Abstract
We have previously isolated a new actinomycete strain from Tunisian soil called Streptomyces sp. US24, and have shown that it produces two bioactive molecules including a Cyclo (L-Phe, L-Pro) diketopiperazine (DKP). To identify the structural genes responsible for the synthesis of this DKP derivative, a PCR amplification (696 bp) was carried out using the Streptomyces sp. US24 genomic DNA as template and two degenerate oligonucleotides designed by analogy with genes encoding peptide synthetases (NRPS). The detection of DKP derivative biosynthetic pathway of the Streptomyces sp. US24 strain was then achieved by gene disruption via homologous recombination using a suicide vector derived from the conjugative plasmid pSET152 and containing the PCR product. Chromatography analysis, biological tests and spectroscopic studies of supernatant cultures of the wild-type Streptomyces sp. US24 strain and three mutants obtained by this gene targeting disruption approach showed that the amplified DNA fragment is required for Cyclo (L-Phe, L-Pro) biosynthesis in Streptomyces sp. US24 strain. This DKP derivative seems to be produced either directly via a nonribosomal pathway or as a side product in the course of nonribosomal synthesis of a longer peptide.
1. INTRODUCTION
Peptides are involved in a wide number of physiologic and biochemical processes, including metabolism, pain, reproduction, and the immune response. While many naturally occurring peptides are linear compounds, nature also provides us with a wide variety of monocyclic and polycyclic peptides and proteins of all different shapes and sizes [1]. Among them, diketopiperazines (DKP), cyclic dipeptides, are the smallest head-to-tail cyclic peptides and form a very attractive family because of their many potential uses. In fact, useful biological properties have been demonstrated for some of them such as antibacterial, fungicidal, herbicidal, antiviral, immunosuppressor, and antitumour activities [2, 3]. Although the number of newly isolated naturally occurring DKPs has increased during the last few years, the biosynthetic pathways of these molecules remain largely unexplored. Generally, DKP derivatives seem to be produced by three different ways. The first one is nonribosomal pathway and product formation takes place under the catalytic control of a large multimodular enzyme complex, termed NRP synthetase (NRPS). Molecular characterization of NRPS genes has revealed a modular organisation [4] in which each module within this biosynthetic assembly line is responsible for the recognition, activation, and incorporation of a certain amino acid into the nascent peptide product. Such catalytic unit (module) is composed of functionally specific and independent domains, each of them responsible for catalyzing one single reaction. Much of the attention in the field of NRPSs is attracted by the potential to obtain new peptide products with novel biological activities by directed engineering of the biosynthetic genes. In fact, the modular organisation of NRPSs lends itself to biocombinatorial approaches, such as domain and module swapping, as well as fusion and truncation of existing catalytic units [5].
DKP derivatives can be also obtained as a side product in the course of nonribosomal synthesis. It is the case of the Cyclo (Phe-Pro) formation during the nonribosomal synthesis of tyrocidine A and gramicidin S in certain strains of Bacillus brevis. Tyrocidine A, (Phe-Pro-Phe-DPhe-Asn-Gln-Tyr-Val-Orn-Leu-)cyc, is produced at the onset of the stationary phase of growth of Bacillus brevis ATCC 8185 strain via a nonribosomal pathway and possesses antibiotic activities against several gram positive bacteria [6]. Gramicidin S, a cyclic decapeptide consisting of two identical pentapeptides (Phe-Pro-Val-Orn-Leu)2, is produced via a nonribosomal pathway by Bacillus brevis ATCC 9999 and presents antibacterial activities [7].
The third DKP biosynthesis way has been described from Streptomyces noursei. However, very little is known about the DKP derivatives from Streptomyces species and their biosynthetic pathways. The only isolated and studied DKP biosynthetic gene cluster from this bacterium genus is that of albonoursin of the Streptomyces noursei [8]. It has been reported by these authors that the biosynthesis of the albonoursin diketopiperazine is independent of nonribosomal peptide synthetases.
We have previously reported the isolation from Tunisian soil of a new actinomycete strain called Streptomyces sp. US24 producing divers biological activities, the purification, and the structure elucidation of two active molecules from this bacterium [9, 10]. The first one is the 3-indolethanol and the second active molecule is a DKP derivative, the Cyclo (L-Phe, L-Pro) diketopiperazine. The aim of this work was to detect the biosynthetic pathway of this DKP derivative. Our strategy is based on gene disruption approach by homologous recombination via a suicide vector derived from pSET152 [11] containing a 696 bp DNA fragment amplified by PCR using the Streptomyces sp. US24 genomic DNA as a template. The detection of the biosynthetic pathway of the Cyclo (L-Phe, L-Pro) diketopiperazine (DKP) derivative from the Streptomyces sp. US24 strain will permit the study and the comprehension of the natural biosynthesis of diketopiperazines from Streptomyces genus and consequently the production of hybrid DKP molecules having interesting biological and pharmacological activities.
2. MATERIALS AND METHODS
2.1. Bacterial strains, plasmids, and primers
Bacterial strains, plasmids, and primers used in this work are listed in Table 1.
Table 1.
