Abstract
Arsenic trioxide (ATO) is an effective cancer therapeutic drug for acute promyelocytic leukemia and has potential anticancer activity against a wide range of solid tumors. ATO exerts its effect mainly through elevated oxidative stress, but the exact molecular mechanism remains elusive. The thioredoxin (Trx) system comprising NADPH, thioredoxin reductase (TrxR), and Trx and the glutathione (GSH) system composed of NADPH, glutathione reductase, and GSH supported by glutaredoxin are the two electron donor systems that control cellular proliferation, viability, and apoptosis. Recently, the selenocysteine-dependent TrxR enzyme has emerged as an important molecular target for anticancer drug development. Here, we have discovered that ATO irreversibly inhibits mammalian TrxR with an IC50 of 0.25 μM. Both the N-terminal redox-active dithiol and the C-terminal selenothiol-active site of reduced TrxR may participate in the reaction with ATO. The inhibition of MCF-7 cell growth by ATO was correlated with irreversible inactivation of TrxR, which subsequently led to Trx oxidation. Furthermore, the inhibition of TrxR by ATO was attenuated by GSH, and GSH depletion by buthionine sulfoximine enhanced ATO-induced cell death. These results strongly suggest that the ATO anticancer activity is by means of a Trx system-mediated apoptosis. Blocking cancer cell DNA replication and repair and induction of oxidative stress by the inhibition of both Trx and GSH systems are suggested as cancer chemotherapeutic strategies.
Keywords: glutathione, reactive oxygen species, oxidative stress, cell death
Arsenic trioxide (ATO) has been used as an anticancer drug for several thousands of years in traditional medicine and recently has been shown to be efficient in the treatment of the newly diagnosed and relapsed acute promyelocytic leukemia (1–4). More interestingly, this anticancer efficiency of ATO was extended to many solid tumors (3). Accumulating evidence suggests that protein sulfhydryl (-SH) groups could be the main targets of the drug (5). However, the exact mechanism of ATO is still vague and incompletely understood. For example, it is known that glutathione (GSH) is a predominant thiol molecule in the cell that plays a critical role in the detoxification of arsenicals (6–9). The sensitivity of cancer cell lines is inversely correlated with their GSH content (6, 10). However, the arsenic cytotoxicity is not caused by the direct inhibition of GSH-related enzymes, including glutathione peroxidase (GPx), glutathione reductase (GR), and glutathione transferase (GST). The arsenic concentrations required to inhibit these enzymes are much higher than the physiologically relevant concentrations (11). Therefore, it is important to clarify the molecular target to elucidate the cell death mechanism induced by ATO.
The thioredoxin (Trx) system, composed of thioredoxin reductase (TrxR), Trx, and NADPH, is one of the main thiol-dependent electron donor systems in the cell and plays critical roles in the regulation of the cellular redox environment and a wide range of cellular activities, such as cell viability and proliferation (12, 13). Mammalian cells have a homodimeric TrxR1 in the cytosol/nucleus (14, 15), TrxR2 in mitochondria (16, 17), and TGR in the testis (18). The three-dimensional crystal structure reveals that TrxR is structurally similar to GR with the FAD and NADPH-binding domains (19). Besides the N-terminal -Cys-Val-Asn-Val-Gly-Cys-dithiol/disulfide, mammalian TrxRs contain a 16-residue C-terminal elongation with the active-site sequence -Gly-Cys-Sec-Gly-OH. The selenocysteine (Sec) (U) in the open C terminus is essential for the reducing activity of TrxR (20). Selenol has a low pKa (5.3) value, and thus selenolate is the predominant form under physiological conditions. This property and the accessible location of C-terminal active site account for the wide substrate specificity of TrxR (21).
The Trx system, especially TrxR, has been suggested as a new target for anticancer drug development because TrxR and Trx are overexpressed in many aggressive tumors, and the tumor cells appear more dependent on a Trx system perhaps for the constant requirement of DNA synthesis (22, 23). More recently, TrxR knockdown by a stable transfection with a small interfering RNA construct changed the mouse lung carcinoma cell morphology and anchorage-independent growth property to be similar to those of normal cells. Furthermore, the tumor progression and metastasis were dramatically reduced when these TrxR knockdown cells were injected into mice (24). These results strongly suggest that TrxR is essential for tumor cell growth in vivo. Considering the extensive involvement of Sec-containing TrxR in the redox regulations and the close link with cancer (25), we have investigated the effect of ATO on TrxR in vitro and in MCF-7 cells. ATO exhibited excellent inhibition of TrxR and thus induced the Trx-mediated cellular redox changes in cells.
Results
Inhibition of TrxR by ATO in Vitro.
