Abstract
Molecularly targeted kinase inhibitor cancer therapies are currently administered sequentially rather than simultaneously. We addressed the potential long-term impact of this strategy in patients with chronic myelogenous leukemia (CML), which is driven by the fusion oncogene BCR-ABL. Analysis of BCR-ABL genotypes in CML patients who relapsed after sequential treatment with the ABL inhibitors imatinib and dasatinib revealed evolving resistant BCR-ABL kinase domain mutations in all cases. Twelve patients relapsed with the pan-resistant T315I mutation, whereas 6 patients developed novel BCR-ABL mutations predicted to retain sensitivity to imatinib based on in vitro studies. Three of these patients were retreated with imatinib (or the chemically related compound nilotinib) and responded; however, selection for compound mutants (2 or 3 BCR-ABL mutations in the same molecule) can substantially limit the potential effectiveness of retreating patients with inhibitors that have previously failed. Furthermore, drug-resistant mutations, when compounded, can increase oncogenic potency relative to the component mutants in transformation assays. The Aurora kinase inhibitor VX-680, currently under clinical evaluation based on its activity against the T315I mutation, is also effective against the other commonly detected dasatinib-resistant mutation in our analysis, V299L. Our findings demonstrate the potential hazards of sequential kinase inhibitor therapy and suggest a role for a combination of ABL kinase inhibitors, perhaps including VX-680, to prevent the outgrowth of cells harboring drug-resistant BCR-ABL mutations.
Introduction
An important issue in the management of human malignancies with kinase inhibitors relates to the timing of therapies. The current strategy, best exemplified in chronic myelogenous leukemia (CML), is sequential treatment. Newly diagnosed patients receive the first-generation ABL inhibitor imatinib (Gleevec), followed by the second-generation ABL inhibitor dasatinib (SPRYCEL) at time of resistance or intolerance. The rationale for this approach is partly historical, since imatinib was approved for CML therapy prior to dasatinib on the basis of a very high single-agent response rate, and partly based on a molecular understanding of resistance mechanisms that led to the evaluation of dasatinib in imatinib-resistant CML. Most imatinib-resistant BCR-ABL point mutations (of more than 50 distinct examples reported clinically) impair drug binding by restricting flexibility of the enzyme, destabilizing the inactive conformation required for imatinib binding (1, 2). Dasatinib binds the BCR-ABL kinase domain in the active conformation and is therefore effective against nearly all imatinib-resistant mutations (3–5). The primary exception is the T315I “gatekeeper” mutation at the base of the ATP-binding pocket, which confers resistance to imatinib, dasatinib, and the imatinib-related compound nilotinib (AMN107) through steric hindrance (3, 5–7). Clinically, dasatinib is approved as second-line CML therapy after failure of imatinib, as it was initially evaluated in imatinib-resistant or -intolerant patients. As predicted from preclinical studies, the clinical activity of dasatinib is genotype dependent. All patients with pretreatment T315I mutations failed to respond, whereas wild-type BCR-ABL and all other represented imatinib-resistant genotypes were associated with clinical responses (8).
As clinical experience with dasatinib grows, it is becoming clear that patients can also relapse on treatment after an initial response, particularly in the setting of advanced-phase CML. One mechanism of acquired resistance anticipated from preclinical studies and from the genotype/response studies of patients prior to treatment is selection for rare subclones harboring the T315I mutation. Indeed, we previously recovered T315I mutations commonly in an in vitro saturation mutagenesis screen for dasatinib-resistant BCR-ABL point mutations (9). We also identified a small number of additional mutations that confer resistance to dasatinib but not imatinib, none of which had been previously detected in imatinib-treated patients. These novel mutations map to critical drug contact residues in the ABL-dasatinib cocrystal structure and presumably cause resistance through steric hindrance mechanisms, similar to T315I (9). It is also plausible that the broad activity of dasatinib against other kinases such as SRC family members may play a role in its clinical activity; therefore, dysregulation of targets other than BCR-ABL may play a role in clinical resistance.
Now that patients are beginning to experience clinical relapses on dasatinib, we assessed whether T315I or any of the uniquely dasatinib-resistant mutations may play a role. The latter are of particular interest as these clones should retain sensitivity to imatinib and raise the possibility that such patients might benefit from retreatment with that drug. Therefore, we determined the BCR-ABL genotype of CML patients who initially responded and relapsed on imatinib, then responded and relapsed on dasatinib. All patients had new BCR-ABL kinase domain mutations at second relapse, each of which were previously shown to confer dasatinib resistance in vitro. Twelve patients had T315I and 1 patient had F317L, whereas 6 had imatinib-sensitive, dasatinib-resistant mutations (V299L, T315A, or F317I) that matched those recovered from the earlier in vitro screen. Notably, the 2 patients with isolated V299L mutation responded to retreatment with imatinib or the imatinib-related compound nilotinib. However, this strategy of cycling between kinase inhibitors is potentially limited by the emergence of compound mutations that, when paired in the same molecule, confer resistance to both drugs and enhance BCR-ABL oncogenicity. We also show that the Aurora kinase inhibitor VX-680, currently in clinical development as a third-generation ABL inhibitor due to its activity against BCR-ABL/T315I (10), is also effective against the BCR-ABL/V299L mutation. These findings make a case for up-front therapy with a cocktail of kinase inhibitors that collectively cover a broad range of mutations and prevent the emergence of resistance.
