Abstract
Dasatinib is a small-molecule kinase inhibitor used for the treatment of imatinib-resistant chronic myelogenous leukemia (CML). We have analyzed the kinases targeted by dasatinib by using an unbiased chemical proteomics approach to detect binding proteins directly from lysates of CML cells. Besides Abl and Src kinases, we have identified the Tec kinases Btk and Tec, but not Itk, as major binders of dasatinib. The kinase activity of Btk and Tec, but not of Itk, was inhibited by nanomolar concentrations of dasatinib in vitro and in cultured cells. We identified the gatekeeper residue as the critical determinant of dasatinib susceptibility. Mutation of Thr-474 in Btk to Ile and Thr-442 in Tec to Ile conferred resistance to dasatinib, whereas mutation of the corresponding residue in Itk (Phe-435) to Thr sensitized the otherwise insensitive Itk to dasatinib. The configuration of this residue may be a predictor for dasatinib sensitivity across the kinome. Analysis of mast cells derived from Btk-deficient mice suggested that inhibition of Btk by dasatinib may be responsible for the observed reduction in histamine release upon dasatinib treatment. Furthermore, dasatinib inhibited histamine release in primary human basophils and secretion of proinflammatory cytokines in immune cells. The observed inhibition of Tec kinases by dasatinib predicts immunosuppressive (side) effects of this drug and may offer therapeutic opportunities for inflammatory and immunological disorders.
Keywords: kinase inhibitor, leukemia, Tec kinases
Expression of Bcr-Abl, the oncogenic counterpart of the tyrosine kinase c-Abl, is the basis for chronic myelogenous leukemia (CML) (1). The small-molecule kinase inhibitor imatinib (Gleevec, Novartis) binds to the active site of the Bcr-Abl kinase domain and inhibits its constitutive tyrosine kinase activity (2, 3). Imatinib represents a paradigmatic case for targeted cancer therapy and is now the first-line treatment for CML (4). Imatinib is a relatively weak inhibitor of Bcr-Abl, with an IC50 in the upper nanomolar range, but has remarkable specificity, targeting only particular conformations of very few other kinases, mainly Abl family members, c-Kit and PDGF-R (5–8). A major drawback of imatinib therapy stems from the fact that many patients develop secondary resistance, leading to patient relapse and disease progression. Resistance is predominantly caused by point mutations in the kinase domain of Bcr-Abl (9, 10). Therefore, kinase inhibitors targeting imatinib-resistant variants of Bcr-Abl have been developed. Of these, only dasatinib (Sprycel, BMS-354825, Bristol-Myers Squibb) has so far been approved for the treatment of adults with CML and Bcr-Abl-positive acute lymphocytic leukemia with imatinib resistance or intolerance to previous therapy (11–13). Dasatinib inhibits Bcr-Abl kinase activity in the low-nanomolar range and inhibits all clinically relevant imatinib-resistant forms with the exception of the common T315I (gatekeeper) mutation (12, 14). Dasatinib has been developed as a Src/Abl inhibitor but subsequently has been shown to affect a wider array of kinases (11). In addition, dasatinib has recently been used in a drug-target profiling approach by using a library of ≈150 selected recombinant kinases expressed as phage particle fusion proteins, of which 67 were bound by dasatinib (15).
To identify native targets of dasatinib in CML cells, we used a chemical proteomics approach (16–20). We immobilized dasatinib and used the resultant resin as an affinity reagent to identify binding proteins from cell lysates by SDS/PAGE and liquid chromatography coupled to tandem MS (LC-MSMS). We found that the Tec kinases Btk and Tec are among the most prominent targets of dasatinib in a variety of cell lines and primary cells. Tec kinases are the second largest group of nonreceptor tyrosine kinases and are closely related to the Src and Abl kinases (21, 22). Functionally, Tec kinases play pivotal roles in the development and signaling of hematopoietic cells (23). Most notably, loss of Btk function is associated with the occurrence of X-linked agammaglobulinemia (XLA) characterized by an impaired development of B cells. As a consequence, XLA patients are severely immunocompromised (24).
By performing structure-based mutagenesis experiments, we have identified the gatekeeper as a critical residue that controls the selectivity of dasatinib. In line with the role of Tec kinases in lymphoid and myeloid cells, dasatinib inhibited the secretion of several immunomodulators.
Results
Btk and Tec Are Among the Most Prominent Interactors of Dasatinib in K562 Cells.
The crystal structure of the Abl kinase domain complexed with dasatinib indicates that the hydroxyethyl side chain on the piperazine ring of dasatinib extends to the protein surface [ref. 25; supporting information (SI) Fig. 6A]. Therefore, it was chosen as an anchor to immobilize dasatinib. To facilitate the coupling, an aminoethyl derivative of dasatinib was synthesized (c-dasatinib; SI Fig. 6B). c-dasatinib and acetylated c-dasatinib displayed the same potency against Abl as unmodified dasatinib (O.H. and U.R., unpublished results). c-dasatinib was coupled to Sepharose beads and incubated with cell extracts from the CML cell line K562. Bound proteins were resolved by SDS/PAGE, bands excised from the gel and the proteins from these bands analyzed by LC-MSMS (Fig. 1A and SI Table 1).
