Abstract
The control of mRNA stability is crucial to the regulation of cytokine expression. We describe here a novel, potent destabilizing element found in the 3′ untranslated region of granulocyte colony-stimulating factor mRNA. This element, which appears to require at least one stem–loop structure, we term the stem–loop destabilizing element (SLDE). Functionally equivalent elements appear to also exist in the interleukin 2 and interleukin 6 mRNAs. The SLDE is functionally distinct from the A+U-rich elements, which are also present in these and other cytokine mRNAs, because it destabilizes a chimeric mRNA in a tumor cell line in which A+U-rich elements do not function. In addition, the effect of the SLDE is insensitive to calcium ionophore and is therefore regulated independently of A+U destabilizing elements. The existence of two distinct mRNA-destabilizing elements provides an additional mechanism for the differential regulation of cytokine expression.
The control of mRNA degradation rate is a major factor in the regulation of expression of many cytokines and proto-oncogenes. The mRNAs of hemopoietic cytokines such as granulocyte colony-stimulating factor (G-CSF), granulocyte/macrophage colony-stimulating factor (GM-CSF), interleukin 1 (IL-1), IL-2, IL-3, IL-6, and tumor necrosis factor α are unstable in unstimulated cells but become stabilized during inflammatory and immune responses (1, 2, 3, 4, 5). A common feature of these and numerous other mRNAs is the presence in the 3′ untranslated region (3′UTR) of multiple copies of the sequence UUAUUUA(U/A)(U/A), which has been shown to act as a destabilizing element (6, 7, 8, 9). This A+U-destabilizing element (AUDE) is sufficient to confer regulation of mRNA turnover in response to calcium (10, 11), but the AUDE alone is not sufficient for stabilization in response to natural inducers of cytokines such as IL-1 and tumor necrosis factor α, or in response to phorbol ester (12, 13, 14).
It is unlikely that a single cis-acting element such as the AUDE would be sufficient to engender the various modes of regulation that individual mRNAs can be subject to. For example, in T cells stimulated with anti-CD3 and anti-CD28, the IL-2, interferon-γ, tumor necrosis factor α, and GM-CSF mRNAs are stabilized, but the c-fos mRNA, which also contains AUDEs, is not (2). Similarly, in monocytes stimulated with lipopolysaccharide, the stability of IL-10 mRNA (which also contains AUDEs) is regulated differently to the G-CSF and GM-CSF mRNAs (15). Furthermore, vascular endothelial growth factor mRNA also contains multiple AUDEs, but this mRNA, which can be expressed by most cell types, is stabilized in response to hypoxia (16).
We had previously established a system for identifying the functional destabilizing element within A+U-rich sequences (8) and sought to apply the system to search for additional regulatory elements in the G-CSF mRNA. We describe here the identification of a novel, potent destabilizing element in the 3′UTR of the G-CSF mRNA that functions independently of A+U-rich sequences. The novel element appears to involve at least one stem–loop structure. We also present evidence that functionally similar destabilizing elements occur in the 3′UTRs of IL-2 and IL-6 mRNAs.
MATERIALS AND METHODS
Plasmid Construction.
Plasmids were constructed by insertion of the sequences described below into the SacI site, or between the KpnI and SacI sites, of pfGH (8). pfGH-AU1 contains the sequence (see in Fig. 1) generated by annealing of chemically synthesized oligonucleotides. pfGH-AU2 and pfGH-GCSF contain the sequence shown in Fig. 1 and the full 3′UTR of human G-CSF, respectively, generated by PCR amplification from a plasmid containing the human G-CSF cDNA (kindly provided by F. Shannon, Hanson Centre for Cancer Research, Adelaide, Australia). We constructed the fGH variants SL1, SL2, SL3, SL6, and SL7 by insertion of PCR amplification products into the SacI site of pfGH. The inserted sequences were as shown in Fig. 4, except that AAGCTT was also included at the 5′ end of the insert and TGTTCCCC was included at the 3′ end. The PCR amplification to generate the insert for fGHSL5 included KpnI and SacI sites immediately flanking the sequence shown in Fig. 4. fGHSL2D was created by insertion of annealed oligonucleotides generating the sequence shown in Fig. 4 immediately flanked by KpnI and SacI sites. To prepare pfGH-IL2 and pfGH-IL6, the full 3′UTRs of human IL-2 and IL-6 cDNAs, respectively, flanked by 5′-KpnI and 3′-SacI sites, were PCR-amplified from plasmids pAT153hIL2 (kindly provided by W. Fiers, Laboratory of Molecular Biology, University of Gent, Belgium) and pBSF2.3.8.1 (kindly provided by T. Hirano, Osaka University Medical School, Japan).
