Abstract
Intermediate states play well established roles in the folding and misfolding reactions of individual RNA and protein molecules. In contrast, the roles of transient structural intermediates in multi-component ribonucleoprotein (RNP) assembly processes and their potential for misassembly are largely unexplored. The mammalian signal recognition particle SRP19 protein is unstructured but forms a compact core domain and two extended RNA-binding loops upon binding the SRP RNA. The SRP54 protein subsequently binds to the fully assembled SRP19-RNA complex to form an intimate three-fold interface with both SRP19 and the SRP RNA and without significantly altering the structure of SRP19. We show, however, that the presence of SRP54 during SRP19-SRP RNA assembly dramatically alters the folding energy landscape to create a non-native folding pathway that leads to an aberrant SRP19-RNA conformation. The misassembled complex arises from the surprising ability of SRP54 to bind rapidly to an SRP19-RNA assembly intermediate and to interfere with subsequent folding of one of the SRP19 RNA-binding loops at the three-way protein-RNA interface. An incorrect temporal order of assembly thus readily yields a non-native three-component ribonucleoprotein particle. We propose there may exist a general requirement to regulate the order of interaction in multi-component RNP assembly by spatial or temporal compartmentalization of individual constituents in the cell.
Keywords: Ribonucleoprotein assembly and misassembly, compartmentalization, energy landscape
Introduction
Assembly of most ribonucleoprotein (RNP) complexes is accompanied by significant conformational changes in both protein and RNA components.1,2 In some cases, these conformational changes may occur in a progressive fashion such that early interactions in assembly lead to further conformational rearrangements that uniformly facilitate later steps. Several structurally linked steps in assembly of the 30S ribosomal subunit appear to proceed in this manner.3–5 However, in view of the multiplicity of RNA-protein and protein-protein interactions that can potentially occur during assembly of multi-component RNPs, the extent to which incorrect or premature interactions among components might interfere with native complex formation remains to be assessed. The idea that some RNA6,7 and protein8,9 molecules readily adopt stable, but misfolded, states is now well established. We have recently identified a striking example of robust misassembly in the mammalian signal recognition particle.10 Our work extended the concept of kinetically trapped, non-native conformational states to multi-component RNP complexes. In this report we analyze the kinetic mechanism for this misassembly.
The signal recognition particle (SRP) recognizes nascent membrane or secretory proteins emerging from translating ribosomes and directs ribosome-peptide-mRNA complexes to the endoplasmic reticulum, where peptide synthesis continues and the proteins are dispatched to their final intra– or extracellular destinations.11 The mammalian SRP consists of one RNA molecule of ~300 nts (SRP RNA) and six protein cofactors, SRP9, SRP14, SRP19, SRP54, SRP68 and SRP72, (Figure 1A). Structurally, the SRP is comprised of two distinct and functionally separable subunits.12,13 About half of the RNA and four proteins (SRP19, SRP54, SRP68 and SRP72) form the ‘large’ subunit. The other half of the RNA and SRP9 and SRP14 form the ‘Alu’ subunit. In both mammalian14–16 and yeast17,18 cells, SRP appears to assemble via an ‘SRP54-late’ pathway in which all SRP proteins except SRP54 are imported into the nucleolus where they bind the SRP RNA (green and gray proteins, Figure 1B). This partially assembled complex is then exported to the cytoplasm where SRP54 binds in a distinct step (in purple, Figure 1B) to form the fully functional SRP holocomplex.
Figure 1.

The signal recognition particle (SRP). (A) Architecture of the mammalian SRP. SRP proteins are shown as colored and gray ovals; the RNA is yellow. (B) ‘SRP54-late’ model for cellular assembly of the mammalian SRP. Five of six SRP proteins (SRP9, SRP14, SRP19, SRP68, and SRP72) (gray and green ovals) enter the nucleus or nucleolus to assemble with the SRP RNA. The partially assembled complex is then transported back into the cytoplasm to bind SRP54 (purple) and form the native SRP holocomplex. (C) In vitro scheme for native SRP19-SRP54-SRP RNA ternary complex formation via the SRP54-late assembly pathway. (D) ‘SRP54-early’ pathway that leads to formation of a non-native ternary complex, termed the non-compartmentalized complex, in which two RNA binding loops in SRP19 do not fold to their native conformation.10 (E) Three-fold protein-RNA interface in the native SRP ternary complex. SRP19 loop 2 (gray) is positioned in a cleft formed by SRP54 and the RNA. The figure shows the same complex20 as panel C, but has been rotated ~90° to afford a view from the ‘top’ of the three-component interface.
Mammalian SRP19 is unstructured in its unbound state and assembles with the SRP RNA via formation of multiple obligatory intermediate complexes.10,19 Assembly of the SRP19-RNA complex is an example of mutually-induced fit because both SRP19 and the SRP RNA undergo significant conformational rearrangements during assembly.10,19–22 A native ternary complex forms in vitro when SRP54 binds to the stable and preformed SRP19-SRP RNA complex (Figure 1C) and is the same as the complex visualized in crystallographic studies.10,21 This ‘SRP54-late’ in vitro assembly pathway mimics the two-step, or compartmentalized, in vivo assembly pathway.14–18
In contrast, if SRP19 and SRP54 are allowed simultaneous access to the SRP RNA in vitro, these three components interact robustly to form a distinct, non-native, complex (Figure 1D).10 In this complex, two RNA binding loops in SRP19 remain misfolded (black dashed lines, Figure 1D). We term this assembly pathway the ‘SRP54-early’ pathway and the misfolded ternary complex the ‘non-compartmentalized’ complex. Here we show that, beginning with identical sets of protein and RNA components, the ‘non-compartmentalized’ structure arises from the surprising ability of SRP54 to bind an SRP19-RNA assembly intermediate: the resulting cleft-like interface that forms between SRP54 and the SRP RNA (Figure 1E) is sufficiently narrow that it disfavors insertion of an RNA-binding loop from SRP19 that normally occurs in the native complex.
Results
Strategy for monitoring assembly of SRP19 with the SRP RNA
Free SRP19 is a natively unstructured protein. Upon binding to the SRP RNA, SRP19 spans three structural motifs, a small globular core domain and two RNA binding loops that extend out from the core (Figure 2A).10,20,21,23 We monitored assembly-induced conformational changes in each motif by attaching an environmentally sensitive fluorophore at four sites in SRP19. Each of the four positions is solvent accessible in both SRP19-SRP RNA binary and SRP19-SRP54-SRP RNA ternary complexes.10,20,21 Fluorophores were placed individually in each of the two RNA binding loops (positions 31 and 72) and in the core domain (positions 93 and 106) (Figure 2A). Control experiments showed that RNA binding affinities for each fluorophore-derivatized SRP19 were either identical or very similar to the unlabeled wild type SRP19 protein (see Materials and Methods). Site-directed cleavage experiments also demonstrated that derivatizing SRP19 at these positions with Fe(II)-EDTA groups retains the native conformation of the complex.10
Figure 2.