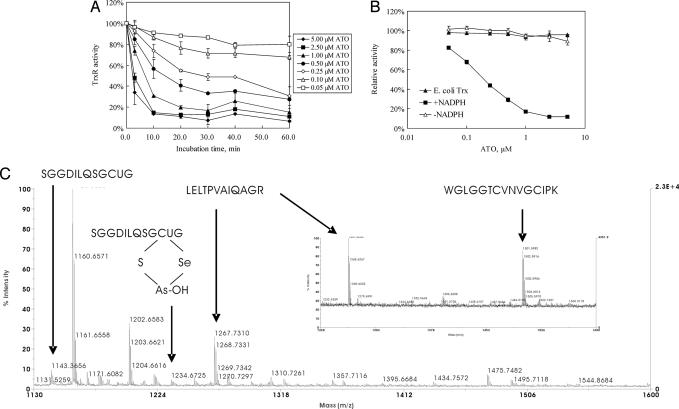
ATO inhibited the activity of TrxR in a time- and concentration-dependent manner as shown by an activity assay using direct reduction of 5 mM 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB) (Fig. 1A). In the range of 0.05–5 μM ATO, the activity of 50 nM recombinant rat TrxR decreased with increasing ATO concentration. When the ATO concentration was >1 μM, TrxR activity decreased to a plateau value in 10 min, whereas for ATO concentrations <1 μM, it took 20–30 min to obtain a constant TrxR activity. The calculated IC50 was ≈0.25 μM after a 30-min incubation. Moreover, the whole Trx system was inhibited by ATO with an IC50 value similar to that shown by an insulin reduction assay (26). The presence of 2 μM human Trx in the incubation solution did not result in a big change of the inhibitory efficiency, suggesting that ATO attacked the TrxR specifically (data not shown).
Fig. 1.
Inhibition of TrxR by ATO in vitro. (A) A 50 nM concentration of NADPH-reduced recombinant rat TrxR was incubated with different concentrations of ATO for different times, and then TrxR activity was assayed by DTNB reduction assay. Error bars represent the standard deviation of duplicate experiments. (B) A 50 nM concentration of recombinant rat TrxR in the presence (■) or absence (▵) of 200 μM NADPH was incubated with different concentrations of ATO for 30 min, and then TrxR activity was assayed by DTNB reduction assay. NADPH (200 μM), E. coli TrxR (25 nM), and E. coli Trx (2 μM) were incubated with different concentrations of ATO for 30 min, and then the activity was measured by the insulin reduction method (▴). (C) Peptide mass of TrxR–arsenic by MALDI mass spectrometry. Here 1 μM NADPH-reduced recombinant rat TrxR was incubated with 10 μM ATO, denatured in 8 M urea, and digested by trypsin and then subjected to mass spectra analysis.
The inhibition of mammalian TrxR by ATO was irreversible. When a completely inhibited sample of TrxR was passed through a desalting Sephadex G-25 column to remove the ATO, no activity was recovered in the eluted enzyme. This property is the same as for the inhibition of TrxR by other inhibitors such as flavonoids and curcumin (27, 28). Moreover, no increased NADPH oxidase activity was induced for the inhibition of TrxR by ATO, and the inhibition was not affected by the presence of superoxide dismutase and catalase (data not shown).
The inhibition by ATO depended on the redox state of TrxR because without the presence of NADPH, TrxR could not be inhibited by ATO (Fig. 1B). In TrxR both the N-terminal disulfide and C-terminal active-site Cys-Sec are oxidized, and they are reduced by NADPH. However, NADPH did not participate directly in the reaction between TrxR and ATO because after removing the coenzyme by a desalting column, the isolated reduced TrxR with free thiol and selenol groups was inhibited by ATO (data not shown). Additionally, there is a remarkable difference between the active sites of mammalian and bacterial TrxR. Both enzymes have a redox-active disulfide plus NADPH- and FAD-binding domains, but mammalian TrxR possesses its active site in an elongation containing a penultimate Sec residue (29). ATO (0–5 μM) exhibited no inhibitory effect on the Escherichia coli Trx system, indicating that the active site Cys-Sec, especially the Sec is critical for the inhibitory effect (Fig. 1B).
In fact, the critical role of Sec in the C-terminal active site for the high affinity of arsenic with TrxR was shown by the association of TrxR with phenylarsine oxide (PAO)–Sepharose, which has been used to purify the recombinant TrxR (30). The recombinantly expressed rat enzyme from E. coli cell lysate with an enzymatic specific activity of 15 units/mg protein is composed of nearly half of full-length active TrxR and half as a truncated TrxR-inactive form lacking the last two Sec-Gly amino acid residues. The active full-length TrxR with its C-terminal GCUG motif is binding much stronger than the truncated form with PAO–Sepharose, a resin containing arsenic. Full-length active TrxR with a specific activity of 35 units/mg protein is obtained after the PAO–Sepharose purification (30).