Figure 1.
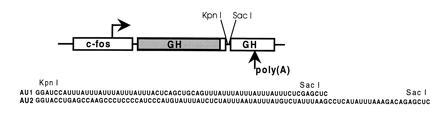
The fGH gene and two of the sequences inserted into the 3′UTR. Boxes indicate regions derived from the chicken c-fos gene and growth hormone cDNA, and lines indicate vector-derived sequences. The translated region is shaded and the polyadenylation site is indicated.
Figure 4.
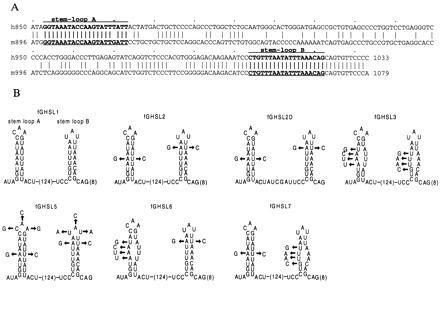
(A) The sequence of the SL1 region of human G-CSF cDNA aligned with the homologous region from the murine G-CSF cDNA. The alignment of GenBank entries HSGCSFR (21) and MUSGCSF (22) was made using the GCG program gap. (B) Various sequences inserted into the 3′UTR of the fGH gene. SL1, from the 3′UTR of human G-CSF, includes two putative stem–loops that are conserved in the mouse and human mRNAs. The 124-base sequence between the stems and 8 bases 3′ of stem–loop B are not shown. The other sequences were derived from SL1 as shown.
Cell Culture and Transfection.
Polyclonal NIH 3T3 cultures transfected by electroporation with variants of the pfGH plasmid were established and cultured as described (8). Cultures of stably transfected 5637 cells were prepared similarly, but electroporation settings of 270 V and 500 μF were used. Serum stimulation was performed as described (8) except that, where indicated, 2 μM A23187 was included in the medium at the time of serum stimulation with 15% fetal calf serum.
RNA Isolation and Quantitation.
RNA was isolated at various times after serum stimulation by the method of Chomczynski and Sacchi (17). Specific transcripts were quantitated by an RNase protection assay using an RNA probe complementary to the growth hormone translated region. Glyceraldehyde-3-phosphate dehydrogenase probe was used as an internal quantitation standard, as described (8). Signals were quantitated by phosphorimaging (Molecular Dynamics).
RESULTS
The 3′UTR of G-CSF mRNA Contains a Destabilizing Element That Is Insensitive to Calcium Ionophore.
To identify regulatory elements in cytokine mRNAs, we have used a reporter mRNA, termed fGH (Fig. 1) (8), which is composed mostly of growth hormone sequences. Transcription of fGH is driven by the fos promoter, so that a brief transcriptional pulse can be generated by serum stimulation, without the use of transcription inhibitors. The fGH mRNA is normally stable (8) but can be made unstable by the insertion of a destabilizing element at unique restriction sites in the 3′UTR, as is shown by the insertion of the synthetic A+U-rich sequence in fGH-AU1 (Figs. 1 and 2). Similarly, insertion of the A+U-rich region from the G-CSF 3′UTR rendered the reporter mRNA unstable (fGH-AU2; Figs. 1 and 2). As expected for instability conferred by A+U-rich elements, both mRNAs could be stabilized by addition of the calcium ionophore A23187 (Fig. 2). To our surprise, however, when the entire 3′UTR of G-CSF was inserted into the reporter gene, the resulting mRNA (fGH-GCSF) remained unstable in cells treated with the calcium ionophore (Fig. 3). This result suggests either that the G-CSF 3′UTR contains sequences outside the main A+U-rich region that influence the regulation of A+U-mediated degradation, or that the G-CSF 3′UTR contains additional instability elements whose function is not regulated by calcium.
Figure 2.