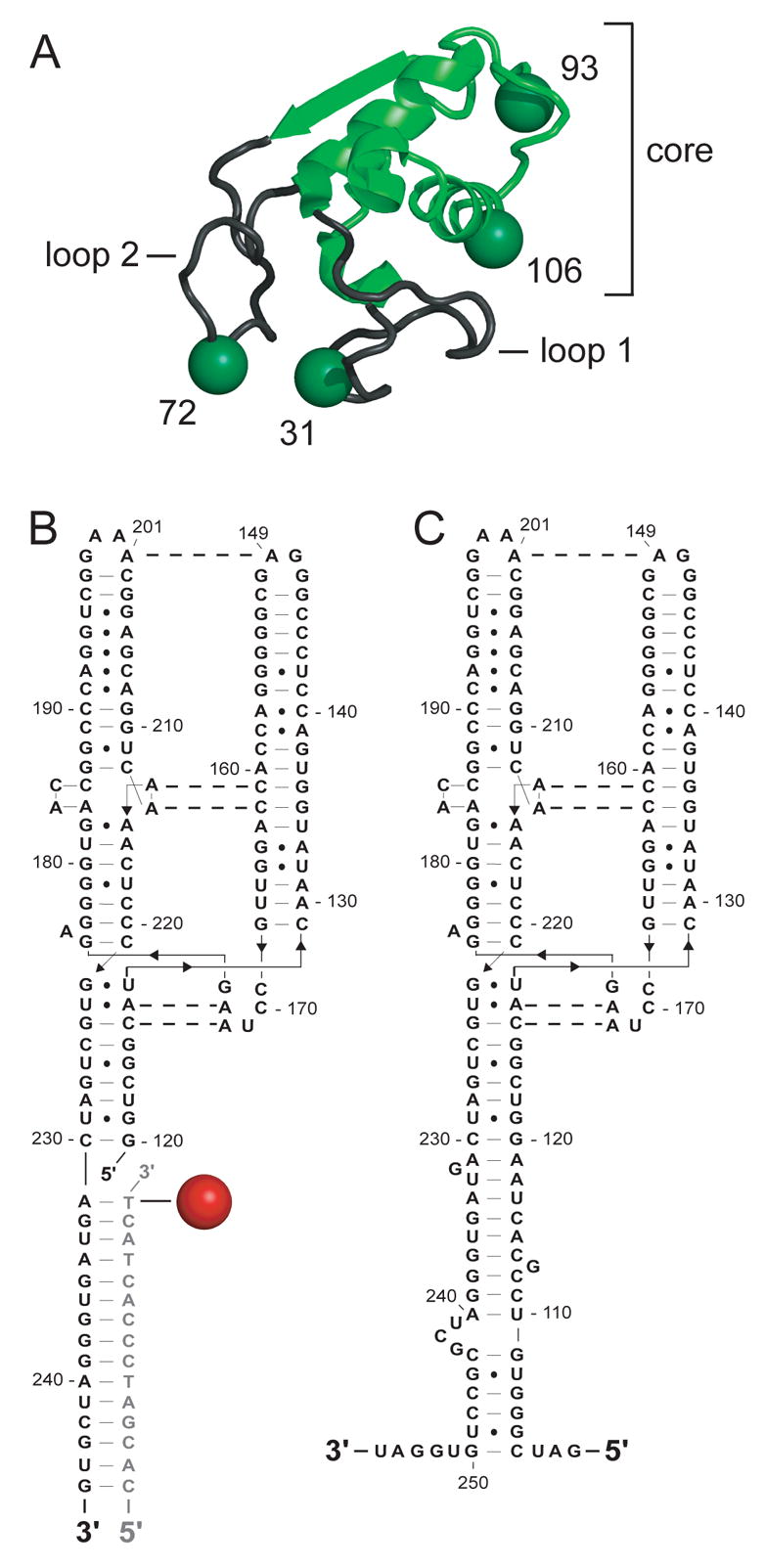
SRP19 and the large subunit SRP RNA (LS RNA). (A) SRP19 structural motifs when bound to the SRP RNA. SRP core is green and the two RNA binding loops are gray. Sites of fluorophore attachment are shown as spheres. (B) Alexa 555 derivatized RNA:DNA hybrid. RNA and DNA are shown in black and gray, respectively. (C) LS RNA.
Conformational changes specific to each position were monitored by either of two approaches. We detected RNA-protein assembly using fluorescence resonance energy transfer (FRET). Energy transfer occurred between a donor Alexa 488 fluorophore tethered to SRP19 and an acceptor Alexa 555 fluorophore tethered to the large subunit of the SRP RNA (LS RNA) via a hybridized DNA oligo (Alexa 555-LS RNA) (Figure 2B). Control experiments showed that SRP19 binding to the Alexa 555-LS RNA is indistinguishable to that of the native LS RNA. In the second approach, SRP19 assembly with the SRP RNA was monitored in single fluorophore experiments using the environmentally sensitive fluorophores Alexa 488 or BODIPY-FL tethered to one of the four SRP19 labeling sites. Assembly of SRP54 with the SRP19-RNA complex could also be monitored as the effect that SRP54 binding has on the environmentally sensitive SRP19-tethered fluorophores.
FRET based analysis of SRP assembly
Assembly of the SRP19-RNA binary and the SRP19-SRP54-RNA ternary complexes was initially monitored as illustrated in Figure 3A. In this experiment, Alexa 488 functioned as both the FRET donor and also as an environmentally sensitive fluorophore. Thus, RNA binding and subsequent conformational changes in SRP19 were detected both as the initial change in the resonance energy transfer efficiency to the RNA-tethered Alexa 555 acceptor fluorophore and also as the local change in donor fluorophore environment (Figure 3A).
Figure 3.