Bacterial strains, plasmids, and primers used in this work.
| Strain, plasmid, and primer | Description | Reference or Source |
| Strains | ||
| Streptomyces sp. US24 | Cyclo (L-Phe, L-Pro) diketopiperazine producer, used in this work as the source of chromosomal DNA to amplify the 696 pb DNA fragment as recipient for intergeneric conjugation and targeted gene disruption | [10] |
| E. coli DH5α | General cloning host (F−φ80 dlacZΔM15Δ(lacZYA-argF) U169 endA1 recA1hsdR17(rk−, mk+) deoR thi-1 susE 44 λ−gyrA96 relA1) | [15] |
| E. coli ET12567 | methylation-defective (dam-13:: Tn9 dcm-6 hsdM Cmr) used for conjugation between E. coli and Streptomyces | [16] |
| Micrococcus luteus | Indicator microorganism | LB 14110 |
| Plasmids | ||
| pSET152 | Integrative vector carrying ϕC31 integrase gene and ϕC31 attP site, oriT and an apramycin resistance gene for selection in Streptomyces and E. coli | [11] |
| pUZ8002 | RK2 derivative with a mutation in oriT. | [17] |
| pGEM-T Easy vector | TA cloning vector AmpR (PCR products cloning) | Promega |
| pSS1 | Derivative of pGEM-T Easy vector carrying a 696 pb DNA fragment involved in the biosynthetic pathway of Cyclo (L-Phe, L-Pro) DKP of the US24 strain | This work |
| pSS2 | Suicide vector derivative of the pSET152 | This work |
| pSS3 | pSS2 carrying the EcoRI DNA fragment insert from pSS1 | This work |
| Primers | ||
| [P1/P2] | Primers used for the amplification of the 696 pb DNA fragment | This work |
| P1 | 5′ATCTACAC(G/C)AGCGGGACGAC(G/C)GGC3′ | |
| P2 | 5′(G/C)AGGTCGCC(G/C)GTGCGGTACAT3′ | |
2.2. Media and culture conditions
E. coli strains were grown in Luria broth (LB) medium. Ampicillin (50 μgmL−1), apramycin (50 μgmL−1), chloramphenicol (25 μgmL−1), nalidixic acid and kanamycin (50 μgmL−1) were added to growth media when required and 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (40 μgmL−1) when appropriate [12]. Transformation of E. coli DH5α with pGEM-T derivatives was carried out according to the manufacture's instructions (Promega, Madison, USA).
Streptomyces sp. US24 strain was grown in Tryptone soya broth medium (TSB: 30 g tryptic soy broth plus 5 g Yeast extract per 1000 mL distilled water) for the preparation of genomic DNA [13] and on R2YE [14] plates for the preparation of spore stocks. TSB supplemented with 30 μg of apramycin mL−1 was used to grow exconjugants.
AS1 medium [18]: 1 g Yeast extract, 5 g Soluble starch, 0.2 g L-alanine, 0.2 g L-arginine, 0.5 g L-asparagine, 2.5 g NaCl, 10 g Na2SO4, and 20 g agar, pH adjusted at 7.5 with KOH, was used for conjugation experiments, and cultureswere grown at 37°C.
For biological activity production, Streptomyces sp. US24 strain and the corresponding exconjugants were grown in TSB medium, supplemented at 1% (w/v) with starch and 0.1% (v/v) of a trace element solution: (0.4 gL−1 ZnCl2; 2 gL−1 FeSO4 · 7H2O; 0.065 gL−1 H3BO3, and 0.135 gL−1 MoNa2O4 · 2H2O).
For antibacterial activities determination, indicator microorganism Micrococcus luteus LB14110 was grown overnight in LB medium at 30°C, then diluted 1:100 in LB medium and incubated for 5 hours under constant agitation of 200 rpm at the appropriate temperature.
2.3. Intergeneric conjugation
Intergeneric conjugation between E. coli and Streptomyces sp. US24 strainwas performed as described previously by Flett et al. [19], with some modifications: E. coli ET12567 (pUZ8002/pSET152) was grown to an absorbance of 0.4–0.6 at 600 nm. The cells were pelleted by centrifugation, washed twice in an equalvolume of LB, pelleted again, and finally resuspended in 1/10 volume of LB. Aliquots of Streptomyces sp. US24 spore suspension store at −20°C were used as recipients. Spores (eq108) were washed in 2x Yeast extract Tryptonemedium [20], resuspended in 500 μL of 2x Yeast extract Tryptone medium, and incubated at 50°C for 10 minutes to induce germination. Donor cells (500 μL approximately 108 cells) were added to the treated spores, the mixture was pelleted by centrifugation, and finally the pellet was resuspended in the residual liquid and the undiluted mixture was plated on AS1 plates containing 10 mmol MgCl2 and incubated at 37°C for 18 hours. The plates were then overlaid each with 1 mL of water containing 500 μg of nalidixic acid and 1 mg of apramycin, incubated further for 5 days at 37°C. The exconjugants were then counted. Cultures prepared from some arbitrary chosen clones were used for furtherstudies.
2.4. DNA isolation and manipulation
Total DNA preparation was carried out from Streptomyces strains (sp. US24 and the corresponding exconjugants) according to Hopwood et al. [21]. Small-scale plasmid preparations from E. coli were performed as described by Sambrook et al. [12].