Results
Acquired resistance to dasatinib is associated with novel BCR-ABL kinase domain mutations.
By sequence analysis of blood samples at the time of relapse, we determined the BCR-ABL kinase domain mutation status of 17 consecutive consenting patients treated at UCLA who developed resistance to dasatinib following imatinib. All patients had new BCR-ABL mutations at the time of relapse on dasatinib that were not detected at the time of relapse on imatinib (Figure 1 and Table 1). The pan-resistant T315I mutation was observed in 12 cases and the relatively resistant F317L mutation was observed in 1 case. Six patients (1 of whom also had T315I and F317L) had mutations previously isolated in our in vitro screen for dasatinib resistance that retain sensitivity to imatinib in vitro. Four patients had V299L, 1 patient had T315A, and 1 had F317I. The fact that all 17 patients acquired new ABL mutations provides evidence that CML cells remain dependent on BCR-ABL function even after multiple prior anti-ABL therapies and suggests that BCR-ABL–independent mechanisms of relapse are rare.
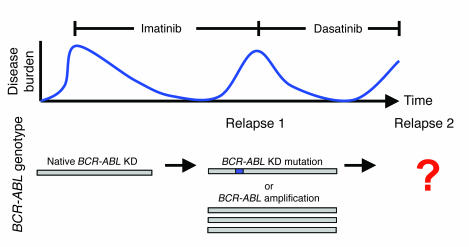
Figure 1. Sequential use of tyrosine kinase inhibitors can select for cells harboring compound kinase domain mutations.
Schematic of sequential kinase inhibitor therapy and predicted BCR-ABL kinase domain (KD) genotypes of dasatinib-resistant cases treated after imatinib failure. Blue box represents imatinib-resistant, dasatinib-sensitive mutation.
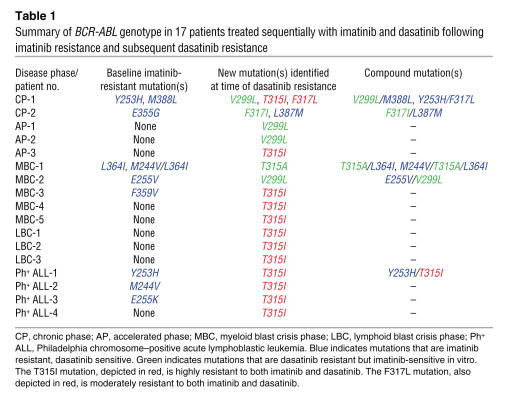
Table 1 .
Summary of BCR-ABL genotype in 17 patients treated sequentially with imatinib and dasatinib following imatinib resistance and subsequent dasatinib resistance
Compound BCR-ABL kinase domain mutations develop in patients treated sequentially with kinase inhibitor therapy.
Through subcloning and sequencing of individual clones, we determined that 5 of the 17 patients had compound mutations (2 or more codon changes in the same BCR-ABL mRNA) that were not detected prior to dasatinib treatment. Serial analysis of samples from 5 of these patients (with chronic-phase CML [designated CP-1 and CP-2]; myeloid blast crisis–phase CML [MBC-1 and MBC-2], and Philadelphia chromosome–positive acute lymphoblastic leukemia [Ph+ ALL-1]) revealed that while imatinib-resistant mutations were present at the initiation of dasatinib therapy, a compound mutation containing the original imatinib-resistant substitution plus a new dasatinib-resistant mutation was detected at the time of relapse on dasatinib. Therefore, clones expressing these compound mutants most likely arose through selective pressure for a second dasatinib-resistant mutation on the genetic background of an earlier imatinib-resistant mutation. Two patients (CP-1, MBC-1) had more than 1 mutation following imatinib failure, but in distinct clones (polyclonal resistance).
Patients with the dasatinib-resistant V299L mutation in isolation can respond to retreatment with imatinib or nilotinib.