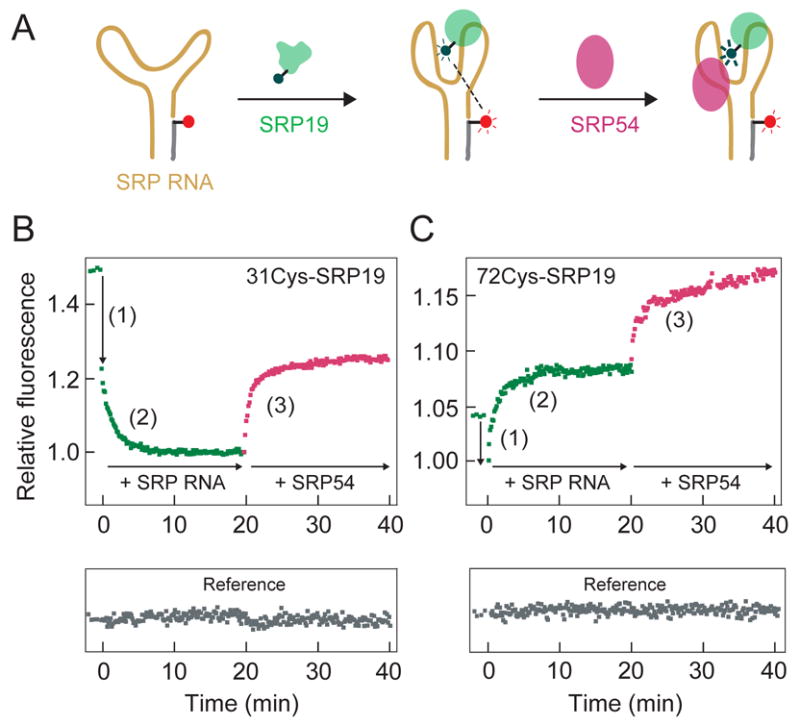
FRET-based analysis of SRP assembly. (A) Scheme for monitoring SRP assembly using Alexa 488-derivatized SRP19 (green) and the Alexa 555-LS RNA (yellow and gray). SRP54 (purple) binding is detected by its affect on the SRP19-RNA complex. (B, C) Three distinct assembly steps are detected during native SRP19-SRP54-SRP RNA ternary complex formation. (1) A rapid burst phase quenching of fluorescence during initial SRP19-RNA assembly (green). (2) Well-resolved increase or decrease in fluorescence emission intensity as the SRP19-RNA complex matures to the native structure (green). (3) An increase in fluorescence intensity due to SRP54 binding to the pre-formed SRP19-RNA complex (purple). Free Alexa 647 reference fluorophore is gray.
When SRP19, labeled at position 31, was added to the Alexa 555-LS RNA, we observed a clear burst phase in which the donor emission intensity was partially quenched, followed by a well-resolved slow phase during which the emission intensity decreased further (phases 1 and 2 in the green trace in Figure 3B). This change in fluorescence was specific to SRP19-RNA interactions because the emission of a free reference fluorophore (Alexa 647) present in the solution remained unchanged over the course of the experiment (gray trace in Figure 3B). Similar overall behavior was observed when assembly was monitored using SRP19 labeled at position 72. Fluorescent emission initially decreased in a rapid burst phase followed by a well-resolved slow phase in which the fluorescence emission intensity increased (green trace, Figure 3C.). These data indicate that both RNA-binding loops in SRP19 assemble with the LS RNA in at least two well-resolved steps.
Assembly of SRP54 with the preformed SRP19-RNA complex was monitored as the effect that SRP54 binding has on emission of the fluorophore attached to SRP19 (Figure 3A). Addition of SRP54 to a SRP19-RNA complex, labeled at either SRP19 position 31 or 72, yielded a clear increase in fluorescence emission intensity due to the perturbation of the local fluorophore environment upon SRP54 binding (purple traces in Figures 3B and C).
Burst phase of SRP19-RNA assembly corresponds to Encounter complex formation
To assign the binding event that corresponds to the rapid burst phase change in fluorescence for SRP19, labeled at either position 31 or 72, we monitored assembly using an A149U mutant RNA. Nucleotides A149 and A201 form a non-canonical base pair that links helices III and IV in the SRP RNA (Figure 2C).20,21 SRP19 forms a labile Encounter complex with the A149U mutant RNA but cannot assemble beyond this step to form the native SRP19-RNA complex.19
SRP19 binding to the A149U mutant RNA proceeded via a rapid burst phase whose magnitude was comparable to that observed for the native sequence RNA as monitored at either position 31 or 72 (Figures 4A,B). In contrast, neither of the fluorescently labeled SRP19 proteins showed the second, slow, phase characteristic of assembly with the native sequence RNA (compare open and solid symbols in Figures 4A,B). We infer that the burst phase corresponds to rapid formation of an Encounter complex between SRP19 and the SRP RNA; whereas, the second, slower phase (visualized in Figures 3B,C) corresponds to slow conformational rearrangements in the nascent SRP19-RNA complex that ultimately requires formation of the native RNA tertiary interaction between nucleotides A149 and A201.
Figure 4.

Burst phase assembly step corresponds to Encounter complex formation between SRP19 and the RNA. (A,B) SRP19-RNA assembly was monitored for the native sequence RNA or an A149U mutant that cannot form native RNA-RNA interactions (solid and open symbols, respectively). Arrows indicate burst phases observed with both RNAs. (C) Addition of SRP54 has no effect on the Encounter complex formed between SRP19 and the A149U RNA.
We then evaluated whether SRP54 interacts with the SRP19-RNA Encounter complex. A stalled SRP19-RNA Encounter complex intermediate was first formed using the A149U mutant RNA and SRP19 derivatized at position 72. As expected, we observed the burst phase fluorescence change upon formation of the initial SRP19-RNA complex (open circles, Figure 4C). When SRP54 was added to this complex, no additional fluorescence change occurred (solid symbols, Figure 4C). These results contrast strongly with the ability of SRP54 to induce conformational changes in the native RNA-SRP19 complex (Figure 3C). We infer that SRP54 does not bind the Encounter complex intermediate.
SRP19 loop and core structures bind the SRP RNA via distinct mechanisms
SRP19 is comprised of distinct core and extended loop motifs (Figure 2A). We detected local conformational changes specific to the two RNA binding loops (positions 31 and 72) and to the SRP19 core domain (positions 93 and 106) using single fluorophore experiments (Figure 5A). RNA binding caused an increase in Alexa 488 fluorescence emission efficiency for SRP19 proteins labeled at positions 72, 93 and 106 and a decrease for the protein labeled at position 31 (Figure 5B).
Figure 5.