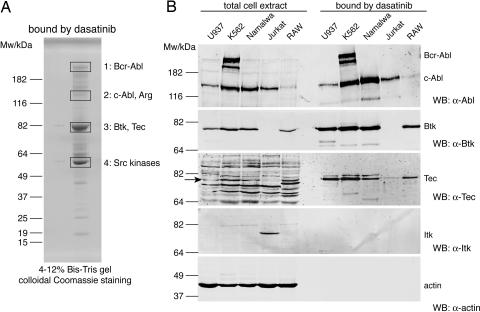
Fig. 1.
Dasatinib binds Btk and Tec, but not Itk. (A) Immobilized dasatinib was incubated with K562 total cell extract and bound proteins eluted by boiling in SDS sample buffer, resolved on a 4–12% NuPAGE gel followed by colloidal Coomassie staining. Bands 1–4 were excised, digested with trypsin, and analyzed by LC-MSMS (see SI Table 1). Main proteins identified in bands 1–4 are indicated. (B) Total cell extracts from U937, K562, Namalwa, Jurkat, and RAW cells and proteins that were bound by immobilized dasatinib were immunoblotted for Abl and the Tec kinases Btk, Tec, and Itk, as well as for actin as a loading and specificity control. Fifty micrograms of total cell extract and 5% of the fractions that were bound by immobilized dasatinib were loaded.
Besides Bcr-Abl, c-Abl, Arg (ABL2), and several Src kinases, two Tec kinases (Btk and Tec) were identified as the most prominent interactors of dasatinib (Fig. 1A and SI Table 1). A panel of 12 unrelated compounds was chosen as specificity control. None of them bound to Btk or Tec (data not shown). In addition, a comprehensive analysis of all proteins bound by dasatinib led to the identification of a large number of Ser/Thr- and Tyr-kinases (O.H. and U.R., unpublished results).
Dasatinib Binds Btk and Tec but Not Itk.
We performed drug pulldown experiments using dasatinib with a panel of hematopoietic cell lines and analyzed bound proteins by immunoblotting (Fig. 1B). The dasatinib resin retrieved both Bcr-Abl and c-Abl from K562 cells and c-Abl in Bcr-Abl-negative immortalized cell lines, such as U937 (human monocyte), Namalwa (human Burkitt lymphoma), Jurkat (human T lymphocyte), and RAW264.7 (mouse macrophage). We could confirm binding of Btk and Tec to immobilized dasatinib in K562, U937, Namalwa, and RAW264.7 cells. In contrast, exposure to extracts derived from Jurkat cells that express low levels of Tec and high levels of Itk, another member of the Tec kinases, resulted in small amounts of bound Tec and no detectable binding of Itk (Fig. 1B).
This suggests that Itk, unlike Btk and Tec, is not targeted by dasatinib, despite the high sequence homology and the conserved structure of the three kinases (refs. 26 and 27; SI Fig. 7).
Dasatinib Inhibits Btk and Tec Kinase Activity in Vitro and in Cells.
Next, we investigated the impact of dasatinib on the kinase activity of Btk, Tec, and Itk. In vitro kinase assays using full-length purified recombinant kinases showed that dasatinib inhibited Btk with an IC50 of 5 nM, a potency in the same range as for Abl (IC50 = 14 nM; Fig. 2A). Tec was also inhibited by dasatinib but with lower potency (IC50 = 297 nM), whereas Itk was completely resistant to dasatinib inhibition (Fig. 4C).
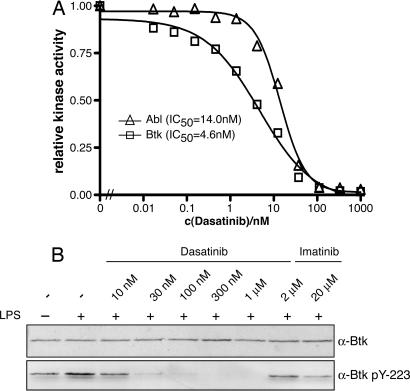
Fig. 2.
Dasatinib is a very potent inhibitor of Btk. (A) Full-length recombinant c-Abl and Btk were assayed in parallel for catalytic activity by in vitro kinase assays at the indicated concentrations of dasatinib and an optimal Abl substrate peptide as substrate. The graph shows the mean of one representative experiment done in triplicate and relative inhibition, in which kinase activity in the absence of dasatinib is set to 1.0. (B) Vitamin D3-differentiated U937 cells were either left untreated or stimulated with 1 μg/ml LPS for 6 h and treated with the indicated concentrations of dasatinib or imatinib for 1 h. Total cell extracts were prepared, and equal amounts of total protein were immunblotted with the indicated antibodies.
Fig. 4.