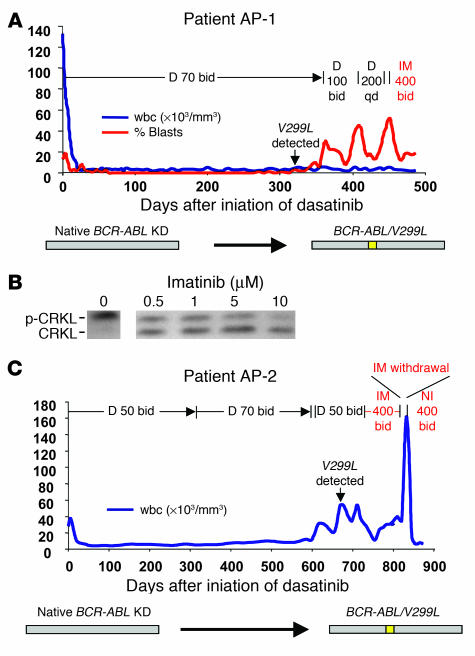
Two patients with accelerated-phase CML (AP-1 and AP-2) developed the V299L mutation in the absence of other detectable BCR-ABL kinase domain mutations. Because this mutation remains sensitive to imatinib in vitro (9), we explored the possibility of clinical benefit from retreatment with imatinib or the more potent imatinib analog nilotinib. At the time of BCR-ABL/V299L–driven relapse, patient AP-1 responded briefly to an escalated dose of dasatinib, then progressed with a rise in the percentage of peripheral blood myeloblasts. Therefore, dasatinib was discontinued and imatinib was started. The percentage of myeloblasts fell from 52% to 12% over 2 weeks after imatinib retreatment and was sustained another 2 weeks prior to starting chemotherapy for a planned allogeneic marrow transplant (Figure 2A). Consistent with this partial clinical response, ex vivo exposure of cells from this patient to imatinib reduced BCR-ABL kinase activity by greater than 50%, as measured by decreased phosphorylation of the direct BCR-ABL substrate CRKL (Figure 2B). At the time of BCR-ABL/V299L relapse, patient AP-2 had a transient response to high doses of imatinib, which was evident when it was withdrawn for 7 days, during which time the patient’s wbc count rapidly rose from 34,000 to 158,000. The patient then began investigational therapy with nilotinib, a more potent investigational derivative of imatinib (7), and achieved a complete hematologic response within 3 weeks, which has been sustained for at least 4 months (Figure 2C).
Figure 2. The dasatinib-resistant V299L mutation is clinically sensitive to imatinib and nilotinib.
(A) wbc count (blue) and peripheral blood myeloblast percentage (red) in an accelerated-phase CML patient (AP-1) who developed dasatinib resistance associated with the V299L mutation in BCR-ABL and responded to imatinib. (B) CRKL immunoblot performed after exposure of leukocytes isolated from patient AP-1 (harboring BCR-ABL/V299L) to varying concentrations of imatinib. (C) wbc count in a second accelerated-phase CML patient (AP-2) who developed resistance to dasatinib due to the V299L mutation and was sequentially treated with imatinib and nilotinib. D, dasatinib; I, imatinib; NI, nilotinib.
Compound BCR-ABL mutant alleles harboring both imatinib- and dasatinib-specific resistant mutations can respond to a combination of these agents.
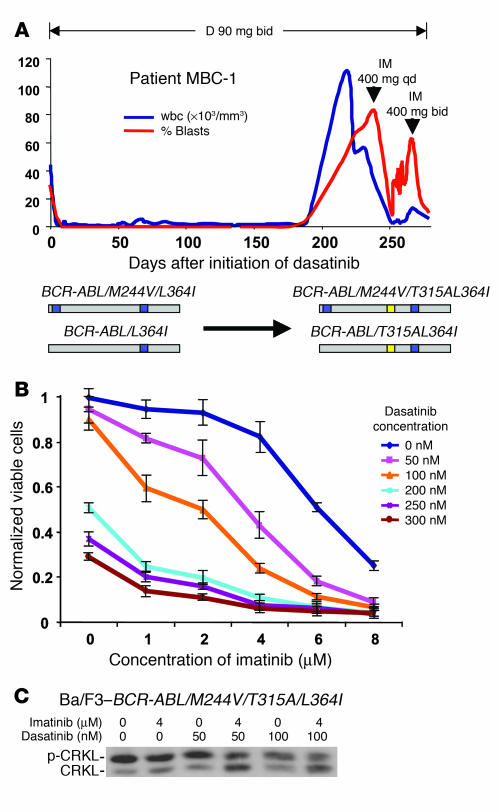
Patient MBC-1 was of particular interest due to evolution of a T315A mutation after a 6-month response to dasatinib (Figure 3A). Whereas BCR-ABL/T315I confers complete resistance to both imatinib and dasatinib, the T315A substitution remains sensitive to imatinib in vitro. Subcloning revealed that T315A developed in the context of 2 previously described imatinib-resistant mutations (M244V/L364I) that we detected in this patient at the time of initial relapse on imatinib, thereby creating a triple compound mutant (M244V/T315A/L364I). Despite the obvious concern that the compound mutant would negate any chance of T315A conferring sensitivity to imatinib retreatment, the patient elected to undergo a trial of combined imatinib and dasatinib therapy due to the lack of other treatment options. Remarkably, the wbc cell count fell from more than 50,000/mm3 to less than 1,000/mm3, and the percentage of peripheral blood myeloblasts dropped from 66% to 10% after addition of imatinib to the ongoing dasatinib dose (Figure 3A). This partial hematologic response was sustained for 2 months after the imatinib dose was adjusted to control a rise in blast percentage.
Figure 3. Additive BCR-ABL inhibitory properties of 2 drugs can lead to clinical responses in cases harboring compound mutations resistant to both agents.