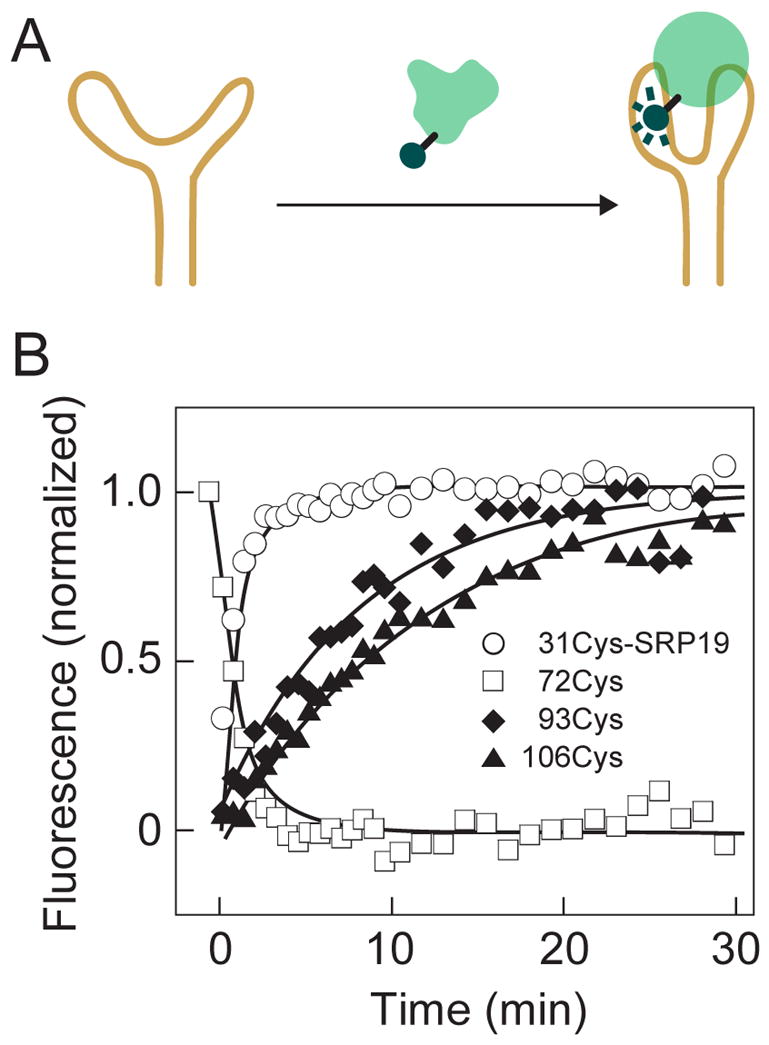
Conformational rearrangements at distinct SRP19 structural motifs during RNP assembly. (A) Scheme for monitoring SRP19-RNA assembly using single Alexa 488 fluorophore experiments. (B) Fluorescence-detected assembly at the RNA-binding loops (open symbols) proceeds significantly faster than assembly of the SRP19 core domain (solid symbols).
Strikingly different time-resolved behavior was observed depending on where the fluorophore was tethered to SRP19. When the fluorophore was linked to either of the RNA-binding loops in SRP19, the observed change in fluorescence intensity was 10-fold faster than when attached to the SRP19 core (compare open and solid symbols, Figure 5B).
We characterized local assembly at each SRP19 motif by following the rate of change in fluorescence intensity as a function of RNA concentration. For all four Alexa 488-labeled SRP19 proteins, rates increased with RNA concentration (Figure 6). SRP19 derivatized at positions 31, 93 and 106 showed good linear behavior in rate versus RNA concentration plots: the slopes of these lines yield the second-order rate constants for complex formation (squares, Figures 6A,C,D).
Figure 6.

SRP19 structural motifs assemble with the RNA via distinct mechanisms. (A–D) Rate of change in fluorescence intensity as a function of RNA concentration for SRP19 variants. Slopes (dashed lines) yield second order rate constants for assembly as detected in distinctive SRP19 loop and core structural motifs. Most experiments were performed with Alexa 488-labeled SRP19 (squares). For the 72Cys and 106Cys variants, assembly was also monitored using the BODIBY-FL fluorophore (circles).
In contrast, SRP19 derivatized with Alexa 488 at position 72 exhibited a distinctive behavior. At RNA concentrations below 100 nM, the observed rate increased with increasing RNA concentration, as expected for a single rate-limiting binding step (squares, Figure 6B). However, at RNA concentrations above 100 nM, the apparent rate of complex formation became independent of concentration and leveled off at 0.9 min−1 (solid line, Figure 6B).
These distinctive kinetic behaviors are independent of the nature of the reporter fluorophore. The SRP19 variants labeled at positions 72 and 106 were also monitored using the BODIPY-FL fluorophore, which is chemically and sterically dissimilar to Alexa 488. The distinctive slow and fast time-resolved behaviors were identical as detected by either fluorophore (compare circles and squares, Figures 6B,D).
These experiments thus illustrate two distinctive features for assembly of the SRP19-RNA complex. First, second order rate constants vary by an order of magnitude and fall into two classes. SRP19 derivatized at either of the two RNA binding loops (positions 31 and 72) have similar second-order rate constants (2–8 × 106 M−1min−1). SRP19 variants derivatized in the core (positions 93 and 106) also have similar rate constants (3–5 × 105 M−1min−1) that are an order of magnitude slower than those observed at the RNA-binding loops. Second, the SRP19 variant derivatized at position 72, has a distinctive biphasic kinetic behavior such that a concentration-independent process with rate constant 0.9 min−1 limits assembly in the vicinity of RNA binding loop 2.
These experiments support a complex three-step mechanism for assembly of SRP19 with the SRP RNA. Free RNA and protein rapidly interact to form the Encounter complex. Encounter complex formation is observed directly as the fast phase in FRET-based assembly experiments (Figure 3). This step is followed by two kinetically separable concentration dependent conformational changes to form a Stable intermediate complex. In these steps, the RNA binding loops in SRP19 undergo RNA-induced conformational changes more rapidly than does the core domain (Figure 5). Finally, regions near loop 2 participate in an additional concentration independent structural rearrangement step that converts the Stable complex into the native SRP19-RNA complex (Figure 6B).
Assembly of SRP54 with the preformed SRP19-RNA complex
Assembly of SRP54 with the preformed SRP19-LS RNA complex was monitored by the effect that SRP54 binding had on the environmentally sensitive fluorophore attached to SRP19. These experiments were similar to those described in Figure 3A, except that only SRP19 was fluorescently labeled. For SRP19 variants derivatized at either position 31 or 72, fluorescence emission intensity increased upon SRP54 binding (Figure 7A,B). SRP54 binds to the preformed SRP19-RNA complex with a Kd = 12 nM10 and, as expected, the net amount of complex formed increased as the SRP54 concentration was increased from 15 to 200 nM. Over a broad range of SRP54 concentrations, second order rate constants were identical, at 1.1 × 107 M−1 min−1 (Figures 7C,D), independent of the site of derivatization in SRP19. SRP54 thus binds in a single kinetically significant step.
Figure 7.