To see the exact binding site of TrxR with ATO, particularly whether the N-terminal disulfide or C-terminal active site reacts with ATO, we examined the trypsin-digested peptide mass of TrxR–ATO with a matrix-assisted laser desorption/ionization mass spectrometry (MALDI). Compared with the control mass spectra of TrxR, a new mass peak at 1,234.6 appeared, which fit the calculation value 1,234.3 of a C-terminal fragment SGGDILQSGCUG plus one AsOH and minus two H (1,144.4 + 74.9 + 17.0 − 2.0). This result suggests that arsenic binds with sulfur and selenium in the motif GCUG of TrxR directly (Fig. 1C). The mass fit SGGDILQSGCUG alone was also observed in the ATO-treated sample; it may be because of the high reactivity of Cys-Sec, and part of the C-terminal fragment loses its arsenic modification during the measurement by a retro Michael addition, which was observed in the interaction between TrxR with 4-hydroxynonenal (31). Peptide mass 1,501.7, which is assigned to oxidized N-terminal fragment W53GLGGTCVNVGCIPK67, was observed in the control sample without ATO treatment but not in the ATO-treated sample, indicating that the N-terminal active site participates in the reaction with ATO. Unfortunately, we did not get the mass that could be assigned to the complex of the fragment W53GLGGTCVNVGCIPK67 with ATO (Fig. 1C Inset). As a control, an adjacent peak 1,267.7, which is assigned to the peptide L340ELTPVAIQAGR351, was present in both ATO-treated and untreated samples. It is known that the two subunits in dimeric TrxR have a head-to-tail arrangement, and the N-terminal redox-active dithiol in one subunit and the C-terminal selenothiol-active site of the adjacent subunit form a redox active center (19). Thus, the above results suggest that both the N-terminal and the C-terminal active sites may react with ATO coincidently.
Inhibition of TrxR in Cells.
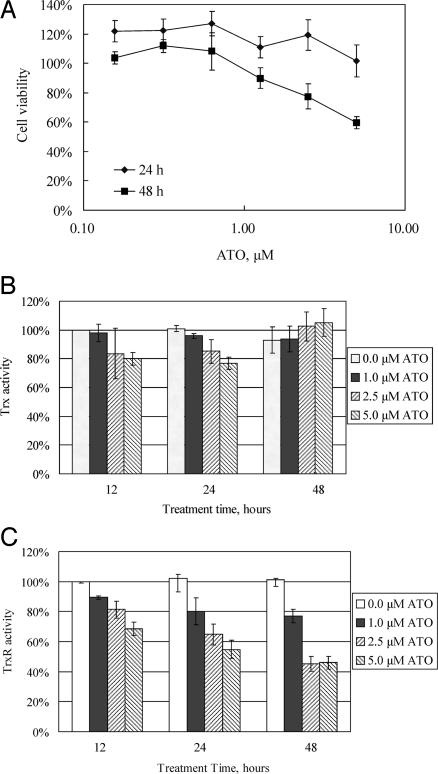
Human breast MCF-7 cancer cells, which are sensitive to ATO, were selected to investigate the effect of ATO on TrxR (32, 33). The cell proliferation of MCF-7 cells that were exposed to various concentrations of ATO was determined by 2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) assay. The growth of MCF-7 was inhibited by ATO in a dose- and time-dependent manner. As illustrated in Fig. 2A, 24-h treatment did not show any obvious effect on the cell growth, whereas 48-h treatment by 2.5 and 5.0 μM ATO resulted in a significant reduction of cell growth. Both TrxR and Trx activity were measured in cell extract with an endpoint method and represented by the percentage of enzyme activity of the lysates from control cells without ATO treatment. TrxR activity was shown to be inhibited by ATO in a time- and dose-dependent manner with 2.5 and 5.0 μM ATO, which induced a marked decrease in TrxR activity (Fig. 2C), whereas Trx activity did not show significant changes with ATO treatment even at the highest concentration (Fig. 2B).
Fig. 2.
Inhibition of TrxR in the MCF-7 cells by ATO. (A) MCF-7 cells were treated with different concentrations of ATO for 24 and 48 h. Cell proliferation and viability were determined by using the XTT method. (B and C) MCF-7 cells were treated with different concentrations of ATO for 12, 24, and 48 h. After the treatment, cells were lysed, and Trx (B) and TrxR (C) activities were determined with an endpoint insulin assay. Error bars represent the standard deviation of three independent experiments.
Trx Redox State Change Induced by ATO.
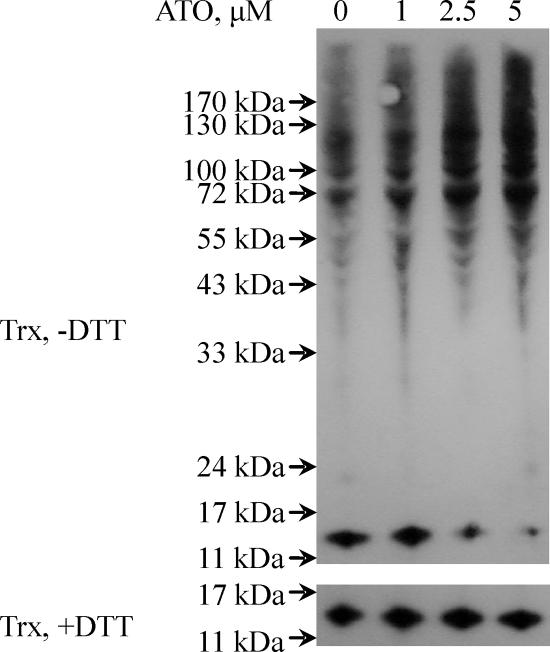
Although overall Trx content in the cell lysates appeared unaffected, as shown by TrxR-dependent insulin reduction assay, Trx could be reversibly inactivated by a change of its redox state because of the inhibition of TrxR, which is the only electron donor for Trx. We used iodoacetamide to alkylate free thiols under denaturing conditions, and subsequently the alkylated samples were analyzed by running a nonreducing SDS/polyacrylamide gel to detect the Trx redox state by Western blotting. In the control sample without ATO treatment, part of Trx was observed in the monomer form, and some formed oligomers or complexes with other proteins, but little was found in dimeric or trimeric forms (Fig. 3). Treatment with 2.5 and 5.0 μM ATO for 48 h resulted in a decrease of the 12-kDa monomer part of Trx, whereas the high molecular mass part of oligomer or complex increased. We present direct evidence to show that Trx can exist in the form with mixed protein disulfides in vivo. This protein–protein interaction by interdisulfide bond is distinct from the noncovalent hydrophobic interaction between reduced E. coli Trx and T7 DNA polymerase, the other well known Trx–protein interaction pattern (34). In the loading control for the SDS/polyacrylamide gel, the Trx amount was shown to be equal in all of the samples. This result showed that ATO treatment induced loss of TrxR activity and subsequent dramatic oxidation of Trx.