(A) wbc count (blue) and peripheral blood myeloblast percentage (red) in MBC-1, who developed resistance associated with the compound mutation M244V/T315A/L364I in BCR-ABL in 22 of 27 clones sequenced, then responded to a combination of dasatinib and imatinib. This patient initially relapsed on imatinib with 2 subclones, 1 with the mutant L364I and a second clone bearing the M244V/L364I compound mutant (both M244V and L364I are known imatinib-resistant mutants). (B) Dasatinib and imatinib cooperatively inhibit the growth of Ba/F3 cells harboring BCR-ABL/M244V/T315A/L364I in vitro. Combination therapy had no effect on parental IL-3–dependent Ba/F3 cells (data not shown). (C) CRKL Western immunoblot of Ba/F3–BCR-ABL/V299L exposed to clinically relevant concentrations of imatinib and dasatinib.
To characterize the mechanism of this clinical response despite the compound M244V/T315A/L364I mutation, we compared the sensitivity of Ba/F3 cells engineered to express the patient’s initial M244V/L364I double mutation and the triple compound M244V/T315A/L364I mutation to imatinib, dasatinib, or the combination. M244V/L364I- and M244V/T315A/L364I-expressing cells were resistant to single-agent imatinib and dasatinib, respectively, at concentrations consistent with clinical relapse (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI30890DS1). However, imatinib plus dasatinib in combination suppressed the growth of M244V/T315A/L364I-expressing cells (Figure 3B) and inhibited BCR-ABL kinase activity (Figure 3C) at easily achievable clinical concentrations (50–100 nM dasatinib; 1–4 μM imatinib). This cooperativity is consistent with previous observations in studies of the efficacy of imatinib plus dasatinib against wild-type BCR-ABL (9, 11).
Compounding of 2 drug-resistant alleles with reduced oncogenic fitness can restore oncogenic potency.
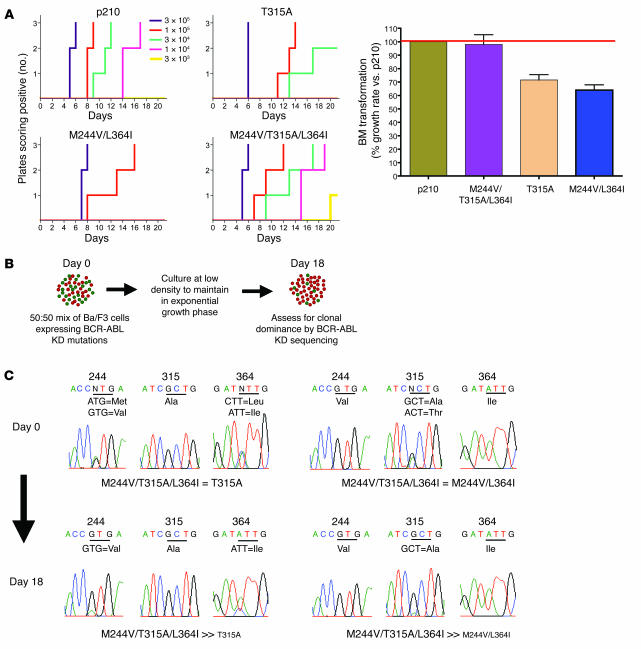
Kinase domain mutations associated with drug resistance can alter the leukemogenic potency of BCR-ABL, with some alleles showing increased oncogenic fitness, whereas others have weaker transformation potential (12, 13). The T315A mutation is notable because it displays severely attenuated oncogenicity in vitro (13), yet patient MBC-1 with the compound T315A mutant had a fulminant clinical relapse, raising the possibility that T315A in the context of other mutations might behave differently. Therefore, we compared the oncogenic potency of T315A to those of M244V/L364I and the triple mutant M244V/T315A/L364I using a modified Whitlock-Witte pre–B cell transformation assay, which can distinguish subtle and quantifiable differences in potency between different BCR-ABL isoforms (13). Potency relative to wild-type BCR-ABL is measured by comparing the number of positive (transformed) wells across a range of limiting dilutions. As expected, T315A had reduced oncogenicity relative to wild-type BCR-ABL, scoring only at the highest cell concentrations plated (Figure 4A). Surprisingly, the compound imatinib-resistant mutant M244V/L364I scored even more weakly; however, the triple compound mutant M244V/T315A/L364I was as potent as wild-type BCR-ABL. This finding was confirmed in cell growth competition assays (Figure 4B). Cells expressing BCR-ABL/M244V/T315A/L364I successfully outgrew both BCR-ABL/T315A and BCR-ABL/M244V/L364I (Figure 4C). Autophosphorylation of BCR-ABL was increased in these 3 mutants in comparison with wild-type BCR-ABL, but there were no obvious differences between the mutants that correlated with oncogenic fitness. (The ability to phosphorylate the exogenous peptide substrate Abltide was impaired in the T315A-containing mutants, consistent with previous observations [ref. 13], thereby precluding more precise determination of relative enzyme efficiency.) An alternative explanation for enhanced fitness of the triple mutant may be altered substrate recognition, as reported for BCR-ABL/T315I through phosphoproteome profiling studies (13). In either case, this example demonstrates that a multiply drug-resistant compound mutant evolving through sequential kinase inhibitor therapy may generate alleles with increased oncogenic potency.