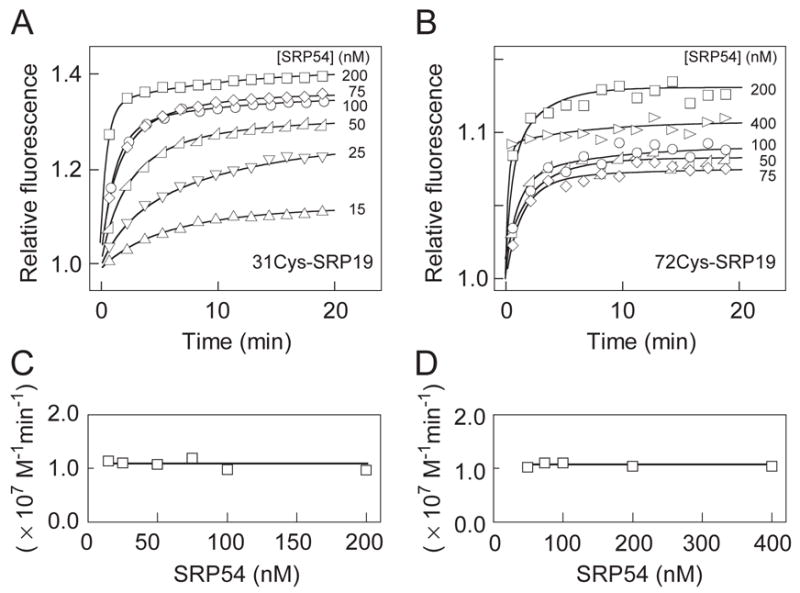
One step assembly of SRP54 to the pre-formed SRP19-RNA complex. (A, B) The rate constant for SRP54 binding to the pre-formed Alexa 488-labeled SRP19-RNA complex was obtained by fitting the change in fluorescence intensity over time to a second order rate equation. (C,D) All SRP54 concentrations yielded identical second-order rate constants.
Concentration dependence of non-compartmentalized ternary complex formation
As described above, if SRP19 and SRP54 assemble with the RNA simultaneously, these three components interact to form the non-compartmentalized complex whose structure is significantly different from the native complex (Figure 1D). We used single fluorophore experiments to characterize the kinetic step that gates whether SRP54 assembles to form the native or non-compartmentalized complex (Figure 8A,D).
Figure 8.
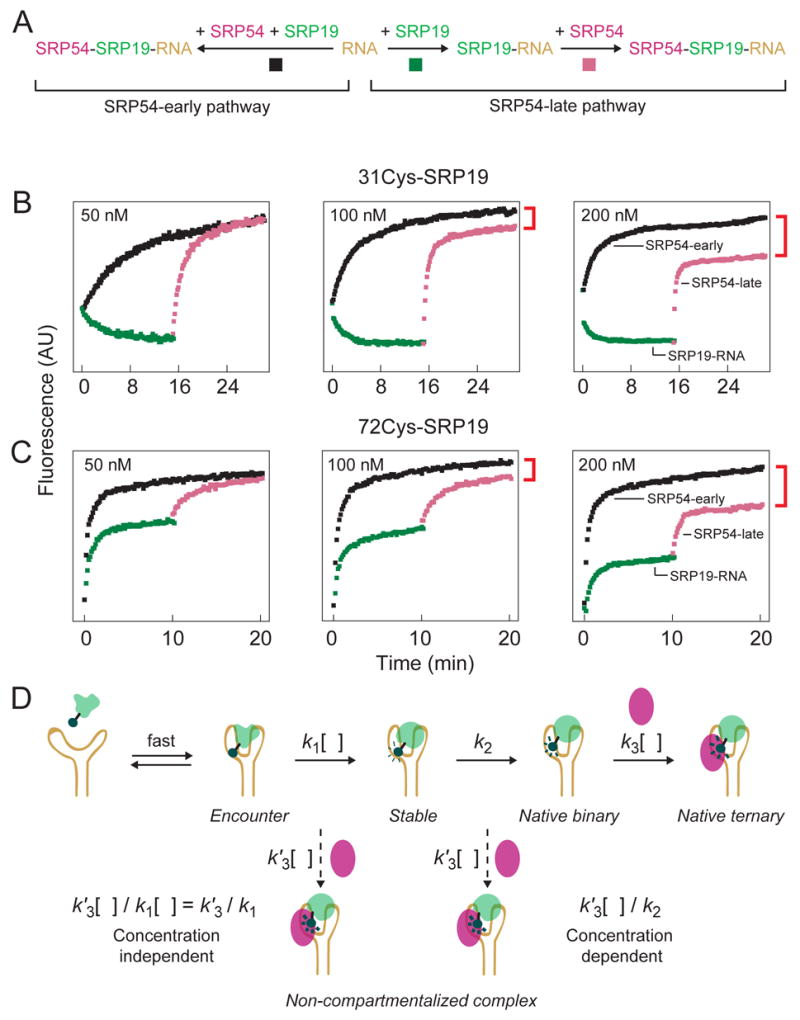
The extent of non-compartmentalized complex formation is concentration dependent. (A) Scheme for monitoring native vs. non-compartmentalized assembly using single fluorophore experiments. Native assembly (the SRP54-late pathway) involves ordered binding by SRP19 (green traces) and SRP54 (purple traces). For non-compartmentalized assembly (the SRP54-early pathway), both proteins assemble simultaneously (black traces). (B,C) Visualization of native and non-compartmentalized ternary complex formation using single fluorophore experiments. Ratios of SRP19, SRP54 and RNA components were held constant at 1:1:2 in all experiments; protein concentrations are given for each panel. Non-compartmentalized complex formation is reported directly as the intensity difference between purple and black traces (see red brackets). (D) Expected concentration dependence for SRP54 binding post-Encounter versus post-Stable complex formation. k1 is a compound rate constant reflecting formation of the Stable complex via the kinetically linked Encounter complex. Dashed arrows show putative, concentration-dependent (k′3[ ]), steps for SRP54-mediated misassembly.
We followed formation of the native complex by sequential addition of SRP19 and SRP54 (green and purple traces, Figures 8B,C). This is the SRP54-late pathway. As monitored using the 31Cys-SRP19 protein, fluorescence emission intensity decreased as free SRP19 bound to the RNA. The rate of binding increased as the protein concentration was increased from 50 to 200 nM (green traces in Figure 8B). Fluorescence intensity then increased upon subsequent addition of SRP54. The rate of SRP54 binding also increased with protein concentration, as expected (purple traces in Figure 8B). These data are consistent with the SRP19 and SRP54 binding experiments outlined in Figures 6A and 7A, respectively.
We then monitored assembly of the non-compartmentalized ternary complex by adding SRP54 prior to adding SRP19. This is the SRP54-early pathway (black traces in Figure 8B). These experiments were performed under the identical component concentrations used to monitor native complex formation. Thus, the experiments represented by the green and purple versus black traces (Figures 8A,B) differed only the order in which SRP components interact but not in the identity or amounts of SRP19, SRP54 or SRP RNA.
Strikingly, when assembly was monitored for component concentrations of 200 nM, complexes formed via the SRP54-early pathway had significantly higher fluorescence than complexes formed via the SRP54-late pathway (compare the endpoints for purple and black traces in Figure 8B). This difference in fluorescence intensity directly reports formation of the non-compartmentalized complex via the SRP54-early pathway. A similar, but smaller, difference in fluorescence was observed at 100 nM protein concentrations; whereas, the difference disappeared when component concentrations were reduced to 50 nM (Figure 8B).