Digestion with restriction endonucleases, separation of DNA fragments by agarose gel electrophoresis, dephosphorylation with alkaline calf intestinal phosphatase, ligation of DNA fragments and transformation were done according to Sambrook et al. [12] for E. coli and Hopwood et al. [21] for Streptomyces.
For Southern blot manipulations [22], DNA was transferred to Hybond N membranes (Amersham, Buckinghamshire, UK). The hybridization conditions and subsequent detection were in accordance with the manufacturer's instructions. 32P-labeled probes were prepared using the random prime labelling system (Amersham).
PCR amplification of the 696 bp DNA fragment, involved in the biosynthesis pathway of Cyclo (L-Phe, L-Pro) DKP of the Streptomyces sp. US24 strain, was performed using two degenerate primers (P1, P2) as described in Table 1. Approximately 150 ng genomic DNA was used as template with 100 pmol of each primer per 50 μL reaction volume. To improve the denaturation of the DNA, 5% (v/v) DMSO was added to the reaction mixture. Amplification was performed in a Gene Amp PCR System 2700 (Applied Biosystems, Foster city, USA) using 1U Pfu DNA polymerase (Stratagene, Amsterdam, The Netherlands) and the recommended buffer system according to the following amplification profile: 94°C (5 minutes) followed by 45 cycles of denaturation at 94°C (30 seconds), annealing at 60°C (60 seconds) and extension at 72°C (90 seconds). The PCR products were analysed by agarose gel electrophoresis and fragment of the expected size was purified then cloned into pGEM-T Easy vector yielding pSS1 plasmid.
Nucleotide sequences were determined on both strands using the dideoxy chain-termination method [23]. Reactions were performed with a Thermo sequenase cycle sequencing kit (Amersham) and specific primers. Homology search was performed using Blast Search algorithm [24].
2.5. Extraction, purification of active compounds, biological assay of antimicrobial activities, LC/MS and LC/MS/MS analysis
For the purification of the active molecules from the Streptomyces sp. US24 strain and three corresponding mutants (obtained after transfer of the pSS3 “see Table 1” into the Streptomyces sp. US24 strain), spores at 107 mL−1 were used to inoculate 500 mL Erlenmeyer flasks with four indents, containing 100 mL TSB medium supplemented at 1% (w/v) with starch and 0.1% (v/v) of a trace element solution [9]. After incubation at 37°C for 24 hours in an orbital incubator with shaking at 250 rpm, this preculture was used to inoculate (5% v/v) a total volume of 1 L culture medium having the same composition of the preculture. After three days incubation at 37°C in an orbital incubator with shaking at 250 rpm, each culture broth was filtered to separate mycelium and supernatant. The supernatant was extracted twice with an equal volume of ethyl acetate and then evaporated on a Rotavapor (Laborata 4000). The crude extract, approximately 100 mg for each culture, was dissolved in 500 μL dichloromethane-methanol 1:1 and fractioned by HPLC. This analysis was performed with an analytical column (C18 column 7 μm, 4.6 mm inner diameter 25 cm length). The elution was at a flow rate of 1 mL min−1 with a gradient of two solutions A (water, 0.1% formic acid) and B (acetonitrile, 0.1% formic acid). After injection of the sample, the column was eluted with a linear gradient from 100% buffer A to 50% buffer A and 50% buffer B over the first 40 minutes, followed by a linear gradient to 100% buffer B from 40 to 60 minutes, and then a steady flow of 100% buffer A through 10 minutes [10]. Elution was monitored using a UV detector at 220 nm.
LC/MS and LC/MS/MS analyses were performed using an LC/MSD Trap XCT-Electrospray (Agilent Technologies), operatedin positive ionisation mode, equipped with an HPLC Agilent 100 DAD detector (C18 column Zorbax 300 2.1 × 150 mm). After injection of 5 μL of the corresponding sample, the column was eluted with a linear gradient 95% buffer A (water, 0.1% formic acid) and 5% buffer B (acetonitrile, 0.1% formic acid) through 2 minutes, followed by a linear gradient to 80% buffer B and then a steady flow of 20% buffer A, 80% buffer B through 3 minutes. The elution was at a flow rate of 250 μL min−1.
For biological assay of antimicrobial activity, a paper disk was impregnated with 80 μL of the corresponding sample, and then laid on the surface of an agar plates containing 3 mL of top agar inseeded by 40 μL of a 5-hour-old culture of M. luteus LB 14110. After 2 hours at 4°C, plates were incubated at 30°C for overnight and then examined for evidence of antimicrobial activities represented by a growth inhibition zone of the indicator micro-organism around the paper disk.