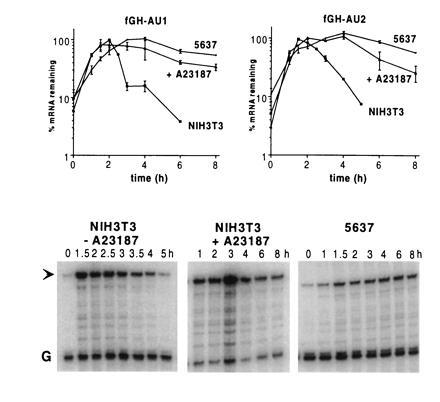
AUDE-containing mRNAs are stable in 5637 bladder carcinoma cells and in NIH 3T3 cells treated with the calcium ionophore A23187. RNA was isolated at various times after serum stimulation of cells stably transfected with chimeric fGH genes containing either a synthetic A+U-destabilizing sequence (fGH-AU1) or the A+U-rich region from G-CSF (fGH-AU2), inserted in the 3′UTR. Specific mRNAs were quantitated by an RNase protection assay. Both mRNAs were unstable in NIH 3T3 cells but stable in 5637 cells, and both were stabilized in NIH 3T3 cells treated with A23187. The graphs show data as mean ± SEM from three experiments. RNase protection gels from a single representative fGH-AU2 experiment are shown, with an arrow indicating the fGH-AU2 signal and the signal from the glyceraldehyde-3-phosphate dehydrogenase internal standard indicated by a G.
Figure 3.
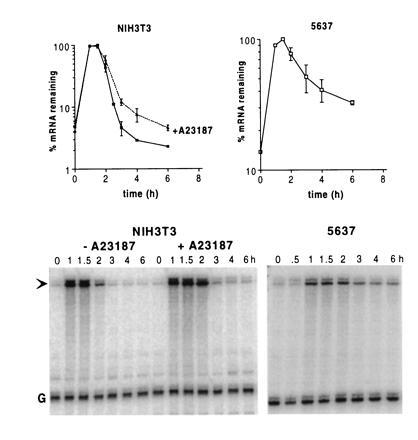
An mRNA containing the entire 3′UTR of G-CSF is unstable in both NIH 3T3 and 5637 cells, and is not stabilized in NIH 3T3 cells treated with the calcium ionophore A23187. RNA was isolated at various times after serum stimulation of cells stably transfected with the fGH-GCSF gene and subjected to an RNase protection assay. The graphs show data as mean ± SEM from two experiments, and a representative RNase protection gel is shown with an arrow indicating the probe protected by fGH-GCSF and G indicating the glyceraldehyde-3-phosphate dehydrogenase internal standard.
AUDEs Do Not Function in 5637 Bladder Carcinoma Cells, but the G-CSF Destabilizing Element Does.
Many tumor cells constitutively express one or more cytokines, due at least in part to the abnormal stability of the cytokine mRNA in these cells (18). We transfected 5637 bladder carcinoma cells with the AUDE-containing reporter genes fGH-AU1 and fGH-AU2 and found that these mRNAs were constitutively stable (Fig. 2), indicating that the A+U-mediated system of mRNA degradation does not function in these cells. However, the fGH-GCSF mRNA, which contains the entire 3′UTR of G-CSF, was relatively unstable in 5637 cells (Fig. 3). These results, together with the experiments with calcium ionophore, suggest that the 3′UTR of G-CSF contains a destabilizing element that functions independently of the A+U-mediated degradation system.
Identification and Characterization of the Stem–Loop Destabilizing Element (SLDE).