We performed comparable experiments with the 72-Cys SRP19 variant. As expected, the rate of SRP19-RNA complex formation was concentration-independent (green traces, Figure 8C) because the conformational rearrangement involving loop 2 binding plateaus at 0.9 min−1 (recall Figure 6B). The rate of SRP54 binding increased with increasing SRP54 concentration, consistent with simple one-step assembly with the preformed SRP19-RNA complex (purple traces, Figure 8C). Similar to assembly as monitored at position 31, the fluorescence of the final 72Cys-complex was again strongly concentration dependent and the difference in fluorescence intensity in the final complexes was larger at 200 nM components than at 100 or 50 nM concentrations (compare purple and black traces in Figure 8C).
In summary, as monitored at either of two locations within SRP19, there exists a kinetic competition such that misassembly is specifically favored at higher component concentrations. As outlined under the Discussion, this concentration-dependent behavior provides strong evidence regarding the physical step that gates misassembly.
Discussion
Assembly of the SRP19-RNA complex via distinct folding stages for the loop and core domains
Our physical model for the alternating interactions that lead to the native SRP19-RNA complex is shown in the first four panels of Figure 9A. Both SRP19 and the SRP RNA are in non-native conformations prior to forming a specific ribonucleoprotein complex. The SRP RNA forms a relaxed but base paired state; whereas, free SRP19 exists in an unstructured coil conformation.
Figure 9.

Mechanisms for native and non-compartmentalized SRP assembly. (A) Native and non-compartmentalized assembly pathways are gated at the Stable complex. (B,C) Folding energy landscapes for SRP19 during SRP ternary complex formation via SRP54-late and SRP54-early pathways.
SRP19 and the RNA initially interact in a rapid, approximately diffusion limited, step to form an Encounter complex. This step occurs with similar, very rapid, kinetics for both the native sequence SRP RNA and a mutant (A149U) RNA that is incapable of maturing to the native complex (Figures 4A,B). We infer that this Encounter complex largely reflects a fast forming heterogeneous electrostatic interaction between the RNA and SRP19.
Further assembly from the Encounter complex involves a series of complex SRP19 folding steps to form the Stable intermediate complex (Figure 9A). SRP19 loops 1 and 2 undergo, RNA-induced, second order conformational changes with rate constants that are an order of magnitude faster than folding of the SRP19 core domain (Figures 5B and 6). Folding of both the core and loop motifs are concentration-dependent (below 100 nM) even though they follow formation of the Encounter complex. This observation suggests that the Encounter complex establishes a rapid pre-equilibrium with the free RNA and SRP19.
An additional SRP19 folding step is revealed by the concentration-independent folding behavior of loop 2. Below 100 nM, folding at loop 2 shows the same simple concentration-dependent behavior as loop 1 (Figures 6A,B); whereas, at higher concentrations, the rate of the conformational change associated with loop 2 plateaus at 0.9 min−1. Thus, a first-order conformational rearrangement in the still-immature RNA-protein complex limits overall assembly at SRP19 loop 2 (Figure 9A). Completion of the first order rearrangement of loop 2 yields the Native SRP19-RNA complex.
The three-fold protein-RNA interface involving SRP19 loop 2 gates native assembly of the SRP ternary complex
SRP54 does not bind stably to the free SRP RNA.10,24,25 Therefore, SRP54-mediated misassembly via the SRP54-early pathway must involve SRP54 binding to an intermediate SRP19-RNA complex. The two candidate classes of intermediate complexes could be either (i) after formation of the Encounter complex or (ii) after formation of the Stable intermediate complex and prior to maturation of the loop 2-RNA interaction (illustrated by dashed arrows in Figure 8D).
Two independent experiments support a model in which SRP54 binds after formation of the Stable complex. First, direct binding experiments show that SRP54 does not modulate the fluorescence of the complex formed between SRP19 and the A149U RNA, which is trapped at the Encounter complex stage (Figure 4C). Second, formation of the non-compartmentalized complex is strongly dependent on the total concentration of SRP protein and RNA components (Figures 8B,C). This is exactly the concentration dependence expected if SRP54-mediated misassembly occurs after Stable complex formation but not if SRP54 binds after Encounter complex formation (summarized in Figure 8D). The governing kinetic competition involves both concentration–independent (k2) and –dependent (k′3[ ]) steps such that the fraction of non-compartmentalized complex formed (k′3[ ]/k2) increases at higher SRP54 concentrations, if SRP54 binds after the Stable complex forms.
Folding of Loop 2, which occurs after formation of the Stable SRP19-RNA intermediate, has exactly the correct slow concentration-independent behavior required to gate native versus non-compartmentalized assembly. SRP19 loop 2 is packed in a cleft-like interface formed by SRP54 and helix IV of the RNA (Figure 1E).21 Thus, loop 2 both exhibits an intrinsically slow local folding behavior and also lies in a highly constrained three-fold interface in the native particle. These observations support a model in which formation of the non-compartmentalized complex involves rapid RNA binding by SRP54 to a nearly native SRP19-RNA interface but in which folding of loop 2 is still incomplete (Figure 9A). Early binding by SRP54 then results in a conformation in which loop 2 lies outside the correct three-fold interface and further insertion of this loop into its native interaction cleft is kinetically unfavorable.
SRP54-RNA interaction alters the SRP19 folding energy landscape
SRP19 has an almost identical structure in both the SRP19-RNA20 binary and native SRP19-SRP54-RNA21 ternary complexes. SRP54 thus has no role in the native folding of SRP19. In contrast, SRP54-induced misfolding of SRP19 indicates that interactions between SRP54 and the RNA alter the folding energy landscape for SRP19. In the absence of SRP54, SRP19 folds to reach its global energy minimum in a landscape whose most important features include formation of the Encounter and Native complexes and a single major intermediate, the Stable complex (Figure 9B). SRP54 binding to the preformed SRP19-RNA complex does not significantly alter the SRP19-RNA complex but further stabilizes it, creating a deeper well in the landscape (Figure 9B).
The presence of SRP54 alters the folding landscape for SRP19 to create a second, deep, low energy minimum corresponding to the non-compartmentalized complex (Figure 9C). Non-compartmentalized complex formation is concentration dependent indicating that the alternative pathway is kinetically driven and leads to a local rather than a new global minimum relative to the native ternary complex.
Broad implications for multi-component RNP assembly reactions
A simple three-component RNP, derived from the mammalian signal recognition particle, has the ability to form a stable alternative, non-native complex. The native and non-compartmentalized complexes differ only in the order by which the three components assemble. The structural requirements for this order-of-interaction driven misassembly are very modest. Alternative complex formation requires only that the three components communicate structurally and that a portion of one binary interface be modulated by a third component. In the SRP example, structural communication involves a direct three-fold interface, but indirect interactions would suffice, as well.