3. RESULTS AND DISCUSSION
3.1. PCR amplification of a Streptomyces sp. US24 DNA fragment potentially involved in the Cyclo (L-Phe, L-Pro) DKP derivative biosynthesis
Although the biosynthetic pathways of DKP derivatives remain largely unexplored, in bacteria and fungi these active molecules generally seem to be produced by nonribosomal pathways (NRPS: nonribosomal peptide synthetases). In analogy to type-I polyketide synthases (PKSs), NRPSs exhibit a modular architecture and a module can be further subdivided into different domains, each responsible for a certain biochemical reaction. The adenylation domain (A-domain) controls the entry of the substrates into the peptide as it recognizes and adenylates its cognate substrate. The thiolation domain (T-domain) is referred to as peptidyl carrier protein (PCP), from where it is condensed with the aminoacyl or peptidyl moieties at the neighboring modules. The latter reaction is catalyzed by the condensation domain (C-domain). These three domains, C, A, and T, constitute a minimal elongation module, the basic repetitive unit of multimodular NRPSs. Accordingly, the first module of an NRPS complex lacks a C-domain, whereas the last module usually contains also a termination domain (Te-domain) to release the product. This modular architecture offers a real potentiality to construct new hybrid peptides with novel biological activities by directed engineering of the biosynthetic genes. The aim of this work was to detect the biosynthetic pathway of the Cyclo (L-Phe, L-Pro) active molecule produced by Streptomyces sp. US24. For that purpose, two degenerate oligonucleotides (P1 and P2) were prepared, according to the typical Streptomyces codon usage, based on the amino acid sequences alignment of two synthetases 1 of the Tyrocidine A and Gramicidin S, respectively. For these two decapeptides, Phe and Pro are the residues number one and two, respectively. Three genes (tycA, tycB, and tycC) coding for three proteins of 124, 405, and 724 kDa, respectively, are involved in the biosynthesis of the Tyrocidine A. TycA (synthetase 1) comprises only one module corresponding to the phenylalanine activase (phenylalanine activing domain) and racemase [25]. In the biosynthesis of the Gramicidin S, three open reading frames organised in one transcriptional unit were identified and the corresponding genes are called grsT, grsA, and grsB. The GrsA (Gramicidin S synthetase 1) is an enzyme with a molecular weight of 126.6 kDa which activates and racemize the amino acid phenylalanine as the first step in nonribosomal biosynthesis of the peptide antibiotic gramicidin S [7]. Comparison of the amino acid sequences of GrsA and TycA revealed an extensive degree of homology over 56%. If similar amino acids are also considered, the degree of homology increases to over 70% [26]. This amino acid comparison lets us find two conserved regions IYTSGTTG and MYRTGDL belonging to the adenylation domains of the two synthetases 1 GrsA and TycA. These two conserved regions are part of the known highly conserved core motifs of the adenylation domains of all known NRPSs. Accordingly, two degenerate PCR primers forward P1 and reverse P2 were designed (Table 1) and used in a PCR reaction with the Streptomyces sp. US24 genomic DNA as template. A PCR product with an expected size of approximately 700 bp was obtained. After purification, this DNA fragment was cloned in the pGEM-T Easy vector, yielding the pSS1 plasmid. The insert of four randomly chosen clones was sequenced. All obtained nucleotide sequences are identical and showed a typical Streptomyces GC content (about 76%). This identity can be explained by the high-stringency PCR conditions and the low degree of degeneration of the two used primers. Frame analysis of the nucleotide sequence (696 bp, accession no. AM282973) revealed the presence of a unique internal open reading frame. The alignment of the corresponding amino acid sequence (aa) with all available protein sequences using the Gapped BLAST and PSI-BLAST program [24] showed as expected important aa identities with different NRPS adenylation domains. This identity reaches 50% and 48% with the adenylation domains of the pristinamycin I (PI) synthetase 2, and actinomycin (ACM) synthetase III, respectively (Figure 1). These two adenylation domains activate the two amino acids L-threonine and L-aminobutyric of the PI produced by Streptomyces pristinaespiralis, and the three amino acids proline, N-methylglycine and N-methyl-L-valine of the ACM produced by Streptomyces chrysomallus, respectively [27, 28].
Figure 1.
Alignment of amino acid sequences of the 696 bp DNA fragment of Streptomyces sp. US24 with pristinamycin I synthase 2 (SnbC) of Streptomyces pristinaespiralis, and actinomycin synthase III (AcmC) of Streptomyces chrysomallus. Consensus sequence (cons) corresponds to the amino acids conserved. The two conserved regions used to design the two degenerate oligonucleotides (P1 and P2) are highlighted.
3.2. Construction of suicide vector derived from pSET152
Streptomyces sp. US24 was refractory to classical polyethylene glycol transformation and electroporation procedures (data not shown). Therefore, we decided to use another approach involving intergeneric conjugation from E coli into Streptomycetes [29]. As part of this approach, we have constructed a conjugative suicide vector derived from the integrative conjugative pSET152 vector. This plasmid was chosen because it is successfully transferred from E. coli ET12567/pUZ8002 into Streptomyces sp. US24 strain by intergeneric conjugation, and corresponding directed-site specific recombination has been characterised and analysed (data not shown). To obtain a nonintegrative form of pSET152, the plasmid was partially digested by the restriction endonuclease HindIII and a resulting 4.8 kb DNA fragment was purified, in which, a part of the integrase encoding gene of the pSET152 has been deleted. After ligation and transformation of E. coli cells, recombinant clones were obtained harbouring the desired pSET152 suicide derivative vector called pSS2.