Fig. 3.
Trx redox state change in MCF-7 cells by the treatment of MCF-7. MCF-7 cells were treated with different concentrations of ATO for 48 h. The ATO-treated MCF-7 cells were lysed in guanidine lysis buffer containing 30 mM iodoacetamide so that protein SH groups were alkylated. Then samples in the presence or absence of DTT were separated on a 12% Bistris gel with Mes running buffer, and Trx was detected with goat anti-human Trx antibodies.
Effect of Glutathione on the Inhibition of TrxR.
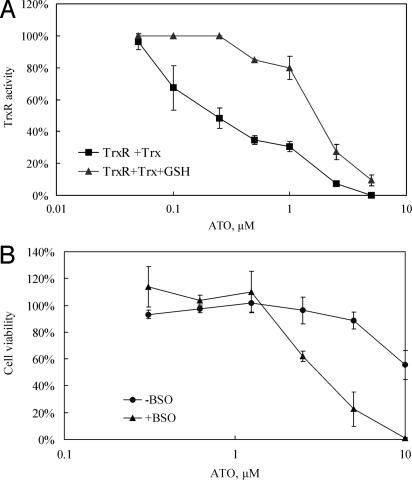
As shown in Fig. 4A, the in vitro TrxR inhibition by 0.1 and 0.25 μM ATO was attenuated by 1 mM GSH, but at >0.5 μM ATO, TrxR was inhibited in the presence of GSH. Consistent with these observations, a protective role of GSH against ATO was found in cell experiments. We used buthionine sulfoximine (BSO), an inhibitor of GSH synthesis, to deplete the intracellular GSH pool. It has been reported that 80% reduction of GSH in MCF-7 cells can be achieved by the treatment of 25 μM BSO without influence on cell viability (35). Thus we pretreated MCF-7 cells 25 μM BSO for 24 h before the addition of ATO. The pretreatment with BSO indeed dramatically increased the sensitivity of MCF-7 cells to ATO (Fig. 4B).
Fig. 4.
Effects of glutathione on the inhibition of TrxR and MCF-7 growth. (A) 10 nM concentration of NADPH-reduced recombinant rat TrxR in the presence of 2 μM human Trx or with 1 mM GSH was incubated with different concentrations of ATO for 30 min. Then the Trx system activity was assayed by the insulin reduction method. (B) MCF-7 cells were treated with 25 μM BSO for 24 h and then with different concentrations of ATO for another 48 h. The control cells were not treated with BSO.
Discussion
The human selenoproteome contains 25 selenoproteins, of which a majority play a critical role in cellular redox regulation (36). The low pKa 5.3 value of a selenocysteine residue makes the selenol group of a selenoprotein such as TrxR highly reactive, especially when it is located in an open C-terminal active site. The unique property of TrxR to reduce Trx makes this enzyme one of the predominant regulators in redox control. Recently, many of the frequently used clinical electrophilic anticancer drugs such as cyclophosphamide (37, 38), nitrosoureas, cisplatin (39), diaziquone, doxorubicin (40), motexafin gadolinium (41), and potential cancer chemoprevention agents such as curcumin (28), or flavonoids quercetin and myricetin (27) have been shown to be excellent inhibitors of TrxR, suggesting that inhibition of TrxR is probably a very common outcome in the cancer chemotherapeutic process.