To identify candidate sequences for such an additional destabilizing element in the G-CSF 3′UTR, we compared the sequences of the human and mouse mRNAs. We noticed that the 3′UTRs (approximately 850 nt) were rather dissimilar, except for 3 segments of 50, 40, and 20 nt. The 50-nt segment contains the A+U-rich region, which we had inserted to make fGH-AU2. The conserved 40- and 20-nt regions each contained sequences capable of forming a stem–loop structure, which we call stem–loop A and stem–loop B, respectively (Fig. 4). We inserted a 183-nt segment (SL1), encompassing both the 40-nt and the 20-nt conserved sequences, into fGH to create fGHSL1. The fGHSL1 mRNA, like the fGH-GCSF mRNA, was unstable (Fig. 5), suggesting that the 183-nt SL1 region contained the additional instability element. However, the interpretation was slightly complicated by the fact that the SL1 region includes two sequences that almost match the AUDE sequence, UUAUUUA(U/A)(U/A), one in the putative stem of stem–loop A, the other in the stem and loop of stem–loop B. We therefore made point mutations to ensure these potential AUDEs were not functional, creating fGHSL2 (Fig. 4). To maintain the potential for stem–loop formation in fGHSL2, we simultaneously introduced complementary changes to maintain base pairing in the stems. The fGHSL2 mRNA was at least as unstable as fGHSL1, was not stabilized in the presence of ionophore (Fig. 5), and functioned in 5637 cells (Fig. 6), indicating that the instability element(s) were still intact and were not regulated by calcium. On the other hand, disruption of the stem–loops by uncompensated mutations in both stems, but leaving the potential AUDEs intact (fGHSL3; Fig. 4), removed the destabilizing effect (Fig. 5). Thus, the presence of one or both stem–loops, but not of the AUDE-like sequences, is necessary for the destabilizing effect of this region.
Figure 5.
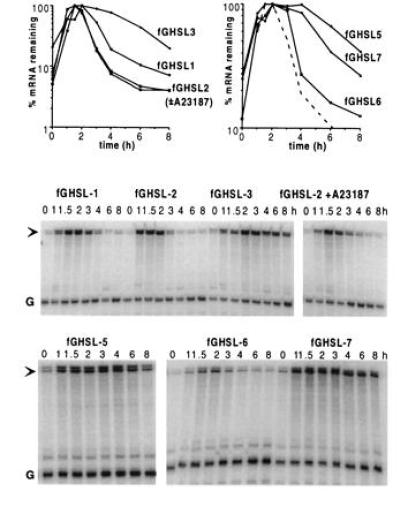
The effects of SL1 and its derivatives on the stability of the fGH mRNA. The sequences inserted into fGH are described in Fig. 4. RNA was isolated at various times after serum stimulation of NIH 3T3 cells stably transfected with variants of the fGH gene containing the indicated sequences inserted in the 3′UTR. The stability of the fGHSL2 mRNA was unchanged by treatment of the cells with the calcium ionophore A23187. The dashed line in the graph at right shows the fGHSL1 data replotted for comparison.
Figure 6.
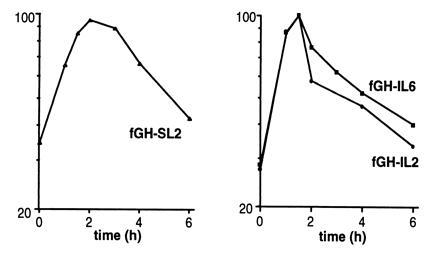
SL2 and the 3′UTRs of IL-2 and IL-6 destabilize the fGH mRNA in 5637 cells. RNA was isolated at various times after serum stimulation of 5637 cells stably transfected with the fGH-SL2, fGH-IL2, or fGH-IL6 genes and analyzed by RNase protection assay.
In fGHSL5, we changed the sequence of both loops, changing loop A from CAA to GCG and loop B from UAU to ACA (Fig. 4). This eliminated the destabilizing effect, indicating that at least some part of the loop sequence(s) is essential (Fig. 5).
To determine whether one or both of the conserved stem–loops acts as a destabilizing element, we made fGHSL6 and fGHSL7, in which mutations were made to disrupt stem A and stem B, respectively (Fig. 4). The fGHSL6 mRNA, with stem A eliminated, remained unstable (Fig. 5), indicating that stem–loop A is not necessary for destabilization, although the fGHSL6 mRNA was not quite as unstable as the fGHSL1 mRNA. However, disruption of stem B caused a marked increase in the stability of the mRNA (fGHSL7; Fig. 5), indicating that stem–loop B is essential for instability. Finally, to test whether the region between the stem–loops is part of the destabilizing element, we made fGHSL2D, in which the 130-nt interloop region of fGHSL2 was replaced by just 12 nt. The fGHSL2D mRNA was stable (data not shown), indicating that part of the interloop region, or at least some sequence flanking the stem–loop, is required for the destabilizing effect.
Taken together, these results show that a region of the G-CSF 3′UTR that includes the conserved stem–loop B functions as a destabilizing element and is responsible for the instability of fGH-GCSF mRNA in ionophore-treated cells and in 5637 cells.