3.3. Integration by homologous recombination
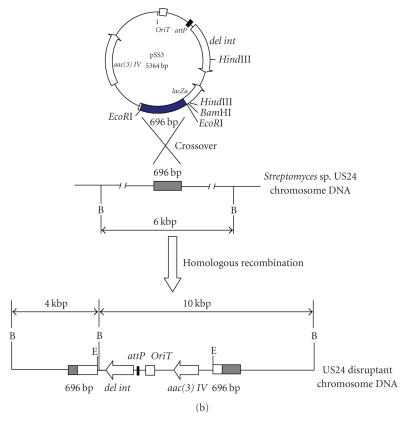
The 696 bp DNA fragment was purified from pSS1 after its digestion by EcoRI and then cloned into pSS2 digested by the same enzyme giving pSS3. This plasmid can be integrated into the chromosome of the Streptomyces sp. US24 strain only via homologous recombination. After transfer of the pSS3 plasmid into the E. coli ET12567 (pUZ8002) strain, we have realized an intergeneric conjugation between E. coli ET12567 (pUZ8002/pSS3) and Streptomyces sp. US24 strain. Streptomyces sp. US24 AprR (apramycin resistant) exconjugants were obtained with a frequency of 10−9. This frequency is a thousand times less comparing to the transconjugation efficiency (10−6) (data not shown) of the native pSET152 which integrates into the Streptomyces sp. US24 chromosome by site-specific recombination between its attP and the chromosomal attB site. This observation lies well with the previously reported finding illustrating that integration via homologous recombination usually occurs 102–103 times less frequently than attB/P. mediated integration. All obtained exconjugants grew well and there are no morphological differences comparing to the wild-type strain. Homologous integration of pSS3 was verified by Southern hybridisation using the 696 bp DNA fragment and the pSS2 as probes (Figure 2(a)). When the 696 bp DNA fragment is used as probe, BamHI digested chromosomal DNA of the wild-type strain Streptomyces sp. US24 hybridized with only one 6 kb DNA fragment, whereas with the BamHI digested chromosomal DNA of one analysed exconjugant, two hybridization signals of about 4 and 10 kb were obtained. Using as probe the pSS2 plasmid linearized by BamHI, there is single BamHI site within the multiple cloning site of this vector, one hybridization signal of about 10 kb was obtained for the chromosomal DNA of the analysed mutant strain and no signal for the wild type. Knowing that the 696 bp amplified DNA fragment has no BamHI site, obtained hybridization results are in concordance with a homologous recombination of the 696 bp DNA fragment via a single crossover into the chromosome of the Streptomyces sp. US24 strain (Figure 2(b)).
Figure 2.
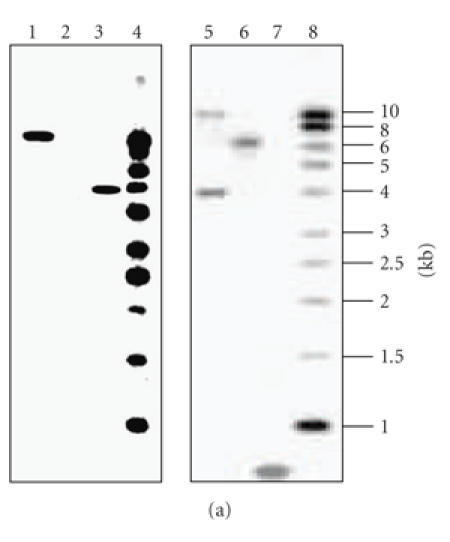
(a) Southern blot analysis confirming insertion of pSS3 plasmid into the Streptomyces sp. US24 chromosome via homologous recombination. Blots were probed with the 696 bp DNA fragment (right panel) or the whole linearized pSS2 (left panel). Lanes 4 and 8, 1 kb ladder used as molecular marker, lanes 2 and 6, wild-type genomic DNA cut with BamHI, lanes 1 and 5, apramycin resistant (AprR) exconjugant mutant genomic DNA cut with BamHI, and lanes 3 and 7, the used probes linearized pSS2 and 696 bp DNA fragment, respectively. (b) Schematic representation of the integration of plasmid pSS3 in the chromosome of Streptomyces sp. US24 by homologous recombination. The resident copy of the 696 bp locus involved in the biosynthetic pathway of Cyclo (L-Phe, L-Pro) DKP of the US24 strain (shaded boxes) present in the chromosome of Streptomyces sp. US24 is shown. del int: deletion of a part of the integrase-encoding gene by partial digestion HindIII; aac(3)IV: apramycin resistance gene; B: restriction site for BamHI; E: restriction site for EcoRI.