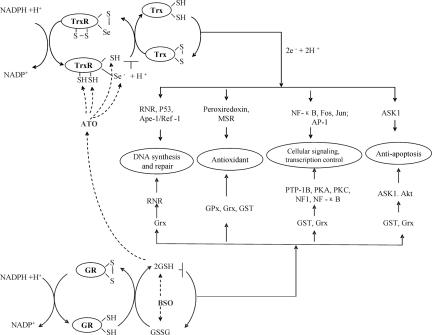
An overview of the two thiol redox systems in a cell is shown in Fig. 5. Trx1 of the Trx system is a ubiquitous 12-kDa protein with a conserved -WCGPC- dithiol active site that reduces many critical proteins such as ribonucleotide reductase (RNR), Trx peroxidases (Prxs), and methionine sulfoxide reductases and thus participates in DNA synthesis and repair, or redox signaling by hydrogen peroxide (26, 42–46). Trx has also been reported to be involved in the regulation of gene expression by mediating the activity of transcription factors such as NF-κB, AP-1, p53 protein, and signaling molecules such as apoptosis signal-regulating kinase 1 (ASK1) and glucocorticoid receptor by thiol redox control (47–49). Therefore, TrxR inhibition will induce the subsequent oxidation of Trx and result in a great change of the activity of related proteins (Fig. 3). For example, many proteins, including ASK1, have been reported to bind only with reduced Trx but not with oxidized Trx. The oxidation of Trx induced by ATO may activate ASK1 and cause ASK-mediated cell death (50). This finding is also supported by the observation that ASK1 downstream kinase JNK was activated by ATO plus BSO even in the presence of N-acetylcysteine and catalase (10). When MCF-7 cells were treated with 5 μM ATO for 24 h, the cell viability was nearly not affected, whereas almost 50% of TrxR was inactivated. After 48 h of treatment, the viability was decreased to 60%, and TrxR activity was reduced by an additional 10% (Fig. 2). The reason for this observation may be that the Trx system lies upstream in cell signaling pathways, and ATO-induced TrxR inhibition occurs as an early cellular event followed by Trx oxidation and then leads to downstream apoptotic events. Twenty-four-hour treatment with ATO will also induce the change of redox state of Trx (data not shown).
Fig. 5.
Trx and GSH pathways interacting with ATO. Trx system including NADPH, TrxR, and Trx coupled with peroxiredoxins and the GSH system composed of NADPH, GR, GSH coupled with Grxs, GPxs, GSTs are the two main electron donor systems that mediate the cellular activity including DNA synthesis, protection against oxidative stress, and thus control cellular proliferation, viability, and apoptosis. The combination inhibition of ATO and BSO will lead to the disruption on both systems and block electron supply for DNA synthesis and give a rise of oxidative stress and thus induce cells into apoptosis.
The other central reducing multifunctional system is glutathione system (NADPH, GR, GSH) coupled with glutaredoxins (Grxs), GPxs, and GSTs (Fig. 5). GSH is the most abundant nonprotein thiol in mammalian cells and plays a critical role in drug resistance with its millimolar cellular concentration (51). GSH/Grxs are involved in the DNA synthesis by providing an electron to RNR and also in redox signal transduction and protein translocation by controlling S-glutathionylation of proteins including PTP-1B, PKA, PKC, NF1, and NF-κB, among others (43, 52, 53). GPxs are well known for catalyzing the reduction of hydrogen peroxide and organic hydroperoxides and thus protecting cells from oxidative damage (54). GSTs participate in the detoxification of electrophilic xenobiotics by catalyzing the conjugation of GSH and regulation of ASK1, NF-κB, etc. (55). As described above, GSH is the major molecule to detoxify ATO (Fig. 4), but GSH-related enzymes are not the target of ATO; the activity of GSH-related enzymes including GPx, GST, and GR was not reduced by ATO treatment in the cell (56). We also did not see a significant change for Grx redox state after a 48-h ATO treatment with the method used here for Trx (data not shown). In contrast, in our in vitro study, in the presence of 1 mM GSH (mimicking the intracellular GSH levels), the Trx system was inhibited by ATO doses >0.5 μM (Fig. 4A). Furthermore, cell lysates of MCF-7 cells treated with ATO showed a reduction in TrxR activity (Figs. 2C and 3). These results demonstrate that the Trx system is a potential mechanistic target for ATO under physiological conditions.
Many agents that can either reduce the content of GSH, such as ascorbic acid (6, 57) and all-trans retinoic acid (58), or act through a different mechanism, such as Trolox (33, 59), enhance the ATO-induced apoptosis. Among these agents, BSO is the most effective. The mechanism behind it can be well explained by our result that TrxR is strongly inhibited by ATO. Both the Trx system and GSH–Grx system may provide electrons to RNR, which is essential for DNA synthesis (43). Moreover, DNA repair activity through Ref1, p53, and RNR is also controlled by Trx, and inactivation of the Trx system will abolish the DNA repair process (31, 60, 61). Trx can provide the electrons to peroxiredoxin for scavenging hydrogen peroxide (45), and moreover, Trx reduces the methionine sulfoxide reductases and thus is involved in the protein repair from the oxidation of protein methionine residues (46). GSH also participates in removing hydrogen peroxide by GPxs (54). The two pathways act in concert to protect cells against oxidative stress. The combination of ATO and BSO should result in the disruption of the electron supply for DNA synthesis and induce the elevated oxidative stress and thus induce cells into apoptosis (Fig. 5). These effects should be the reason for H2O2 accumulation in cells upon ATO treatment (7).