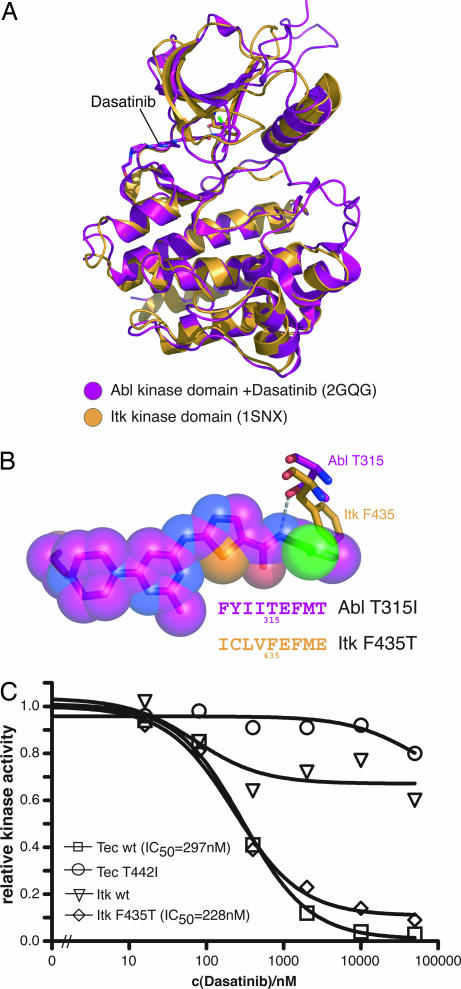
Mutation of the gatekeeper residue in Itk confers sensitivity to dasatinib. (A) Structural superposition of the Itk kinase domain (PDB ID code 1K2P) and the Abl kinase domain in complex with dasatinib (PDB ID code 2GQG) identifies Phe-435 in Itk as structural homologous residue to the gatekeeper residue Thr-315 in Abl. (B) Closeup view of dasatinib bound to the Abl kinase domain and the gatekeeper residues Thr-315 in Abl and Phe-435 in Itk showing hydrogen bonding of the Cβ-OH group of Thr-315 in Abl with dasatinib (gray dashed line) and sterical clash of Phe-435 in Itk with dasatinib. All other residues are omitted for graphical clarity. (C) TAP-tagged Tec WT, Tec T442I, Itk WT, and Itk F435T were purified by IgG Sepharose precipitation, and catalytic activity was determined at the indicated concentrations of dasatinib and an optimal Abl substrate peptide as substrate. The graph shows the mean of two experiments done in duplicate and relative inhibition, in which kinase activity in the absence of dasatinib is set to 1.0.
To assess the activity of dasatinib in cultured cells, vitamin D3-differentiated U937 cells were chosen, which activate Btk in response to LPS stimulation (28). Dasatinib (30 nM) led to a clear reduction of Btk autophosphorylation on Tyr-223, which is used as a marker for in vivo Btk kinase activity (29). Higher concentrations completely abolished autophosphorylation on Tyr-223, indicating complete inhibition of Btk kinase activity by dasatinib at concentrations of ≥100 nM (Fig. 2B).
In conclusion, dasatinib is a very potent inhibitor of Btk and Tec but not of Itk.
Btk T474I and Tec T442I Are Resistant to Dasatinib.
Next, we assessed the structural requirements for the binding of dasatinib to Btk and Tec. The only clinically relevant Bcr-Abl mutation that confers resistance to dasatinib is the T315I (gatekeeper) mutation (12). Structural alignment of the crystal structure of the Btk kinase domain and the structure of the Abl kinase domain complexed with dasatinib indicated that Thr-474 in Btk is structurally homologous to Thr-315 in Abl (refs. 26 and 25; Fig. 3A). We therefore mutated Thr-474 to Ile in Btk and created stable cell lines expressing tandem affinity purification (TAP)-tagged versions of the WT and T474I variant of Btk. We isolated TAP-tagged Btk WT and Btk T474I by IgG Sepharose precipitation and assayed for Btk kinase activity using the immobilized kinase. Btk WT was inhibited by dasatinib with an IC50 of 1.3 nM, whereas Btk T474I was completely resistant to dasatinib (Fig. 3B), supporting the hypothesis that the interaction mode of dasatinib with Btk resembles that of Abl.
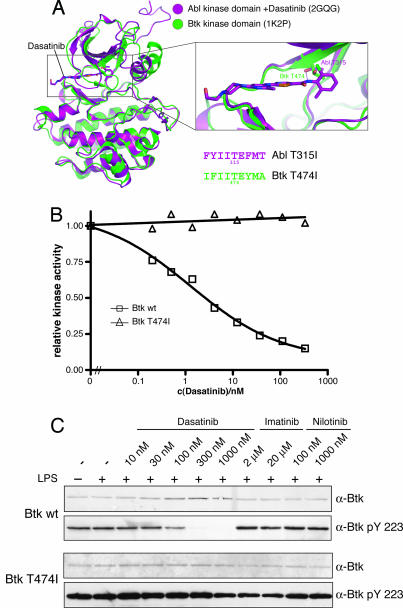
Fig. 3.