Figure 4. Sequential kinase inhibitor therapy can result in compound mutations that increase transformation potency and growth rate in vitro.
(A) Bone marrow transformation assays with select drug-resistant BCR-ABL kinase domain mutants are shown. After infection with mutant or wild-type p210 BCR-ABL retrovirus, primary mouse bone marrow cells were plated at the serial dilutions indicated then monitored for oncogenic transformation. The time to outgrowth in assays performed in triplicate at 5 separate dilutions is shown on the left, and the right panel shows the percentage growth rate of various BCR-ABL isoforms relative to wild-type BCR-ABL. (B) Schematic of cell growth competition experiment. (C) Representative sequencing results following cell growth competition assay reveal BCR-ABL/M244V/T315A/L364I to be capable of conferring a growth advantage over BCR-ABL/M244V/L364I and BCR-ABL/T315A.
The V299L dasatinib-resistant BCR-ABL kinase domain mutation is sensitive to VX-680.
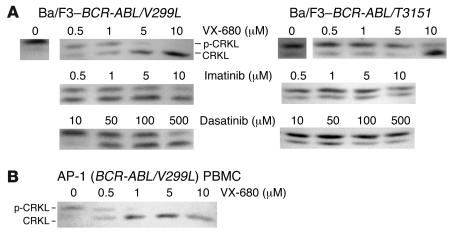
We previously showed that VX-680 can bind the T315I variant of the ABL kinase domain and inhibit BCR-ABL/T315I kinase activity in the low micromolar range (14, 15). Subsequent clinical evaluation of VX-680 in CML patients has shown activity in patients with the T315I mutation (10). Interestingly, V299 is a contact residue for VX-680 (as well as for dasatinib) in ABL, but the corresponding residue in Aurora A is leucine. Therefore, we reasoned that the dasatinib-resistant V299L mutation may actually retain sensitivity to VX-680, since this substitution is mimetic of Aurora A. Indeed, cell-based kinase assays demonstrate that the V299L mutation is sensitive to VX-680 in the low micromolar range (Figure 5A), consistent with the hypothesis that this mutation may actually increase affinity of this compound for BCR-ABL. Additionally, BCR-ABL kinase activity was inhibited in cells from patient AP-1 when exposed ex vivo to low micromolar concentrations of VX-680 (Figure 5B).
Figure 5. The most common dasatinib-resistant mutations are sensitive to the Aurora kinase inhibitor VX-680 in vitro.
(A) CrkL immunoblot of Ba/F3 cells transformed with dasatinib-resistant BCR-ABL isoforms depicted after exposure to varying concentrations of the kinase inhibitors indicated. (B) Primary CML cells harboring the dasatinib-resistant BCR-ABL/V299L mutation retain biochemical sensitivity to VX-680.
Discussion
Although first reported in CML, resistant kinase domain mutations are increasingly common in other cancers treated with kinase inhibitors, such as gastrointestinal stromal tumor (16) and lung cancer (17, 18). Treatment strategies for these diseases, as with CML, are currently following a model of sequential therapy using second-line kinase inhibitors, since acquired resistance is associated with new kinase domain mutations in more than half of such patients. Our results suggest that secondary kinase domain mutations (in 17 of 17 consecutive CML patients studied) may be even more common in relapse to second-line kinase inhibitor therapy, reinforcing the critical role these targeted kinases play in propagating the malignant clone even after multiple rounds of therapy.
Consistent with preclinical predictions, mutations that confer clinical resistance to inhibitors that bind to the active conformation of the target kinase (such as dasatinib) appear to be restricted to contact residues and therefore are less numerous than those conferring resistance to inhibitors that exclusively bind an inactive conformation (such as imatinib). The high frequency of the T315I mutation in our analysis could reflect the high degree of dasatinib resistance conferred by this allele or may simply reflect the presence of an already evolving, but not dominant, T315I clone in imatinib-resistant patients.
Of particular concern is the strong selective pressure for compound drug-resistant mutants in the context of sequential cancer therapy in a substantial proportion of patients. (These mutants could emerge by selection of preexisting clones or generation of new mutations.) Compound mutants not only diminish the chance of retaining sensitivity to a hypothetical “third-generation” inhibitor due to their potential complexity but also carry the risk of creating novel kinase alleles with enhanced oncogenic potency. Prevention of selection for such compound mutants will likely require up-front combination therapy utilizing compounds that can collectively suppress all single point mutations. To this end, we have provided evidence that VX-680, which is undergoing clinical evaluation and has shown early efficacy in T315I-associated cases (10), can inhibit the kinase activity of the most common dasatinib-resistant mutants. Future studies will need to address the tolerability of such combinations and whether the potential benefit of preventing resistance is outweighed by additional toxicities. In addition, our analysis was focused primarily on advanced-phase CML (since these patients were the first to relapse). Although compound mutations were identified at the time of dasatinib failure in both chronic phase patients studied in this analysis, longer follow-up on a larger number of patients is required to discern the impact of sequential ABL kinase inhibitor therapy in early-stage CML.