Arsenic has been normally treated as one of the most toxic and carcinogenic agent (62–64). However, in the treatment of acute promyelocytic leukemia, ATO is known to be well tolerated, and the adverse events are manageable (4). The treatment of ATO plus ascorbic acid shows an acceptable toxicity in phase I clinical trial (57). The combination of ATO and BSO significantly increased the survival rate and decreased the side effects in an orthotopic mouse model, and the cancer cells are more sensitive to the combination treatment than normal cells (9), which may be because the cancer cells require a constant DNA precursor supply for their proliferation and also are more sensitive to oxidative stress (65–68).
In summary, we show here that the clinical anticancer agent ATO is a strong inhibitor of mammalian TrxR with a mechanism involving both the C-terminal and N-terminal redox active sites of the enzyme. The inhibition of TrxR subsequently resulted in the inactivation of whole Trx system. Our results link the mechanism of anticancer activity of ATO with the inhibition of Trx system. The combination inhibition of both the Trx and GSH systems and thus resulting in the blockage of DNA synthesis and oxidative stress may provide a unique chemotherapeutic strategy.
Materials and Methods
Chemicals and Enzymes.
Purified recombinant rat TrxR1 with a specific activity of 15 units/mg was as described previously (69). The enzyme concentration was measured by using ε463 = 11.3 mM−1·cm−1. E. coli TrxR, E. coli Trx, human Trx1, and goat anti-human Trx1 and anti-human Grx1 antibodies were from IMCO, Ltd. [Stockholm, Sweden (www.imcocorp.se)]. ATO were obtained from Sigma (St. Louis, MO). ATO was dissolved in 1 M NaOH as the stock solution. Trypsin was obtained from Promega (Madison, WI). All other reagents were of analytical grade.
Determination of Activity.
Experiments were performed in 96-well plates with 100 μl of 50 mM Tris·HCl/1 mM EDTA, pH 7.5/200 μM NADPH. A 50 nM concentration of recombinant active rat TrxR was incubated with different concentrations of ATO for 30 min. TrxR activity was assayed by the DTNB method in the solution containing 50 mM Tris·HCl, pH 7.5, 200 μM NADPH, 5 mM DTNB, and 1 mM EDTA (26, 69, 70). The A412 was followed with an Ultrospec 3000 UV/visible spectrophotometer (Amersham Pharmacia, Piscataway, NJ) or a VERSA microplate reader (Molecular Devices, Eugene, OR). TrxR activity was calculated by measuring the slope of absorbance change during the initial 2 min.
Trx System Activity.
The mammalian Trx system inhibitory experiment was performed in a 96-well plate with 100 μl of 50 mM Tris·HCl/1 mM EDTA, pH 7.5/200 μM NADPH. A 10 nM concentration of recombinant rat TrxR and 2 μM human Trx1 were incubated with different concentration of ATO for 30 min. Then 100 μl of 200 μM NADPH/160 μM insulin was added to the solution, and the A340 nm was followed. TrxR activity was calculated by the value of ΔA340 per min of the initial 5 min (69). The inhibition of the E. coli Trx system was performed in 50 mM Tris·HCl/1 mM EDTA, pH 7.5/200 μM NADPH/25 nM E. coli TrxR/2 μM E. coli Trx. Other conditions were the same as for the measurements of the inhibition of mammalian Trx system.
Peptide Mass of TrxR–Arsenic by MALDI Mass Spectrometry.
A 1 μM concentration recombinant rat TrxR and 200 μM NADPH in 50 mM Tris·HCl/1 mM EDTA, pH 7.5 were incubated with 10 μM ATO at room temperature for 1 h, and then ATO and NADPH were removed by a PD10 (Sephadex G-25) desalting column. If there was some TrxR activity left, the above procedure was repeated again. Then the enzyme was denatured in 8 M urea at 60°C for 1 h. The denatured protein sample was diluted 10-fold by 50 mM Tris·HCl, pH 7.6/1 mM CaCl2 and cleaved at a trypsin/protein ratio of 1:10 at 37°C for 6 h. A Voyager De-PRO MALDI mass spectrometer (Applied Biosystems, Foster City, CA) at the Protein Analysis Center of Karolinska Institute was used to detect the peptide mass.
Cell Culture.
Human breast MCF-7 cancer cells were cultured in RPMI medium 1640 (GIBCO, Grand Island, NY) supplemented with 2 mM l-glutamine/10% FCS/100 units/ml penicillin/100 μg/ml streptomycin at 37°C in a 5% CO2 incubator.
Cell Viability Assay.
MCF-7 cells were plated at a density of 6 × 103 cells per well in 96-microwell plates and allowed to grow overnight. Then the medium was changed to the growth medium containing different concentrations of ATO, and incubation was conducted for another 24 and 48 h. Then the medium was changed to culture medium containing XTT labeling mixture, and the incubation continued for 3 h. XTT was metabolized to soluble formazan salt by viable cells and yielded an A492 (Roche, Indianapolis, IN). Thus, the cell proliferation and viability were determined by measuring the A492.
Preparation of Cell Lysate.