The gatekeeper residue is a major determinant of dasatinib sensitivity in Btk. (A) Structural superposition of the Btk kinase domain [Protein Data Bank (PDB) ID code 1K2P] and the Abl kinase domain in complex with dasatinib (PDB ID code 2GQG) identifies Thr-474 in Btk as structural homologous residue to the gatekeeper residue Thr-315 in Abl. (B) TAP-tagged Btk WT and Btk T474I were purified by IgG Sepharose precipitation, and catalytic activity was determined at the indicated concentrations of dasatinib and an optimal Abl substrate peptide as substrate. The graph shows the mean of two experiments done in duplicate and relative inhibition, in which kinase activity in the absence of dasatinib is set to 1.0. (C) Vitamin D3-differentiated U937 cells were retrovirally transduced with either Btk WT (Upper) or Btk T474I (Lower) and either left untreated or stimulated with 1 μg/ml LPS for 6 h and treated with the indicated concentrations of dasatinib, imatinib, or nilotinib for 1 h. Total cell extracts were prepared, and equal amounts of total protein were immunoblotted with the indicated antibodies.
To monitor Btk activity in vivo, we generated cell lines expressing untagged Btk WT and Btk T474I and assayed for Btk autophosphorylation on Tyr-223 in the presence of different concentrations of dasatinib, imatinib, and another second-generation Bcr-Abl inhibitor, nilotinib (Tasigna; ref. 30). As expected, Btk autophosphorylation on Tyr-223 is inhibited only by dasatinib but not by imatinib or nilotinib (Fig. 3C Upper). On the contrary, Btk T474I autophosphorylation on Tyr-223 is completely resistant to dasatinib (Fig. 3C Lower). Similar results were obtained for Tec upon mutation of the gatekeeper residue (Thr-442) to Ile (Fig. 4C and data not shown). Furthermore, dasatinib also inhibits Btk-induced tyrosine phosphorylation in Namalwa cells. Tyrosine phosphorylation is restored by introducing the gatekeeper mutation in Btk (SI Fig. 8).
This highlights the importance of the gatekeeper residue in Btk and Tec for dasatinib binding and inhibition.
Mutation of the Gatekeeper Residue in Itk Confers Sensitivity to Dasatinib.
As described above, Itk is neither bound nor inhibited by dasatinib (Figs. 1B and 4C). Closer inspection of the Itk kinase domain structure revealed the presence of a bulky amino acid residue in the gatekeeper position (Phe-435, ref. 27) that may sterically preclude binding of dasatinib to the active site of the kinase (Fig. 4 A and B). Furthermore, a critical hydrogen bond between the gatekeeper residue and dasatinib cannot be formed (Fig. 4B). Because all other residues that would interact with dasatinib are conserved between Itk and Btk, we reasoned that mutation of the gatekeeper residue Phe-435 to Thr could make Itk susceptible to dasatinib inhibition. To test this hypothesis, we created the corresponding mutation in Itk and assayed Itk kinase activity in vitro, as described above for Btk and Tec. Strikingly, mutation of Phe-435 to Thr in Itk renders Itk susceptible to dasatinib inhibition (Fig. 4C). Itk F435T has a similar IC50 for dasatinib as Tec WT (228 nM).
This shows that the gatekeeper residue is a major determinant for dasatinib sensitivity for the Tec kinases.
Dasatinib Blocks the Secretion of Proinflammatory Cytokines.
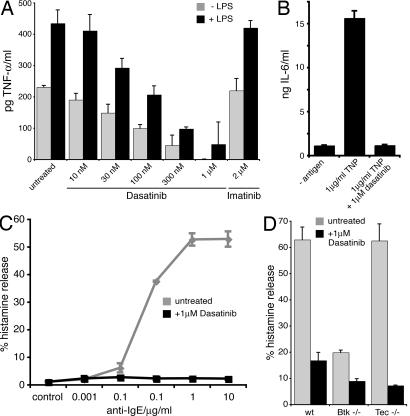
To assess the physiological consequences of Btk inhibition, we determined the impact of dasatinib on the secretion of TNF-α and IL-6, which were described to depended on Btk (28, 31, 32). Firstly, TNF-α release of vitamin D3-differentiated U937 cells was measured in the presence of different concentrations of dasatinib and imatinib. Dasatinib treatment led to a dose-dependent decrease of both basal and LPS-induced TNF-α secretion and almost complete inhibition at 1 μM dasatinib. In contrast, 2 μM imatinib did not have an impact on either basal or LPS-induced TNF-α secretion (Fig. 5A). Similarly, 1 μM dasatinib completely blocked the antigen-induced secretion of IL-6 in murine mast cells (Fig. 5B). This indicates that inhibition of Btk by dasatinib results in the impaired secretion of proinflammatory cytokines.
Fig. 5.