3.4. The 696 bp amplified DNA fragment required for the Cyclo (L-Phe, L-Pro) DKP derivative biosynthesis
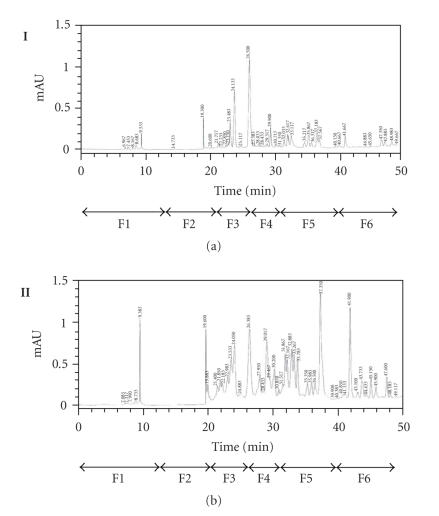
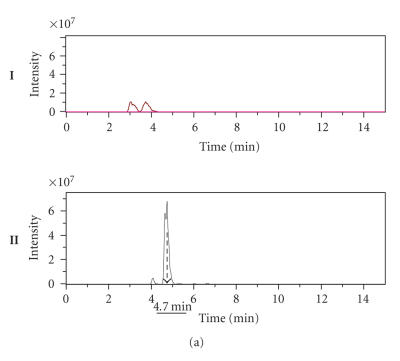
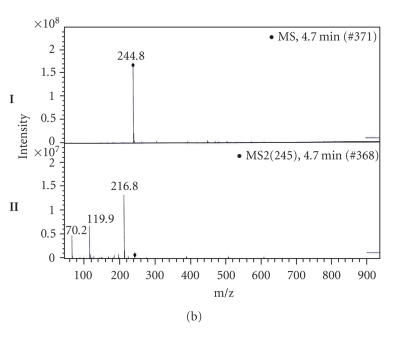
To determine whether the amplified DNA fragment of 696 bp is really involved in Cyclo (L-Phe, L-Pro) biosynthesis, approximately 100 mg of crude extract for each culture, obtained after ethyl acetate extraction from 1 L supernatant culture of the Streptomyces sp. US24 strain and of three different AprR exconjugant mutants (MI, MII, and MIII), was dissolved in 500 μL dichloromethane-methanol 1:1 and fractioned with an analytical HPLC column. For the three analysed exconjugants, obtained chromatograms are identical and present global profiles quite similar to that of the wild type. Six fractions were collected separately between 0 and 50 minutes both for the wild type and one exconjugant (Figure 3). F1 to F6: (F1 “0–13 minutes”; F2 “13–21 minutes”; F3 “21–26 minutes”; F4 “26–31 minutes”; F5 “31–40 minutes,” and F6 “40–50 minutes”). These fractions were concentrated by Rotavapor and then tested for their antibacterial activities against M. luteus. For the Streptomyces sp. US24 strain, an inhibitory activity was detected for the two fractions F3 and F5. However, concerning the three exconjugant mutants, only the F5 fraction was active and the corresponding diameter of inhibition zone was comparable to that of F5 fraction of the Streptomyces sp. US24 strain (Table 2). These microbiological results strongly suggest the presence of the Cyclo (L-Phe, L-Pro) active molecule in the crude extract (fraction F3) only for the Streptomyces sp. US24 wild-type strain. It should be noted that according to our previous work [10], the active molecule Cyclo (L-Phe, L-Pro) was purified from the culture supernatant of Streptomyces sp. US24 strain with a retention time of 25 minutes using the same HPLC conditions used in the present work. This retention time was included in the time interval of the fraction F3 (21–26 minutes). The biological activity observed in the fraction F5 for the four crude extracts (that of the wild type and those of the three mutants) is due to the 3-indolethanol molecule. In fact, Mehdi-Ben Ameur et al. [10] have extracted purified and characterized two active molecules from the supernatant culture of the Streptomyces sp. US24 strain: the Cyclo (L-Phe, L-Pro) “M+ = 244”, and the 3-indolethanol “M+ = 161”. These results are in perfect concordance with those obtained by the determination of the extracted ion chromatograms (EIC), after acetonitrile removing of the four F3 fractions and LC/MS and LC/MS/MS studies. Indeed, for the three exconjugant mutants, we have obtained identical (EIC) which are completely different to that of the wild-type Streptomyces sp. US24. Only this later presents two overlapping important peaks between 4.5 and 5.2 minutes (Figure 4(a)). LC/MS analysis of the different products of these two peaks reveals the presence of an ion corresponding to [M + H]+ = 244.8 at 4.7 minutes. LC/MS/MS analysis of this ion gives three ion fragments at m/z 216.8, m/z 119.9, and m/z 70.2 (Figure 4(b)). These expected fragments have been already obtained in EI/MS analysis during our previous works concerning the Cyclo(L-Phe, L-Pro) of the Streptomyces sp. US24 strain (data not shown). The ion fragments at m/z 120 and m/z 70 correspond to the immonium ion of Phe and Pro, respectively. Otherwise, LC/MS and LC/MS/MS analysis of the four active F5 fractions showed the presence of an ion of [M + H]+ = 162 at 7 minutes for the wild type as well as for the three mutants (MI, MII, and MIII) crude extracts. Corresponding ion fragments were identical for all the four F5 fractions [M + H]+ = 162 “m/z 118.7 and m/z 118.7”. These results are in concordance with those obtained previously (data not shown) concerning the active molecule 3-indolethanol: “M+ = 161”.
Figure 3.
HPLC chromatograms in semipreparative conditions of the crude extracts of 1 L supernatant cultures of the wild-type Streptomyces sp. US24 strain (I) and one exconjugant mutant (II). Six fractions were collected separately between 0 and 50 minutes both for the wild type and one exconjugant. F1 to F6: (F1 “0–13 minutes,” F2 “13–21 minutes,” F3 “21–26 minutes,” F4 “26–31 minutes,” F5 “31–40 minutes,” and F6 “40–50 minutes”).