Mathematical modeling studies have estimated that the probability of a single drug-resistant mutation is 10–6 to 10–7 per cell division (19). Therefore, identification of CML subclones with 2–3 mutations in a single molecule presumably requires an extraordinary number of tumor doublings, as well as the possibility of additional mutations in DNA mismatch repair that might increase the diversity of mutations generated. In addition, the fitness advantage observed with the triple compound mutant studied here may further increase the probability of such gain-of-function alleles expanding within individual patients, even without the selective pressure of kinase inhibitor therapy.
Targeted therapy of human malignancies is still in its infancy. Optimal patient management in the future will likely require periodic genotyping to determine the likelihood of response to a particular kinase inhibitor. Given the critical reliance of several malignancies upon key tyrosine kinases, the full therapeutic potential of small molecule tyrosine kinase inhibitors will not be realized until strategies are developed that can effectively prevent selection for drug-resistant kinase domain point mutations. CML has served as a valuable treatment paradigm for a growing number of malignancies associated with activated tyrosine kinases. Frontline combinations of ABL inhibitors, if proven to be acceptably safe, could substantially improve both the depth and durability of clinical responses in BCR-ABL–associated leukemias, particularly in advanced phases of the disease, where relapse typically occurs rapidly, by their ability to effectively eliminate BCR-ABL kinase domain point mutation as a mechanism of disease resistance. It is likely that other cancers being treated with kinase inhibitor therapy will benefit from a similar treatment strategy.
Methods
BCR-ABL kinase domain sequencing analysis.
Seventeen sequential cases of acquired dasatinib resistance in imatinib-resistant CML and Philadelphia chromosome–positive acute lymphoblast leukemia patients were analyzed. Criteria for determination of response and relapse were as defined in the phase I clinical trial of dasatinib (8). All patients were participating in phase I or phase II clinical trials with dasatinib at UCLA. PBMCs were isolated from patients consenting to a UCLA institutional review board–approved research blood draw protocol prior to initiation of dasatinib therapy and at the time of dasatinib resistance. RNA extraction was performed using an RNeasy kit (QIAGEN) or as previously described (20). A semi-nested RT-PCR approach was used to specifically amplify the BCR-ABL kinase domain (21). In all pre-dasatinib samples, and dasatinib-resistant cases where visual inspection of the sequencing chromatogram failed to identify a single dominant mutation, PCR products were ligated into pBluescript, and a minimum of 10 independent clones were sequenced as previously described (2).
Generation of Ba/F3 cells expressing BCR-ABL kinase domain mutants.
Site-directed mutagenesis of a retroviral vector harboring the native BCR-ABL gene was performed as previously described (2). Cotransfection of 293T cells with BCR-ABL retroviral expression plasmids and the packaging plasmid Ecopack generated replication-defective virus that was used to infect the murine pro–B cell line Ba/F3. Stably infected populations were selected in the presence of puromycin and were capable of growing in the absence of exogenous IL-3 (2).
Analysis of CRKL phosphorylation status.
2.5 × 106 Ba/F3–BCR-ABL cells or patient PBMCs were exposed to kinase inhibitors in RPMI containing 10% FCS for 90 minutes. Whole-cell lysates were prepared as previously described (2) and subjected to Western immunoblotting with anti-CRKL sera (Santa Cruz Biotechnology Inc.) as previously described (6).
Cell growth viability studies of Ba/F3 cells harboring BCR-ABL kinase domain mutations.
Ba/F3 cells (2 × 104 ) were plated in the presence of varying concentrations of imatinib (Gleevec; courtesy of Novartis), dasatinib (SPRYCEL; courtesy of Bristol-Myers Squibb), or VX-680 (courtesy of Ambit Biosciences). Experiments were performed in triplicate. After 48 hours, the number of trypan blue–excluding cells was determined using an automated viable cell counter as previously described (9).
Murine bone marrow transformation assays.