Dasatinib inhibits the secretion of immunomodulators. (A) U937 cells were differentiated with vitamin D3, pretreated with the indicated concentrations of drugs for 1 h and stimulated with LPS for 6 h. TNF-α secretion was measured by ELISA. The graph shows the mean of two experiments done in duplicate. (B) IgE primed WT mast cells were activated with TNP for 24 h in the presence of 1 μM dasatinib. IL-6 secreted into the culture medium was determined by ELISA. (C) Basophils from a healthy donor were preincubated in control medium or 1 μM dasatinib (Bristol-Myers Squibb, Princeton, NJ) for 30 min. Cells were exposed to various concentrations of an anti-IgE antibody (37°C, 30 min) as indicated. Histamine was measured in cell lysates (total histamine) and cell-free supernatants. Histamine release is expressed as percent of total histamine and represents the mean of triplicate determinations in one donor. (D) BMMCs from WT, Btk−/−, and Tec−/− mice were preincubated in medium with or without 1 μM dasatinib for 30 min. Then, cells were washed and exposed to 1 μg/ml TNP (37°C, 30 min). Histamine release was measured as in C.
Dasatinib Inhibits Histamine Release.
It has been demonstrated that Btk plays a major role in histamine release (31, 32). Therefore, we evaluated the functional consequences of Btk inhibition by dasatinib on antigen-induced histamine release from primary human basophils. Cross-linking with an anti-IgE antibody led to a strong induction of histamine release in these cells that was completely abrogated by 1 μM dasatinib (Fig. 5C). In agreement with this, antigen-induced histamine release was significantly decreased in WT bone marrow-derived murine mast cells (BMMCs) by dasatinib. Notably, histamine release was decreased to a similar extent in BMMCs derived from Btk-deficient mice (Fig. 5D). In contrast, histamine release was not impaired in Tec-deficient BMMCs (Fig. 5D; U.S. and W.E., unpublished results). This indicates that Btk plays a predominant role in this process. In conclusion, these experiments suggest that dasatinib-mediated inhibition of Btk may be the cause for the observed reduction in histamine release. This is consistent with the notion that Btk was also identified among the most prominent targets of dasatinib in a dasatinib pulldown from murine BMMCs (data not shown).
Discussion
We performed a chemical proteomics profiling approach that preserves as much as possible the native configuration of endogenous protein kinases, including posttranslational modifications and binding partners. One of the major advantages of the chemical proteomics approach is its unbiased nature (20). Kinases that are difficult to clone or express in full-length form are not included in the profiling panels of screening companies (15, 19). For example, the chemical proteomics approach identified Tec as a dasatinib target, a protein that could not be identified with previously published drug-profiling approaches using recombinant proteins, as Tec was not included in any of the panels used (15). In addition, the chemical proteomics strategy is not only limited to kinases but captures any relevant drug–protein interaction in the relevant cell type or tissue, in which they act in their natural cellular context and bear their essential posttranslational modifications.
Using this approach and biochemical validation, we found the Tec kinases Btk and Tec to be very prominent targets of dasatinib. In addition, we have identified the gatekeeper residue as a critical determinant for dasatinib sensitivity. Small gatekeeper residues, like Thr in Btk and Tec, allow for dasatinib binding, whereas bulky gatekeeper residues, like Phe in Itk, block dasatinib binding. Based on this concept, we predict that the two remaining Tec family members, Bmx and Txk, may also be dasatinib targets, because they have a Thr in the gatekeeper position (SI Fig. 7). Based on our findings for the Tec kinases, we have analyzed the identity of the gatekeeper residue in the kinases that were bound by dasatinib. Among the 94 kinases that have a Thr in the gatekeeper position, so far, 60 kinases have been found to be targeted by dasatinib (ref. 15; U.R., unpublished results). In contrast, only five further dasatinib targets were identified so far: Four with a Met (of 183 kinases in the kinome) and one with a Val (of 15 kinases in the kinome) in the gatekeeper position (see SI Table 2). Considering the conservation of the other residues that contact dasatinib, this strongly implies that the gatekeeper residue can be used as a predictor for dasatinib sensitivity on a kinome-wide scale. This prompts the conclusion that the binding mode of dasatinib to kinases is highly promiscuous in general, but certain selectivity is determined by the identity of the gatekeeper residue.
We show that dasatinib inhibits the secretion of histamine and IL-6 from mast cells and basophils, as well as TNF-α secretion from U937 cells, all of which depend on the kinase activity of Btk (28, 31). Dasatinib is an inhibitor with a broad specificity, also termed multitarget inhibitor. As a consequence, it is very difficult to show that inhibition of a particular physiological readout by dasatinib critically depends on Btk and not on other dasatinib target upstream or downstream of Btk, like Src kinases. This would require a pathway, in which Btk is the only dasatinib target that critically controls the pathway. We expressed Btk wt and Btk T474I in various cellular systems, including Btk-deficient BMMCs and assayed for the effect of dasatinib on histamine release. Although we successfully reconstituted Btk-deficient BMMCs with Btk WT and Btk T474I, biochemically we have not been able to assign an unequivocal role for exclusive targeting of Btk by dasatinib. Introduction of dasatinib-resistant Btk may not be able to restore signaling, as the pathway may be blocked both upstream and downstream.