Table 2.
Antibacterial activities against M. luteus of the crude extracts HPLC fractions of the wild-type Streptomyces sp. US24 and the three studied exconjugant mutant strains.
| Diameter (mm) of inhibition zones | ||||||
| Strain | F1 | F2 | F3 | F4 | F5 | F6 |
| Streptomyces sp. US24 | 0 | 0 | 14 | 0 | 10 | 0 |
| Exconjugant mutants | ||||||
| MI | 0 | 0 | 0 | 0 | 10 | 0 |
| MII | 0 | 0 | 0 | 0 | 10 | 0 |
| MIII | 0 | 0 | 0 | 0 | 10 | 0 |
Figure 4.
(a) Extracted ion chromatograms (EIC) of two fractions F3: one of exconjugant mutant (I) and the second of the wild-type Streptomyces sp. US24 strain (II). (b) LC/MS analysis (I) and LC/MS/MS (II) fragments determination of the product of the peak 4.7 minutes (fraction F3 of the wild-type Streptomyces sp. US24 strain) corresponding to the ion [M + H]+ = 244.8.
Together, all these data (microbiological, chemical, and spectroscopic studies) supported the presence of the active molecule Cyclo (L-Phe, L-Pro) only in the crude extract of the wild-type Streptomyces sp. US24, and the active molecule 3-indolethanol in all the four analysed crude extracts (that of Streptomyces sp. US24 and those of MI, MII, and MIII exconjugant mutants). Consequently, the amplified DNA fragment of 696 bp from the genomic DNA Streptomyces sp. US24 strain is proven to be involved in its Cyclo (L-Phe, L-Pro) biosynthetic pathway.
However, to our knowledge for Streptomyces species, the only isolated and studied DKP biosynthetic gene cluster from this bacterium genus is that of albonoursin of the Streptomyces noursei [8]. It has been reported by these authors that the biosynthesis of the albonoursin diketopiperazine is independent of nonribosomal peptide synthetases. Contrarily, in our case, the biosynthesis of the DKP derivative Cyclo (L-Phe, L-Pro) by the Streptomyces US24 seems to be a nonribosomal pathway. To this level of work, two hypotheses can be proposed. The first one is to consider the direct production by the Streptomyces sp. US24 strain of the Cyclo (L-Phe, L-Pro) via a nonribosomal pathway. The second hypothesis is the obtaining of this active molecule as a side product in the course of nonribosomal synthesis of a longer peptide by the Streptomyces sp. US24. In fact, the Phe-Pro dipeptide can be released from the NRPSs by a nonenzyme-catalysed reaction to give the Cyclo (Phe-Pro). This is due to the high proportion of cis conformation in the Phe-Pro dipeptide, which is induced by N-alkylated amino acids such as Pro. Actually, our study is focused on the construction of a cosmid library from the genomic DNA of the Streptomyces sp. US24 to identify the complete biosynthetic pathway of this DKP derivative.
ACKNOWLEDGMENTS
This work was supported by the Tunisian government (Contract Program CBS-LEMP) and the CMCU project (2006–2008) N° 06/S 0901 “MELLOULI/AIGLE”. The authors are grateful to Dr. R. Mehdi-Ben Ameur and Dr. H. Chouayekh for their generous help and useful conversations.
References
- 1.Brunel FM, Spatola AF. Synthesis of diketopiperazines with on-resin N-methylation and cyclative release. Journal of Peptide Research. 2004;63(3):213–222. doi: 10.1111/j.1399-3011.2004.00130.x. [DOI] [PubMed] [Google Scholar]
- 2.Chu M, Truumees I, Rothofsky ML, et al. Inhibition of c-fos proto-oncogene induction by Sch 52900 and Sch 52901, novel diketopiperazines produced by Gliocladium sp. Journal of Antibiotics. 1995;48(12):1440–1445. doi: 10.7164/antibiotics.48.1440. [DOI] [PubMed] [Google Scholar]
- 3.Rhee K-H. Cyclic dipeptides exhibit synergistic, broad spectrum antimicrobial effects and have anti-mutagenic properties. International Journal of Antimicrobial Agents. 2004;24(5):423–427. doi: 10.1016/j.ijantimicag.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Doekel S, Marahiel MA. Dipeptide formation on engineered hybrid peptide synthetases. Chemistry and Biology. 2000;7(6):373–384. doi: 10.1016/s1074-5521(00)00118-6. [DOI] [PubMed] [Google Scholar]
- 5.Gruenewald S, Mootz HD, Stehmeier P, Stachelhaus T. In vivo production of artificial nonribosomal peptide products in the heterologous host Escherichia coli . Applied and Environmental Microbiology. 2004;70(6):3282–3291. doi: 10.1128/AEM.70.6.3282-3291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mootz HD, Marahiel MA. The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. Journal of Bacteriology. 1997;179(21):6843–6850. doi: 10.1128/jb.179.21.6843-6850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krätzschmar J, Krause M, Marahiel MA. Gramicidin S biosynthesis operon containing the structural genes grsA and grsB has an open reading frame encoding a protein homologous to fatty acid thioesterases. Journal of Bacteriology. 1989;171(10):5422–5429. doi: 10.1128/jb.171.10.5422-5429.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lautru S, Gondry M, Genet R, Pernodet J-L. The albonoursin gene cluster of S. noursei: biosynthesis of diketopiperazine metabolites independent of nonribosomal peptide synthetases. Chemistry and Biology. 2002;9(12):1355–1364. doi: 10.1016/s1074-5521(02)00285-5. [DOI] [PubMed] [Google Scholar]