Functionally Similar Destabilizing Elements Are Present in the IL-2 and IL-6 3′UTRs.
To investigate whether cytokines in addition to G-CSF may contain similar non-AU instability elements, we made a series of chimeric genes in which entire 3′UTRs from various AUDE-containing cytokine genes replaced the 3′UTR of fGH. We compared the stabilities of the resulting mRNAs in NIH 3T3 cells in the presence and absence of calcium ionophore. The fGH-GMCSF and fGH-IL4 mRNAs, containing the 3′UTR from GM-CSF and IL-4, respectively, were significantly stabilized by ionophore (data not shown), suggesting that for these mRNAs instability is primarily due to the A+U-rich regions. In contrast, the fGH-IL2 and fGH-IL6 mRNAs were only modestly stabilized in the presence of ionophore (Fig. 7) and furthermore functioned in 5637 cells (Fig. 6). Thus, the IL-2 and IL-6 mRNAs appear to contain instability elements with functionally similar properties to the stem–loop element of G-CSF.
Figure 7.
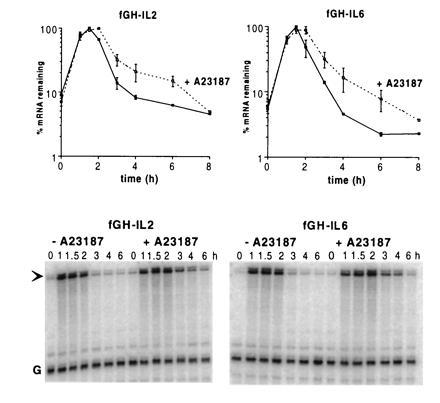
The 3′UTRs of IL-2 and IL-6 destabilize the fGH mRNA in the presence and absence of the calcium ionophore A23187. The entire 3′UTR of IL-2 or IL-6 was inserted near the termination codon of the fGH gene, generating fGH-IL2 and fGH-IL6, respectively. RNA was isolated at various times after serum stimulation of NIH 3T3 cells stably transfected with either gene and quantitated by RNase protection assay. The data shown are means ± SEM from two experiments. Addition of A23187 at the time of serum stimulation resulted in only a modest increase in the survival of the mRNAs compared with the effect on mRNAs containing only A+U-rich insertions (see Fig. 2).
DISCUSSION
Given the variety of mRNAs whose stability is regulated, and the diversity of situations that induce changes in the stability of one or more of these mRNAs, it is to be expected that various different RNA sequence elements will participate in this regulation. The most common instability element is the A+U-rich element, of which there may be several classes (19, 20). This may explain some of the differences between the regulation of cytokine mRNAs, which are typically stabilized during inflammatory and immune responses, and the regulation of proto-oncogene mRNAs, such as the c-fos and c-myc mRNAs, which remain unstable (ref. 2 and unpublished data). However, the A+U-rich elements in most cytokine mRNAs, including the G-CSF, IL-2, and IL-6 mRNAs, appear to belong to a single class, which is composed of multiple copies of the sequence UUAUUUA(U/A)(U/A).
In this report, we show that the 3′UTRs of the G-CSF, IL-2, and IL-6 mRNAs also contain a destabilizing element that is functionally distinct from the AUDE. The element in the G-CSF 3′UTR is located within a 183-nt region and appears to require at least one stem–loop structure (stem–loop B). We consequently refer to the element as the SLDE. Although the sequence within stem–loop B is identical in the human and mouse mRNAs, the element remains functional when an AU base pair is replaced by a GC base pair, suggesting that it is the structure of the stem, rather than its sequence, that is important for function. Changing the sequence of the loops in both stem–loop A and stem–loop B inactivated the element, but we do not know which and how many loop nucleotides are essential. Because of this, we cannot predict by inspection of the sequence, or from computer-generated predictions of secondary structure, where the functionally equivalent element is in the IL-2 and IL-6 mRNAs.
Unlike the AUDE, the function of the SLDE is unaffected by the calcium flux generated by the calcium ionophore A23187. In addition, the SLDE functions in a bladder carcinoma cell line, 5637, in which AUDEs do not function. Thus, the SLDE appears to function independently of AUDEs. The presence of the SLDE in some cytokine mRNAs provides a possible mechanism for regulating the stability of these mRNAs independently from other AUDE-containing cytokine mRNAs that may be simultaneously present in the same cell. The details of the regulatory properties conferred by the SLDE remain to be determined.