Experiments involving mice were performed with the approval of the UCLA Animal Research Committee. Retrovirus was prepared by transfecting one 10-cm dish of 293T cells with 15 μg MSCV-BCR-ABL constructs and 15 μg Ecopack packaging plasmid using Lipofectamine 2000 (Invitrogen). Cells were grown overnight in DMEM (Cellgro) plus 10% FCS (Omega Scientific Inc.), then DMEM was replaced with IMDM plus 10% FCS. Thirty-six hours after transfection, viral supernatants were collected and used to infect primary bone marrow cells. Bone marrow cells were obtained by flushing femurs and tibias of 3-week-old BALB/c male mice (Charles River Laboratories) with PBS. Bone marrow cells (1 × 107) were placed in a 12-well plate and 3 ml of each undiluted viral supernatant, supplemented with polybrene (4 μg/ml final) was added, and centrifuged at 1,545 g for 90 minutes. Concurrently, 0.75 × 106 NIH 3T3 cells and 1 × 106 Ba/F3 cells were infected by the same method to confirm equal viral titer between the different samples and to ensure IL-3 resistance, respectively. After spinfection, plates were placed in a 37°C incubator for 4 hours, and the viral supernatant was replaced with 3 ml Whitlock-Witte media (RPMI containing 20% FCS, 55 μM 2-mercaptoethanol, 2 mM glutamine, 100 U penicillin/ml, and 10 μM streptomycin/ml [Invitrogen]). The next morning, cells were counted and serially diluted (3 × 105, 1 × 105, 3 × 104, 1 × 105, and 3 × 103 cells/well) in triplicate into 24-well plates. Untransfected bone marrow cells, grown overnight in Whitlock-Witte media, were added to the wells to total 1 × 106 cells/well to add stromal support. This final plating started the assay as day 0. Cells were grown in a 37°C incubator, and medium was changed every 4 days. Starting on day 5, wells were scored positive when the number of transformed cells first reached or exceeded 1 × 106, and the endpoint of the assay was 21 days. These data were analyzed using a model in which the time to reach 1 × 106 cells is lognormal with the constant coefficient of variation (Cv) and median equal to t0 + [ln(1 × 106/initial cell count)]/k, where k is the exponential growth rate and t0 is a lag time before exponential growth starts. We accounted for both left and right censoring for the day each well was scored positive in maximum likelihood estimation of k for each mutant and the common t0 and Cv. Model fits using this approach were superior to those of an alternate model that attributed any apparent lag to a reduced fraction of cells capable of dividing. Exponential doubling times were estimated by 0.693/k, and growth rates across separate studies were averaged after they were normalized as a percentage of the k for p210. NIH 3T3 cells were grown for 48 hours after infection, and lysates were subjected to gel electrophoresis and Western blotting using anti-Abl antibodies (OP-20; Calbiochem) and actin (AC-15; Sigma-Aldrich) to confirm equal viral titer between the mutants. Each set of bone marrow experiments was repeated more than 3 times, with similar results. Prism software (version 4; GraphPad Software) was used to graphically represent the data.
Cell growth competition assays.
Viable, exponentially growing Ba/F3 cells harboring BCR-ABL kinase domain mutations were counted and mixed in equal proportion at a concentration of 2 × 105 cells/ml in medium containing 1% FCS as previously described (12). Cells were split every 48 hours and replated at 2 × 105 cells/ml to ensure exponential growth phase. After 18 days, genomic DNA was extracted and the BCR-ABL kinase domain was sequenced as previously described (2). Experiments were performed in triplicate.
BCR-ABL kinase activity studies.
MSCV-BCR-ABL constructs were transiently transfected into a 1 × 10–cm dish of 293T cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s directions. Thirty-six hours after transfection, cells were equalized by cell number and lysed in modified radioimmunoprecipitation assay buffer (10 mM Tris, pH 7.3, 150 mM NaCl, 0.4 mM Na3VO4, 0.4 mM EDTA, 10 mM NaF, 1 mM PMSF, 1/100 dilution of protease inhibitor III cocktail [Calbiochem], 1% NP-40, 0.1% SDS, 0.5% deoxycholate). BCR-ABL was immunoprecipitated from lysates with anti–c-Abl antibodies (Calbiochem) prebound to protein A–protein G Sepharose beads (Calbiochem). Immunoprecipitations were washed 3 times in wash buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 0.4 mM Na3VO4, 0.1% NP-40, protease inhibitors) and 2 times in wash buffer without NP-40. Twenty percent of each immunoprecipitate was visualized by SYPRO Ruby (Molecular Probes; Invitrogen) after gel electrophoresis, and BCR-ABL protein levels were equalized using ImageQuant (Amersham Biosciences) compared with a porcine myosin standard (188 kDa). For autokinase assays, 120 ng kinase was used in each reaction. The kinase reaction was performed using 15 μl kinase buffer (KB: 20 mM Tris, pH 7.5, 10 mM MgCl2, 10 mM p-nitrophenylphosphate, 20 mM β-glycerophosphate, 1 mM dithiothreitol, 1 mM BSA, 50 μM ATP, 0.5 μCi [γ-32P]ATP/reaction [Amersham Biosciences]) for 5, 15, or 30 minutes at room temperature. Reactions were stopped by the addition of 15 μl loading buffer and boiled prior to loading on a 7.5% acrylamide gel. After SDS-PAGE, the gels were dried, and phosphorylated BCR-ABL was detected by autoradiography. For transphosphorylation studies, 40 ng kinase (~0.2 μM) was used in each reaction. Various concentrations of biotinylated Abltide (Upstate) were utilized as substrate in KB. Reactions (25 μl) were performed in triplicate for 15 minutes and terminated by the addition of 12.5 μl 7.5-M guanidine hydrochloride. 12.5 ml of the reaction was spotted onto streptavidin-coated filter paper squares (SignaTECT; Promega) and washed per the manufacturer’s directions, and Abltide phosphate incorporation was quantitated by liquid scintillation counting of the squares. ATP-specific activity was determined by spotting 5 μl of 2 random reactions onto SignaTECT paper. These squares were not washed, and the 2 liquid scintillation values were averaged. Background was determined independently for each construct by omitting Abltide in the reaction mix. Prism software was used to determine Michaelis-Menten kinetics by plotting velocity (V; pmol ATP incorporated into Abltide/reaction time in minutes/μg kinase) versus Abltide concentration as well as values for Vmax and Km for each mutant construct (data not shown).
