Crystallogr D. 2004;60:2305–2309. doi: 10.1107/S0907444904024084. [DOI] [PubMed] [Google Scholar]
- 27.Anderson S, Srajer V, Moffat K. Photochem Photobiol. 2004;80:7–14. doi: 10.1562/2004-03-15-RA-115.1. [DOI] [PubMed] [Google Scholar]
- 28.Genick UK, Devanathan S, Meyer TE, Canestrelli IL, Williams E, Cusanovich MA, Tollin G, Getzoff ED. Biochemistry. 1997;36:8–14. doi: 10.1021/bi9622884. [DOI] [PubMed] [Google Scholar]
- 29.van Brederode ME, Gensch T, Hoff WD, Hellingwerf KJ, Braslavsky SE. Biophys J. 1995;68:1101–1109. doi: 10.1016/S0006-3495(95)80284-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Stokkum IHM, Larsen DS, van Grondelle R. Biochim Biophys Acta. 2004;1657:82–104. doi: 10.1016/j.bbabio.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y, Ujj L, Meyer TE, Cussanovich MA, Atkinson GH. J Phys Chem A. 2001;105:5719–5726. [Google Scholar]
- 32.Kim M, Mathies RA, Hoff WD, Hellingwerf KJ. Biochemistry. 1995;34:12669–12672. doi: 10.1021/bi00039a024. [DOI] [PubMed] [Google Scholar]
- 33.Harada I, Takeuchi H. In: Spectroscopy of Biological Systems. Clark RJH, Hester RE, editors. Vol 13. Chichester, UK: Wiley; 1996. [Google Scholar]
- 34.van Thor JJ, Pierik AJ, Nugteren-Roodzant I, Xie AH, Hellingwerf KJ. Biochemistry. 1998;37:16915–16921. doi: 10.1021/bi981170f. [DOI] [PubMed] [Google Scholar]
- 35.Xie A, Kelemen L, Hendriks J, White BJ, Hellingwerf KJ, Hoff WD. Biochemistry. 2001;40:1510–1517. doi: 10.1021/bi002449a. [DOI] [PubMed] [Google Scholar]
- 36.Unno M, Kumauchi M, Sasaki J, Tokunaga F, Yamauchi S. Biochemistry. 2002;41:5668–5674. doi: 10.1021/bi025508o. [DOI] [PubMed] [Google Scholar]
- 37.Groenhof G, Lensink MF, Berendsen HJC, Snijders JG, Mark AE. Proteins Struct Funct Genet. 2002;48:202–211. doi: 10.1002/prot.10136. [DOI] [PubMed] [Google Scholar]
- 38.Ren Z, Perman B, Srajer V, Teng T-V, Pradervand C, Bourgeois D, Schotte F, Ursby T, Kort R, Wulff M, Moffat K. Biochemistry. 2001;40:13788–13801. doi: 10.1021/bi0107142. [DOI] [PubMed] [Google Scholar]
- 39.Yeremenko S, van Stokkum IHM, Moffat K, Hellingwerf KJ. Biophys J. 2006;90:4224–4235. doi: 10.1529/biophysj.105.074765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van der Horst MA, van Stokkum IHM, Dencher NA, Hellingwerf KJ. Biochemistry. 2005;44:9160–9167. doi: 10.1021/bi050237d. [DOI] [PubMed] [Google Scholar]
- 41.Kort R, Hellingwerf KJ, Ravelli RBG. J Biol Chem. 2004;279:26417–26424. doi: 10.1074/jbc.M311961200. [DOI] [PubMed] [Google Scholar]
- 42.Takeshita K, Imamoto Y, Kataoka M, Mihara K, Tokunaga F, Terazima M. Biophys J. 2002;83:1567–1577. doi: 10.1016/S0006-3495(02)73926-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Groenhof G, Bouxin-Cademartory M, Hess B, de Visser SP, Berendsen HJ, Olivucci M, Mark AE, Robb MA. J Am Chem Soc. 2004;126:4228–4233. doi: 10.1021/ja039557f. [DOI] [PubMed] [Google Scholar]
- 44.Imamoto Y, Kataoka M, Liu RS. Photochem Photobiol. 2002;76:584–589. doi: 10.1562/0031-8655(2002)076<0584:mpftpr>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 45.Hendriks J, Gensch T, Hviid L, van der Horst MA, Hellingwerf KJ, van Thor JJ. Biophys J. 2002;82:1632–1643. doi: 10.1016/S0006-3495(02)75514-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marcano AO, Rodriguez L, Alvarado Y. J Opt Soc Am A. 2003;5:S256–S261. [Google Scholar]
- 47.Kurian A, Unnikrishnan KP, George DS, Gopinath P, Nampoori VPN, Vallabhan CPG. Spectrochim Acta Part A. 2003;59:487–491. doi: 10.1016/s1386-1425(02)00192-0. [DOI] [PubMed] [Google Scholar]
- 48.Hoff WD, van Stokkum IHM, van Ramesdonk HJ, van Brederode ME, Brouwer AM, Fitch JC, Meyer TE, van Grondelle R, Hellingwerf KJ. Biophys J. 1994;67:1691–1705. doi: 10.1016/S0006-3495(94)80643-5. [DOI] [PMC free article] [PubMed] [Google Scholar]