- 9.Mellouli L, Ben Ameur-Mehdi R, Sioud S, Salem M, Bejar S. Isolation, purification and partial characterization of antibacterial activities produced by a newly isolated Streptomyces sp. US24 strain. Research in Microbiology. 2003;154(5):345–352. doi: 10.1016/S0923-2508(03)00077-9. [DOI] [PubMed] [Google Scholar]
- 10.Ben Ameur-Mehdi R, Mellouli L, Chabchoub F, Fotso S, Bejar S. Purification and structure elucidation of two biologically active molecules from a new isolated Streptomyces sp. US24 strain. Chemistry of Natural Compounds. 2004;40(5):510–513. [Google Scholar]
- 11.Bierman M, Logan R, O'Brien K, Seno ET, Rao RN, Schoner BE. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116(1):43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 13.Stegmann E, Pelzer S, Wilken K, Wohlleben W. Development of three different gene cloning systems for genetic investigation of the new species Amycolatopsis japonicum MG417-CF17, the ethylenediaminedisuccinic acid producer. Journal of Biotechnology. 2001;92(2):195–204. doi: 10.1016/s0168-1656(01)00360-1. [DOI] [PubMed] [Google Scholar]
- 14.Thompson CJ, Ward JM, Hopwood DA. DNA cloning in Streptomyces: resistance genes from antibiotic-producing species. Nature. 1980;286(5772):525–527. doi: 10.1038/286525a0. [DOI] [PubMed] [Google Scholar]
- 15.Hanahan D. Studies on transformation of Escherichia coli with plasmids. Journal of Molecular Biology. 1983;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 16.MacNeil DJ, Occi JL, Gewain KM, et al. Complex organization of the Streptomyces avermitilis genes encoding the avermectin polyketide synthase. Gene. 1992;115(1-2):119–125. doi: 10.1016/0378-1119(92)90549-5. [DOI] [PubMed] [Google Scholar]
- 17.Paget MSB, Chamberlin L, Atrih A, Foster SJ, Buttner MJ. Evidence that the extracytoplasmic function sigma factor σE is required for normal cell wall structure in Streptomyces coelicolor A3(2) Journal of Bacteriology. 1999;181(1):204–211. doi: 10.1128/jb.181.1.204-211.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baltz RH. Genetic recombination by protoplast fusion in Streptomyces . Developments in Industrial Microbiology. 1980;21:43–54. [Google Scholar]
- 19.Flett F, Mersinias V, Smith CP. High efficiency intergeneric conjugal transfer of plasmid DNA from Escherichia coli to methyl DNA-restricting streptomycetes . FEMS Microbiology Letters. 1997;155(2):223–229. doi: 10.1111/j.1574-6968.1997.tb13882.x. [DOI] [PubMed] [Google Scholar]
- 20.Kitani S, Bibb MJ, Nihira T, Yamada Y. Conjugal transfer of plasmid DNA from Escherichia coli to Streptomyces lavendulae FRI-5. Journal of Microbiology and Biotechnology. 2000;10(4):535–538. [Google Scholar]
- 21.Hopwood DA, Bibb MJ, Chater KF, et al. Genetic Manipulation of Streptomyces. A Laboratory Manual. Norwich, UK: The John Innes Foundation; 1985. [Google Scholar]
- 22.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning, A Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 23.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 1977;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mootz HD, Schwarzer D, Marahiel MA. Construction of hybrid peptide synthetases by module and domain fusions. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(11):5848–5853. doi: 10.1073/pnas.100075897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weckermann R, Fürbaß R, Marahiel MA. Complete nucleotide sequence of the tycA gene coding the tyrocidine synthetase 1 from Bacillus brevis . Nucleic Acids Research. 1988;16(24):11841–11852. doi: 10.1093/nar/16.24.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.De Crécy-Lagard V, Blanc V, Gil P, et al. Pristinamycin I biosynthesis in Streptomyces pristinaespiralis: molecular characterization of the first two structural peptide synthetase genes. Journal of Bacteriology. 1997;179(3):705–713. doi: 10.1128/jb.179.3.705-713.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schauwecker F, Pfennig F, Schröder W, Keller U. Molecular cloning of the actinomycin synthetase gene cluster from Streptomyces chrysomallus and functional heterologous expression of the gene encoding actinomycin synthetase II. Journal of Bacteriology. 1998;180(9):2468–2474. doi: 10.1128/jb.180.9.2468-2474.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mazodier P, Petter R, Thompson C. Intergeneric conjugation between Escherichia coli and Streptomyces species. Journal of Bacteriology. 1989;171(6):3583–3585. doi: 10.1128/jb.171.6.3583-3585.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]