Evidence that assembly of other RNPs requires spatial or temporal regulation is currently circumstantial, but strongly suggestive. Like the SRP, several other cellular RNPs have multi-site assembly phases involving transit through nucleolar and Cajal body compartments.26,27 Examples in which spatial control potentially facilitates RNP assembly include snRNPs, spliceosomal RNPs and telomerase. A second strategy for regulated RNP assembly would be to impart structural specificity through preferred temporal assembly phases. In the small subunit of the bacterial ribosome,28 three proteins, S3, S10 and S14, and the 3′-domain of 16S RNA form an intimate four-fold interface that appears to physically require that proteins S10 and S14 assemble with the RNA prior to protein S3. Consistent with this structural view, S10 assembles with 16S RNA more rapidly than does S3.3,29 We speculate that slow RNA binding by S3 specifically functions to reduce RNP misassembly at this protein-RNA interface. Multi-fold interfaces that may require early or rapid binding by buried components also exist in the large subunit of the ribosome28, including those formed by L14, L19 and 23S RNA and by L23, L29 and 23S RNA, and in the archaeal H/ACA complex formed by L7ae, Cbf5, Nop10, Gar1 and the H/ACA RNA.30
Given that multi-fold interfaces are common in RNP complexes, we propose that avoiding formation of stable, but misassembled, complexes is of fundamental importance for the structural biogenesis of many RNPs. Mechanisms for regulating assembly, including preferred early and late assembly phases and cellular compartmentalization, may play critical regulatory roles in preventing order-of-interaction driven misassembly for multi-component RNPs in the cell.
Materials and Methods
Preparation of SRP RNA and SRP54
Native and A149U mutant RNAs spanning nts 101–255 of the human SRP RNA (LS RNA) were transcribed in vitro from plasmid phR31 and purified by denaturing gel electrophoresis. The 5′-truncated native and A149U mutant LS RNAs were transcribed from PCR-generated templates. All RNAs were refolded by heating to 95 °C (1 min), snap cooling at 0 °C, incubating at 60 °C (10 min) in RNA refolding buffer [300 mM potassium acetate (pH 7.6), 5 mM MgCl2, 20 mM Hepes (pH 7.6), 0.01% (v/v) Triton], followed by slow cooling to room temperature (~40 min). The Alexa 555-labeled LS RNA was generated by co-folding of the appropriate 5′-truncated RNA and a fluorescently labeled DNA oligonucleotide (Trilink) (see Figure 2B). SRP54 was expressed and purified as described.10
BODIPY-FL and Alexa 488 labeled SRP19 variants
Single cysteine (E31C, W72C, L93C or L106C) SRP19 variant proteins were expressed in E. coli strain BL21-CodonPlus(DE3)-RIL (Stratagene) and purified as described,10 except that the final dialysis buffer was pH 7.2 and contained 2 mM TCEP as the reducing agent. All four sites involve residues whose amino acid identity is phylogentically variable32 and are solvent accessible in both the SRP19-RNA binary20 and SRP19-SRP54-RNA ternary21 complexes. SRP19-fluorophore conjugates were generated by treating each protein (~20 μM) with a 50–100 fold molar excess of a Alexa 488 or BODIPY-FL fluorophore maleimide derivative (22 °C, 2 hr; Molecular probes). Unreacted fluorophore was removed by incubating the (His)6-tagged protein with an Ni2+-NTA slurry (Invitrogen) and washing extensively with protein dilution buffer (PDB) [300 mM NaCl, 50 mM sodium phosphate (pH = 8.0), 10 mM 2-mercaptoethanol]. Fluorophore-labeled proteins were eluted in PDB containing 200 mM imidazole, subjected to dialysis to remove imidazole, and stored in PDB supplemented with 50% (v/v) glycerol at −20 °C. Derivatization extent was calculated from UV absorbance at 280 (protein) and 500 nm (fluorophore). For Alexa-labeled SRP19 variants, the extent of derivatization was 95–100% and for the BODIPY-FL-labeled proteins was ~30%. Equilibrium dissociation constants for SRP19-fluorophore derivatives were determined by filter partitioning19 using internally labeled [32P]-LS RNA (0.01 nM) or the Alexa 555 fluorophore RNA-DNA hybrid. Equilibrium dissociation constants for all fluorophore-containing complexes were either identical or very similar to the native SRP19-LS RNA complex (native SRP19, 5 nM; CysSRP19, 4 nM; Alexa 488-31CysSRP19, 11 nM; Alexa 488-72CysSRP19, 1 nM; Alexa 488-93CysSRP19, 3 nM; Alexa 488-106CysSRP19, 11 nM, BODIPY-FL-72CysSRP19, 4 nM; BODIPY-FL-106CysSRP19, 19 nM).
Monitoring the SRP assembly pathway using FRET and single fluorophore experiments
Fluorescent measurements were performed in a Varian/Cary Eclipse Spectrofluorometer. Final reaction mixtures (22 °C, 500 μl) contained RNA refolding buffer supplemented with 1/5 vol PDB. Calibration between different measurements, when required, was performed using an inert Alexa 647 reference fluorophore. Formation of the Encounter complex between SRP19 and the SRP RNA was monitored via fluorescence resonance energy transfer (FRET) between 20 nM Alexa 488-labeled 31Cys or 72Cys SRP19 variants and 25 nM Alexa 555-labeled LS RNA. Conformational changes in SRP19 upon RNA binding were monitored using single-fluorophore experiments using 5 or 10 nM protein labeled with Alexa 488 or BODIPY-FL and varying concentrations of LS RNA (as indicated in Figure 6). Fluorescence intensity change over time was fit to a single exponential equation. Second order rate constants were calculated from the slope of the line obtained by plotting observed rate versus RNA concentration. For SRP54 binding to the preformed SRP19-RNA complex, assembly was monitored using 20 nM Alexa 488-labeled SRP19-RNA complex and the change in fluorescence intensity as a function of time was fit directly to a second order rate equation. Observed fluorescence = A[(exp(kc1t)-(exp(kc2t))(c1c2)/(c1exp(kc1t)-c2(exp(kc2t)] + b, where k is the second-order rate constant, c1 and c2 are the initial concentrations of SRP54 and the SRP19-RNA complex, A is the amplitude of the fluorescence change, and b is the initial fluorescence of the pre-formed SRP19-RNA complex. For experiments comparing native versus non-compartmentalized assembly, protein concentrations are given in Figures 8B,C and the RNA concentration was twice that of the proteins.