Up to now, only Bcr-Abl and Src kinases have been considered as relevant targets of dasatinib in vivo (11). In addition to Tec kinases, we have identified a large number of dasatinib targets (U.R., unpublished results). In agreement with this, many physiological processes are impaired by dasatinib. This may have several consequences: (i) The increased potency of dasatinib versus imatinib in advanced phase and blast crisis CML might highlight the benefit of a kinase inhibitor with a broader specificity that may target additional kinases upstream or downstream of Bcr-Abl, thereby decreasing the overall oncogenic potency of the Bcr-Abl effect and possibly counteract the outgrowth of resistant cell clones (10, 33). (ii) A multitarget inhibitor like dasatinib is more likely to cause adverse effects. Indeed, dasatinib, although well tolerated in most cases, can induce severe side effects in leukemia patients, including pleural effusion, arthralgia, fever and cytopenias (34, 35). Based on our data and studies conducted in Btk deficiency models, we predict that dasatinib acts as a potent immunosuppressor by affecting B and T cell development, as well as leukocyte function, which may be detrimental in long-term treatment of leukemia. (iii) Dasatinib might be of clinical benefit in conditions with excessive or uncontrolled production of immunomodulators. These include a diverse array of diseases ranging from rheumatoid arthritis, Crohn's disease and systemic lupus erythematosus to allergies and allergic asthma. In addition, inhibition of Btk by dasatinib might be of clinical benefit in pre-B lymphoblastic leukemia, in which Btk activity is essential for survival signals (36). Whether dasatinib can counteract basophil-, mast cell-, or/and lymphocyte activation in atopic patients or chronic inflammatory disorders remains to be determined in forthcoming studies.
Materials and Methods
Kinase Inhibitors.
For the basophil histamine release experiments of Fig. 5C, dasatinib (BMS-354825) was kindly provided by Francis Y. Lee (Bristol-Myers Squibb, Princeton, NJ). In all other experiments, dasatinib, c-dasatinib, imatinib, and nilotinib were used that were synthesized by WuXi PharmaTech (Shanghai, China).
Affinity Purification and MS Data Analysis.
Pulldown experiments were performed and analyzed by LC-MSMS by using total lysates of K562 cells. The acquired data were searched using Mascot (MatrixScience, London, U.K.) against the human International Protein Index database, version 3.12 (see SI Text).
DNA Constructs and Mutagenesis.
ESTs for Btk, Tec, and Itk were obtained from German Resource Center for Genome Research (RZPD) and cloned using the Gateway cloning system (Invitrogen, Carlsbad, CA). Point mutations were introduced using the Quick change site-directed mutagenesis Kit (Stratagene, La Jolla, CA).
Inhibition Assays for Recombinant Kinases.
Dasatinib was assayed in vitro for inhibition of recombinant full-length c-Abl and Btk (Upstate Biotechnology, Charlottesville, NC). A peptide with the preferred c-Abl substrate sequence (37) carrying an N-terminal biotin (biotin-GGEAIYAAPFKK-amide) was used as substrate. The terminated reaction was spotted onto a SAM2 Biotin Capture membrane (Promega, Madison, WI) and further treated according to the instructions of the manufacturer.
Inhibition Assays for TAP-Tagged Kinases.
TAP-tagged Tec kinases were stably transduced in Namalwa cells by retroviral gene transfer. Cells were lysed in lysis buffer (50 mM Tris, pH 7.5/125 mM NaCl/5% glycerol/0.2% Nonidet P-40/1.5 mM MgCl2/25 mM NaF/1 mM Na3VO4/protease inhibitors), and the lysate was cleared by centrifugation. The lysate was incubated with rabbit-IgG agarose (Sigma, St. Louis, MO) at 4°C for 2 h. Beads were washed twice with lysis buffer and once in kinase assay buffer (20 mM Tris·HCl, pH 7.5/5 mM MgCl2/5 mM MnCl2/1 mM DTT) and assayed for kinase activity as described above.
TNF-α Cytokine ELISA.
U937 cells were differentiated with 100 nM 1,25-dihydroxyvitamin D3 for 48 h, pretreated with the indicated concentrations of dasatinib and imatinib for 1 h, and stimulated with 1 μg/ml LPS for 6 h. Supernatants were harvested, cleared by centrifugation, and assayed for the production of human TNF-α by ELISA (R&D Systems, Minneapolis, MN) according to the instructions of the manufacturer.
Generation and Activation of BMMCs.
BMMCs of WT or Btk−/− mice (mixed background of C57BL/6 × 129 Sv; 4–6 weeks of age) were generated and assayed for IL-6 and histamine release as described in SI Text.
Histamine Release from Human Basophils.
Dextran-enriched basophils from a healthy donor were incubated in the presence or absence of dasatinib from Bristol-Myers Squibb (1 μM) for 30 min, washed, and incubated in various concentrations of anti-IgE (E.124.2.8, clone Dε2) for 30 min, and the cell-free supernatants were recovered. Histamine was measured in whole-cell suspensions (total histamine) and cell-free supernatants by RIA (Immunotech, Vaudreuil-Dorion, PQ, Canada). Histamine release was expressed as percent of total histamine.