MCF-7 cells were plated at a density of 1 × 106 cells in 100-mm Petri dishes in normal growth medium. After overnight incubation at 37°C, the cells were washed with PBS and treated with different concentrations of ATO for 12, 24, or 48 h. After the treatment, the cells were washed with PBS twice and lysed in lysis buffer (25 mM Tris·HCl, pH 7.5/100 mM NaCl/2.5 mM EDTA/2.5 mM EGTA/20 mM NaF/1 mM Na3VO4/20 mM sodium β-glycerophosphate/10 mM sodium pyrophosphate/0.5% Triton X-100/protease inhibitor cocktails (Roche Diagnostics, Mannheim, Germany). The protein concentration was determined by DC protein assay (Bio-Rad, Hercules, CA).
Activity Assay for TrxR and Trx in the Cell Lysates.
TrxR and Trx activity assay was performed in 96-well plates with an endpoint insulin assay (26, 69). For the measurement of TrxR activity, ≈25 μg of cell lysate was mixed thoroughly with 50 μl of 55 mM Hepes, pH 7.6/0.2 mM insulin/0.4 mM NADPH/2 mM EDTA/2 μM human Trx1 in the VERSA microplate reader. The reaction solutions without human Trx1 were used as the control. After the reaction was performed at 37°C for 20 min, 200 μl of 1 mM DTNB in 6 M guanidine hydrochloride solution was added to stop the reaction. The free thiols of the reduced insulin were determined by DTNB reduction, and the activity of TrxR was represented by the A412, where 1 mol of NADPH reduced 1 mol of disulfide, giving rise to 2 mol of free TNB with the extinction coefficient 13.6 mM−1 ·cm−1.
The Trx activity assay was performed in 50 μl of reaction solution containing 25 μg of cell lysate/55 mM Hepes, pH 7.6/0.2 mM insulin/0.4 mM NADPH/2 mM EDTA/600 nM recombinant rat TrxR. The reaction solutions without TrxR were used as the control, which was subtracted. The other procedures were same as for the TrxR activity assay.
Detection of the Trx Redox State in MCF-7 Cells.
The ATO-treated MCF-7 cells were washed with PBS twice and lysed in guanidine hydrochloride (GnCl) lysis buffer (50 mM Tris·HCl, pH 7.5/1 mM EDTA/6 M GnCl) containing 30 mM iodoacetamide. After removing the cell debris by centrifugation, cells lysates were precipitated by 10% trichloroacetic acid. Precipitates were washed with ice-cold acetone three times and dissolved in the 50 mM Tris·HCl, pH 8.5/1% SDS. Proteins were incubated with SDS-loading buffer with or without 50 mM DTT at 60°C for 30 min and then separated on a 12% [bis(2-hydroxyethyl)amino]tris(hydroxymethyl)methane (Bistris) gel with Mes running buffer. Trx was detected with goat anti-human Trx antibodies at 1:1,000 dilutions, followed by the detection of Chemiluminescence Plus reagent.
Effect of GSH Depletion on the Inhibition of MCF-7 Growth.
MCF-7 cells were plated at a density of 4 × 103 cells per well in 96-microwell plates and allowed to grow overnight. The medium was changed to contain 25 μM BSO, and the incubation was continued for 24 h. Then the cells were treated with different concentrations of ATO, and incubation was conducted for another 48 h. Cell proliferation and viability were determined by using XTT method according to the instructions of the manufacturer (Roche).
Acknowledgments
We thank Dr. Elias Arnér and Mr. Olle Rengby (in our division) for kindly providing recombinant TrxR. This work was supported by Swedish Cancer Society Grant 961, Swedish Research Council Medicine Grant 13x-3529, The K. A. Wallenberg Foundation, and the Karolinska Institutet (to A.H.).
Abbreviations
- ASK
apoptosis signal-regulating kinase
- ATO
arsenic trioxide
- Bistris
[bis(2-hydroxyethyl)amino]tris(hydroxymethyl)methane
- BSO
buthionine sulfoximine
- DTNB
5,5′-dithiobis(2-nitrobenzoic acid)
- GPx
glutathione peroxidase
- GR
glutathione reductase
- Grx
glutaredoxin
- GSH
glutathione
- GST
glutathione transferase
- PAO
phenylarsine oxide
- RNR
ribonucleotide reductase
- Sec
selenocysteine
- Trx
thioredoxin
- TrxR
thioredoxin reductase
- XTT
2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.D. is a guest editor invited by the Editorial Board.