Acknowledgments
We are indebted to Howard Fried for many helpful conversations. This work was supported by the U.S. National Institutes of Health (GM065491 to K.M.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Weeks KM. Protein-facilitated RNA folding. Curr Opin Struct Biol. 1997;7:336–342. doi: 10.1016/s0959-440x(97)80048-6. [DOI] [PubMed] [Google Scholar]
- 2.Williamson JR. Induced fit in RNA-protein recognition. Nat Struct Biol. 2000;10:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 3.Held WA, Ballou B, Mizushima S, Nomura M. Assembly mapping of 30S ribosomal proteins from Escherichia coli. J Biol Chem. 1974;249:3103–3111. [PubMed] [Google Scholar]
- 4.Agalarov SC, Prasad GS, Funke PM, Stout CD, Williamson JR. Structure of the S15, S6, S18-rRNA complex: Assembly of the 30S ribosome central domain. Science. 2000;288:107–112. doi: 10.1126/science.288.5463.107. [DOI] [PubMed] [Google Scholar]
- 5.Agalarov SC, Williamson JR. A hierarchy of RNA subdomains in assembly of the central domain of the 30 S ribosomal subunit. RNA. 2000;6:402–408. doi: 10.1017/s1355838200991945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 7.Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat Rev Mol Cell Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 8.Dill KA, Chan HS. From Levinthal to pathways to funnels. Nat Struct Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 9.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 10.Maity TS, Leonard CW, Rose MA, Fried HM, Weeks KM. Compartmentalization directs assembly of the signal recognition particle. Biochemistry. 2006;45 doi: 10.1021/bi060890g. in press. [DOI] [PubMed] [Google Scholar]
- 11.Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu Rev Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 12.Siegel V, Walter P. Each of the activities of signal recognition particle (SRP) is contained within a distinct domain: Analysis of biochemical mutants of SRP. Cell. 1988;52:39–49. doi: 10.1016/0092-8674(88)90529-6. [DOI] [PubMed] [Google Scholar]
- 13.Halic M, Becker T, Pool MR, Spahn CMT, Grassucci RA, Frank J, Beckmann R. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 2004;427:808–814. doi: 10.1038/nature02342. [DOI] [PubMed] [Google Scholar]
- 14.Jacobson MR, Pederson T. Localization of signal recognition particle RNA in the nucleolus of mammalian cells. Proc Natl Acad Sci USA. 1998;95:7981–7986. doi: 10.1073/pnas.95.14.7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Politz JC, Yarovoi S, Kilroy SM, Gowda K, Zwieb C, Pederson T. Signal recognition particle components in the nucleolus. Proc Natl Acad Sci USA. 2000;97:55–60. doi: 10.1073/pnas.97.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alavian CN, Politz JCR, Lewandowski LB, Powers CM, Pederson T. Nuclear export of signal recognition particle RNA in mammalian cells. Biochem Biophys Res Commun. 2004;313:351–355. doi: 10.1016/j.bbrc.2003.11.126. [DOI] [PubMed] [Google Scholar]
- 17.Ciufo LF, Brown JD. Nuclear export of yeast signal recognition particle lacking Srp54p by the Xpo1p/Crm1p NES-dependent pathway. Curr Biol. 2000;10:1256–1264. doi: 10.1016/s0960-9822(00)00743-0. [DOI] [PubMed] [Google Scholar]
- 18.Grosshans H, Deinert K, Hurt E, Simos G. Biogenesis of the signal recognition particle (SRP) involves import of SRP proteins into the nucleolus, assembly with the SRP-RNA, and Xpo1p-mediated export. J Cell Biol. 2001;153:745–761. doi: 10.1083/jcb.153.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rose MA, Weeks KM. Visualizing induced fit in early assembly of the human signal recognition particle. Nat Struct Biol. 2001;8:515–520. doi: 10.1038/88577. [DOI] [PubMed] [Google Scholar]
- 20.Oubridge C, Kuglstatter A, Jovine L, Nagai K. Crystal Structure of SRP19 in complex with the S domain of SRP RNA and its implication for the assembly of the signal recognition particle. Mol Cell. 2002;9:1251–1261. doi: 10.1016/s1097-2765(02)00530-0. [DOI] [PubMed] [Google Scholar]
- 21.Kuglstatter A, Oubridge C, Nagai K. Induced structural changes of 7SL RNA during the assembly of human signal recognition particle. Nat Struct Biol. 2002;9:740–744. doi: 10.1038/nsb843. [DOI] [PubMed] [Google Scholar]
- 22.Hainzl T, Huang S, Sauer-Eriksson AE. Structural insights into SRP RNA: an induced fit mechanism for SRP assembly. RNA. 2005;11:1043–50. doi: 10.1261/rna.2080205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hainzl T, Huang S, Sauer-Eriksson AE. Structure of the SRP19–RNA complex and implications for signal recognition particle assembly. Nature. 2002;417:767–771. doi: 10.1038/nature00768. [DOI] [PubMed] [Google Scholar]
- 24.Romisch K, Webb J, Herz J, Prehn S, Frank R, Vingron M, Dobberstein B. Homology of 54K protein of signal-recognition particle, docking protein and two E. coli proteins with putative GTP-binding domains. Nature. 1989;340:478–482. doi: 10.1038/340478a0. [DOI] [PubMed] [Google Scholar]
- 25.Gowda K, Chittenden K, Zwieb C. Binding site of the M-domain of human protein SRP54 determined by systematic site-directed mutagenesis of signal recognition particle RNA. Nucl Acids Res. 1997;25:388–394. doi: 10.1093/nar/25.2.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerbi SA, Borovjagin AV, Lange TS. The nucleolus: a site of ribonucleoprotein maturation. Curr Opin Cell Biol. 2003;15:318–325. doi: 10.1016/s0955-0674(03)00049-8. [DOI] [PubMed] [Google Scholar]
- 27.Matera AG, Shpargel KB. Pumping RNA: nuclear bodybuilding along the RNP pipeline. Curr Opin Cell Biol. 2006;18:317–24. doi: 10.1016/j.ceb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 28.Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–42. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 29.Talkington MW, Siuzdak G, Williamson JR. An assembly landscape for the 30S ribosomal subunit. Nature. 2005;438:628–32. doi: 10.1038/nature04261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L, Ye K. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature. 2006;443:302–307. doi: 10.1038/nature05151. [DOI] [PubMed] [Google Scholar]
- 31.Zwieb C. Interaction of protein SRP19 with signal recognition particle RNA lacking individual RNA-helices. Nucl Acids Res. 1991;19:2955–2960. doi: 10.1093/nar/19.11.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenblad MA, Gorodkin J, Knudsen B, Zwieb C, Samuelsson T. SRPDB: Signal Recognition Particle Database. Nucleic Acids Res. 2003;31:363–4. doi: 10.1093/nar/gkg107. [DOI] [PMC free article] [PubMed] [Google Scholar]