Supplementary Material
Acknowledgments
We thank G. Dürnberger for statistical analysis of the MS data; M. Planyavsky for MS sample preparation; H. Kaltenegger and I. Kaupe for skillful technical assistance; M. Sibilia, R. Kralovics, and G. Drewes for the critical reading of the manuscript; and all members of the participating laboratories for help and discussions. This study was supported by the Austrian Academy of Sciences, Grant P18737 from the Fonds zur Förderung der Wissenschaftlichen Forschung, DRAGON Grant GZ 200.142/1-VI/I/2006 of the Gen-AU program of the Austrian Ministry of Research, and Grants BU 2180/1-1 (to T.B.) and Schm 2128/1-1 (to U.S.) from the DeutscheForschungsgemeinschaft.
Abbreviations
- CML
chronic myelogenous leukemia
- LC-MSMS
liquid chromatography tandem MS
- BMMC
bone marrow-derived murine mast cells
- TAP
tandem affinity purification.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702654104/DC1.
References
- 1.Wong S, Witte ON. Annu Rev Immunol. 2004;22:247–306. doi: 10.1146/annurev.immunol.22.012703.104753. [DOI] [PubMed] [Google Scholar]
- 2.Deininger M, Buchdunger E, Druker BJ. Blood. 2005;105:2640–2653. doi: 10.1182/blood-2004-08-3097. [DOI] [PubMed] [Google Scholar]
- 3.Schindler T, Bornmann W, Pellicena P, Miller WT, Clarkson B, Kuriyan J. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 4.Druker BJ, Guilhot F, O'Brien SG, Gathmann I, Kantarjian H, Gattermann N, Deininger MW, Silver RT, Goldman JM, Stone RM, et al. N Engl J Med. 2006;355:2408–2417. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 5.Buchdunger E, Zimmermann J, Mett H, Meyer T, Muller M, Druker BJ, Lydon NB. Cancer Res. 1996;56:100–104. [PubMed] [Google Scholar]
- 6.Nagar B, Bornmann WG, Pellicena P, Schindler T, Veach DR, Miller WT, Clarkson B, Kuriyan J. Cancer Res. 2002;62:4236–4243. [PubMed] [Google Scholar]
- 7.Nagar B, Hantschel O, Young MA, Scheffzek K, Veach D, Bornmann W, Clarkson B, Superti-Furga G, Kuriyan J. Cell. 2003;112:859–871. doi: 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 8.Mol CD, Dougan DR, Schneider TR, Skene RJ, Kraus ML, Scheibe DN, Snell GP, Zou H, Sang BC, Wilson KP. J Biol Chem. 2004;279:31655–31663. doi: 10.1074/jbc.M403319200. [DOI] [PubMed] [Google Scholar]
- 9.Shah N, Nicoll J, Nagar B, Gorre M, Paquette R, Kuriyan J, Sawyers C. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 10.O'Hare T, Corbin AS, Druker BJ. Curr Opin Genet Dev. 2006;16:92–99. doi: 10.1016/j.gde.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Lombardo LJ, Lee FY, Chen P, Norris D, Barrish JC, Behnia K, Castaneda S, Cornelius LA, Das J, Doweyko AM, et al. J Med Chem. 2004;47:6658–6661. doi: 10.1021/jm049486a. [DOI] [PubMed] [Google Scholar]
- 12.Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 13.Kantarjian H, Jabbour E, Grimley J, Kirkpatrick P. Nat Rev Drug Discov. 2006;5:717–718. doi: 10.1038/nrd2135. [DOI] [PubMed] [Google Scholar]
- 14.O'Hare T, Walters DK, Stoffregen EP, Jia T, Manley PW, Mestan J, Cowan-Jacob SW, Lee FY, Heinrich MC, Deininger MW, Druker BJ. Cancer Res. 2005;65:4500–4505. doi: 10.1158/0008-5472.CAN-05-0259. [DOI] [PubMed] [Google Scholar]
- 15.Carter TA, Wodicka LM, Shah NP, Velasco AM, Fabian MA, Treiber DK, Milanov ZV, Atteridge CE, Biggs WH, III, Edeen PT, et al. Proc Natl Acad Sci USA. 2005;102:11011–11016. doi: 10.1073/pnas.0504952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burdine L, Kodadek T. Chem Biol. 2004;11:593–597. doi: 10.1016/j.chembiol.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Guiffant D, Tribouillard D, Gug F, Galons H, Meijer L, Blondel M, Bach S. Biotechnol J. 2007;2:68–75. doi: 10.1002/biot.200600223. [DOI] [PubMed] [Google Scholar]