Acknowledgments
We thank Justin Dibbens, Leeanne Coles, Tom Gonda, and Mathew Vadas for helpful comments on the manuscript. This work was supported by a program grant from the National Health and Medical Research Council of Australia.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. §1734 solely to indicate this fact.
Abbreviations: AUDE, A+U destabilizing element [UUAUUUA(U/A)(U/A)]; G-CSF, granulocyte colony-stimulating factor; GM-CSF, granulocyte/macrophage colony-stimulating factor; IL, interleukin; SLDE, stem–loop destabilizing element; 3′UTR, 3′ untranslated region.
References
- 1.Koeffler H P, Gasson J, Tobler A. Mol Cell Biol. 1988;8:3432–3438. doi: 10.1128/mcb.8.8.3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lindsten T, June C H, Ledbetter J A, Stella G, Thompson C B. Science. 1989;244:339–343. doi: 10.1126/science.2540528. [DOI] [PubMed] [Google Scholar]
- 3.Guba S C, Stella G, Turka L A, June C H, Thompson C B, Emerson S G. J Clin Invest. 1989;84:1701–1706. doi: 10.1172/JCI114352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akashi M, Loussararian A H, Adelman D C, Saito M, Koeffler H P. J Clin Invest. 1990;85:121–129. doi: 10.1172/JCI114401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elias J A, Lentz V. J Immunol. 1990;145:161–166. [PubMed] [Google Scholar]
- 6.Shaw G, Kamen S. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 7.Wilson T, Treisman R. Nature (London) 1988;363:396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- 8.Lagnado C A, Brown C Y, Goodall G J. Mol Cell Biol. 1994;14:7984–7995. doi: 10.1128/mcb.14.12.7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zubiaga A M, Belasco J G, Greenberg M E. Mol Cell Biol. 1995;15:2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wodnar Filipowicz A, Moroni C. Proc Natl Acad Sci USA. 1990;87:777–781. doi: 10.1073/pnas.87.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwai Y, Akahane K, Pluznik D H, Cohen R B. J Immunol. 1993;150:4386–4394. [PubMed] [Google Scholar]
- 12.Akashi M, Shaw G, Hachiya M, Elstner E, Suzuki G, Koeffler P. Blood. 1994;83:3182–3187. [PubMed] [Google Scholar]
- 13.Iwai Y, Bickel M, Pluznik D H, Cohen R B. J Biol Chem. 1991;266:17959–17965. [PubMed] [Google Scholar]
- 14.Akashi M, Shaw G, Gross M, Saito M, Koeffler H P. Blood. 1991;78:2005–2012. [PubMed] [Google Scholar]
- 15.Brown C Y, Lagnado C A, Vadas M A, Goodall G J. J Biol Chem. 1996;271:20108–20112. doi: 10.1074/jbc.271.33.20108. [DOI] [PubMed] [Google Scholar]
- 16.Levy A P, Levy N S, Wegner S, Goldberg M A. J Biol Chem. 1995;270:13333–13340. doi: 10.1074/jbc.270.22.13333. [DOI] [PubMed] [Google Scholar]
- 17.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 18.Ross H J, Sato N, Ueyama Y, Koeffler H P. Blood. 1991;77:1787–1795. [PubMed] [Google Scholar]
- 19.Chen C Y, Shyu A B. Mol Cell Biol. 1994;14:8471–8482. doi: 10.1128/mcb.14.12.8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen C Y, Shyu A B. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 21.Nagata S, Tsuchiya M, Asano S, Kaziro Y, Yamazaki T, Yamamoto O, Hirata Y, Kubota N, Oheda M, Nomura H, Ono M. Nature (London) 1986;319:415–418. doi: 10.1038/319415a0. [DOI] [PubMed] [Google Scholar]
- 22.Tsuchiya M, Asano S, Kaziro Y, Nagata S. Proc Natl Acad Sci USA. 1986;83:7633–7637. doi: 10.1073/pnas.83.20.7633. [DOI] [PMC free article] [PubMed] [Google Scholar]