Acknowledgments
N.P. Shah is the recipient of a Leukemia & Lymphoma Society Career Development Award for Special Fellows, a Clinical Scientist Development Award from the Doris Duke Charitable Foundation, and the UCSF Sandler Research Award. C.L. Sawyers is the recipient of a Doris Duke Charitable Foundation Distinguished Clinical Scientist Award and is an investigator of the Howard Hughes Medical Institute.
Footnotes
Nonstandard abbreviations used: CML, chronic myelogenous leukemia; MBC, myeloid blast crisis.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:2562–2569 (2007). doi:10.1172/JCI30890
References
- 1.Schindler T., et al. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289:1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 2.Shah N.P., et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 3.Shah N.P., et al. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 4.Lombardo L.J., et al. Discovery of N-(2-chloro-6-methyl- phenyl)-2-(6-(4-(2-hydroxyethyl)- piperazin-1-yl)-2-methylpyrimidin-4- ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J. Med. Chem. 2004;47:6658–6661. doi: 10.1021/jm049486a. [DOI] [PubMed] [Google Scholar]
- 5.Tokarski J.S., et al. The structure of dasatinib (BMS-354825) bound to activated ABL kinase domain elucidates its inhibitory activity against imatinib-resistant ABL mutants. Cancer Res. 2006;66:5790–5797. doi: 10.1158/0008-5472.CAN-05-4187. [DOI] [PubMed] [Google Scholar]
- 6.Gorre M.E., et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–880. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 7.Weisberg E., et al. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 2005;7:129–141. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 8.Talpaz M., et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N. Engl. J. Med. 2006;354:2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 9.Burgess M.R., Skaggs B.J., Shah N.P., Lee F.Y., Sawyers C.L. Comparative analysis of two clinically active BCR-ABL kinase inhibitors reveals the role of conformation-specific binding in resistance. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3395–3400. doi: 10.1073/pnas.0409770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giles F.J., et al. MK-0457, a novel kinase inhibitor, is active in patients with chronic myeloid leukemia or acute lymphocytic leukemia with the T315I BCR-ABL mutation. Blood. 2007;109:500–502. doi: 10.1182/blood-2006-05-025049. [DOI] [PubMed] [Google Scholar]
- 11.O’Hare T., et al. Combined Abl inhibitor therapy for minimizing drug resistance in chronic myeloid leukemia: Src/Abl inhibitors are compatible with imatinib. Clin. Cancer Res. 2005;11:6987–6993. doi: 10.1158/1078-0432.CCR-05-0622. [DOI] [PubMed] [Google Scholar]
- 12.Griswold I.J., et al. Kinase domain mutants of Bcr-Abl exhibit altered transformation potency, kinase activity, and substrate utilization, irrespective of sensitivity to imatinib. Mol. Cell. Biol. 2006;26:6082–6093. doi: 10.1128/MCB.02202-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skaggs B.J., et al. Phosphorylation of the ATP-binding loop directs oncogenicity of drug-resistant BCR-ABL mutants. Proc. Natl. Acad. Sci. U. S. A. 2006;103:19466–19471. doi: 10.1073/pnas.0609239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carter T.A., et al. Inhibition of drug-resistant mutants of ABL, KIT, and EGF receptor kinases. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11011–11016. doi: 10.1073/pnas.0504952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Young M.A., et al. Structure of the kinase domain of an imatinib-resistant Abl mutant in complex with the Aurora kinase inhibitor VX-680. Cancer Res. 2006;66:1007–1014. doi: 10.1158/0008-5472.CAN-05-2788. [DOI] [PubMed] [Google Scholar]
- 16.Tamborini E., et al. A new mutation in the KIT ATP pocket causes acquired resistance to imatinib in a gastrointestinal stromal tumor patient. Gastroenterology. 2004;127:294–299. doi: 10.1053/j.gastro.2004.02.021. [DOI] [PubMed] [Google Scholar]
- 17.Pao W., et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi S., et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N. Engl. J. Med. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 19.Michor F., et al. Dynamics of chronic myeloid leukaemia. Nature. 2005;435:1267–1270. doi: 10.1038/nature03669. [DOI] [PubMed] [Google Scholar]
- 20.Branford S., Hughes T. Diagnosis and monitoring of chronic myeloid leukemia by qualitative and quantitative RT-PCR. Methods Mol. Med. 2006;125:69–92. doi: 10.1385/1-59745-017-0:69. [DOI] [PubMed] [Google Scholar]
- 21.Branford S., Hughes T. Detection of BCR-ABL mutations and resistance to imatinib mesylate. Methods Mol. Med. 2006;125:93–106.. doi: 10.1385/1-59745-017-0:93. [DOI] [PubMed] [Google Scholar]