- 18.Wissing J, Godl K, Brehmer D, Blencke S, Weber M, Habenberger P, Stein-Gerlach M, Missio A, Cotten M, Muller S, Daub H. Mol Cell Proteom. 2004;3:1181–1193. doi: 10.1074/mcp.M400124-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Fabian MA, Biggs WH, III, Treiber DK, Atteridge CE, Azimioara MD, Benedetti MG, Carter TA, Ciceri P, Edeen PT, Floyd M, et al. Nat Biotechnol. 2005;23:329–336. doi: 10.1038/nbt1068. [DOI] [PubMed] [Google Scholar]
- 20.Daub H. Biochim Biophys Acta. 2005;1754:183–190. doi: 10.1016/j.bbapap.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 21.Smith CI, Islam TC, Mattsson PT, Mohamed AJ, Nore BF, Vihinen M. BioEssays. 2001;23:436–446. doi: 10.1002/bies.1062. [DOI] [PubMed] [Google Scholar]
- 22.Hantschel O, Superti-Furga G. Nat Rev Mol Cell Biol. 2004;5:33–44. doi: 10.1038/nrm1280. [DOI] [PubMed] [Google Scholar]
- 23.Lewis CM, Broussard C, Czar MJ, Schwartzberg PL. Curr Opin Immunol. 2001;13:317–325. doi: 10.1016/s0952-7915(00)00221-1. [DOI] [PubMed] [Google Scholar]
- 24.Vihinen M, Mattsson PT, Smith CI. Front Biosci. 2000;5:D917–D928. doi: 10.2741/vihinen. [DOI] [PubMed] [Google Scholar]
- 25.Tokarski JS, Newitt JA, Chang CY, Cheng JD, Wittekind M, Kiefer SE, Kish K, Lee FY, Borzillerri R, Lombardo LJ, et al. Cancer Res. 2006;66:5790–5797. doi: 10.1158/0008-5472.CAN-05-4187. [DOI] [PubMed] [Google Scholar]
- 26.Mao C, Zhou M, Uckun FM. J Biol Chem. 2001;276:41435–41443. doi: 10.1074/jbc.M104828200. [DOI] [PubMed] [Google Scholar]
- 27.Brown K, Long JM, Vial SC, Dedi N, Dunster NJ, Renwick SB, Tanner AJ, Frantz JD, Fleming MA, Cheetham GM. J Biol Chem. 2004;279:18727–18732. doi: 10.1074/jbc.M400031200. [DOI] [PubMed] [Google Scholar]
- 28.Horwood NJ, Mahon T, McDaid JP, Campbell J, Mano H, Brennan FM, Webster D, Foxwell BM. J Exp Med. 2003;197:1603–1611. doi: 10.1084/jem.20021845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park H, Wahl MI, Afar DE, Turck CW, Rawlings DJ, Tam C, Scharenberg AM, Kinet JP, Witte ON. Immunity. 1996;4:515–525. doi: 10.1016/s1074-7613(00)80417-3. [DOI] [PubMed] [Google Scholar]
- 30.Weisberg E, Manley PW, Breitenstein W, Bruggen J, Cowan-Jacob SW, Ray A, Huntly B, Fabbro D, Fendrich G, Hall-Meyers E, et al. Cancer Cell. 2005;7:129–141. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 31.Hata D, Kawakami Y, Inagaki N, Lantz CS, Kitamura T, Khan WN, Maeda-Yamamoto M, Miura T, Han W, Hartman SE, et al. J Exp Med. 1998;187:1235–1247. doi: 10.1084/jem.187.8.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawakami Y, Kitaura J, Satterthwaite AB, Kato RM, Asai K, Hartman SE, Maeda-Yamamoto M, Lowell CA, Rawlings DJ, Witte ON, Kawakami T. J Immunol. 2000;165:1210–1219. doi: 10.4049/jimmunol.165.3.1210. [DOI] [PubMed] [Google Scholar]
- 33.Martinelli G, Soverini S, Rosti G, Baccarani M. Leukemia. 2005;19:1872–1879. doi: 10.1038/sj.leu.2403950. [DOI] [PubMed] [Google Scholar]
- 34.Talpaz M, Shah NP, Kantarjian H, Donato N, Nicoll J, Paquette R, Cortes J, O'Brien S, Nicaise C, Bleickardt E, et al. N Engl J Med. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 35.Kantarjian H, Pasquini R, Hamerschlak N, Rousselot P, Holowiecki J, Jootar S, Robak T, Khoroshko N, Masszi T, Skotnicki A, et al. Blood. 2007;109:5143–5150. doi: 10.1182/blood-2006-11-056028. [DOI] [PubMed] [Google Scholar]
- 36.Feldhahn N, Klein F, Mooster JL, Hadweh P, Sprangers M, Wartenberg M, Bekhite MM, Hofmann WK, Herzog S, Jumaa H, et al. J Exp Med. 2005;201:1837–1852. doi: 10.1084/jem.20042101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou S, Carraway KL, III, Eck MJ, Harrison SC, Feldman RA, Mohammadi M, Schlessinger J, Hubbard SR, Smith DP, Eng C, et al. Nature. 1995;373:536–539. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.