References
- 1.Chen GQ, Zhu J, Shi XG, Ni JH, Zhong HJ, Si GY, Jin XL, Tang W, Li XS, Xong SM, et al. Blood. 1996;88:1052–1061. [PubMed] [Google Scholar]
- 2.Hu J, Fang J, Dong Y, Chen SJ, Chen Z. Anticancer Drugs. 2005;16:119–127. doi: 10.1097/00001813-200502000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Gazitt Y, Akay C. Hematology. 2005;10:205–213. doi: 10.1080/10245330500067090. [DOI] [PubMed] [Google Scholar]
- 4.Douer D, Tallman MS. J Clin Oncol. 2005;23:2396–2410. doi: 10.1200/JCO.2005.10.217. [DOI] [PubMed] [Google Scholar]
- 5.Miller WH, Jr, Schipper HM, Lee JS, Singer J, Waxman S. Cancer Res. 2002;62:3893–3903. [PubMed] [Google Scholar]
- 6.Dai J, Weinberg RS, Waxman S, Jing Y. Blood. 1999;93:268–277. [PubMed] [Google Scholar]
- 7.Jing Y, Dai J, Chalmers-Redman RM, Tatton WG, Waxman S. Blood. 1999;94:2102–2111. [PubMed] [Google Scholar]
- 8.Cai X, Shen YL, Zhu Q, Jia PM, Yu Y, Zhou L, Huang Y, Zhang JW, Xiong SM, Chen SJ, et al. Leukemia. 2000;14:262–270. doi: 10.1038/sj.leu.2401650. [DOI] [PubMed] [Google Scholar]
- 9.Maeda H, Hori S, Ohizumi H, Segawa T, Kakehi Y, Ogawa O, Kakizuka A. Cell Death Differ. 2004;11:737–746. doi: 10.1038/sj.cdd.4401389. [DOI] [PubMed] [Google Scholar]
- 10.Chen D, Chan R, Waxman S, Jing Y. Cancer Res. 2006;66:11416–11423. doi: 10.1158/0008-5472.CAN-06-0409. [DOI] [PubMed] [Google Scholar]
- 11.Chouchane S, Snow ET. Chem Res Toxicol. 2001;14:517–522. doi: 10.1021/tx000123x. [DOI] [PubMed] [Google Scholar]
- 12.Arnér ES, Holmgren A. Eur J Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 13.Lillig CH, Holmgren A. Antioxid Redox Signal. 2007;9:25–47. doi: 10.1089/ars.2007.9.25. [DOI] [PubMed] [Google Scholar]
- 14.Gladyshev VN, Jeang KT, Stadtman TC. Proc Natl Acad Sci USA. 1996;93:6146–6151. doi: 10.1073/pnas.93.12.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong L, Arnér ES, Ljung J, Aslund F, Holmgren A. J Biol Chem. 1998;273:8581–8591. doi: 10.1074/jbc.273.15.8581. [DOI] [PubMed] [Google Scholar]
- 16.Lee SR, Kim JR, Kwon KS, Yoon HW, Levine RL, Ginsburg A, Rhee SG. J Biol Chem. 1999;274:4722–4734. doi: 10.1074/jbc.274.8.4722. [DOI] [PubMed] [Google Scholar]
- 17.Biterova EI, Turanov AA, Gladyshev VN, Barycki JJ. Proc Natl Acad Sci USA. 2005;102:15018–15023. doi: 10.1073/pnas.0504218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun QA, Kirnarsky L, Sherman S, Gladyshev VN. Proc Natl Acad Sci USA. 2001;98:3673–3678. doi: 10.1073/pnas.051454398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandalova T, Zhong L, Lindqvist Y, Holmgren A, Schneider G. Proc Natl Acad Sci USA. 2001;98:9533–9538. doi: 10.1073/pnas.171178698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong L, Holmgren A. J Biol Chem. 2000;275:18121–18128. doi: 10.1074/jbc.M000690200. [DOI] [PubMed] [Google Scholar]
- 21.Gromer S, Urig S, Becker K. Med Res Rev. 2004;24:40–89. doi: 10.1002/med.10051. [DOI] [PubMed] [Google Scholar]
- 22.Powis G, Mustacichi D, Coon A. Free Radical Biol Med. 2000;29:312–322. doi: 10.1016/s0891-5849(00)00313-0. [DOI] [PubMed] [Google Scholar]
- 23.Urig S, Becker K. Semin Cancer Biol. 2006;16:452–465. doi: 10.1016/j.semcancer.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 24.Yoo MH, Xu XM, Carlson BA, Gladyshev VN, Hatfield DL. J Biol Chem. 2006;281:13005–13008. doi: 10.1074/jbc.C600012200. [DOI] [PubMed] [Google Scholar]
- 25.Arnér ES, Holmgren A. Semin Cancer Biol. 2006;16:420–426. doi: 10.1016/j.semcancer.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 26.Luthman M, Holmgren A. Biochemistry. 1982;21:6628–6633. doi: 10.1021/bi00269a003. [DOI] [PubMed] [Google Scholar]
- 27.Lu J, Papp LV, Fang J, Rodriguez-Nieto S, Zhivotovsky B, Holmgren A. Cancer Res. 2006;66:4410–4418. doi: 10.1158/0008-5472.CAN-05-3310. [DOI] [PubMed] [Google Scholar]
- 28.Fang J, Lu J, Holmgren A. J Biol Chem. 2005;280:25284–25290. doi: 10.1074/jbc.M414645200. [DOI] [PubMed] [Google Scholar]
- 29.Zhong L, Arnér ES, Holmgren A. Proc Natl Acad Sci USA. 2000;97:5854–5859. doi: 10.1073/pnas.100114897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johansson L, Chen C, Thorell JO, Fredriksson A, Stone-Elander S, Gafvelin G, Arner ES. Nat Methods. 2004;1:61–66. doi: 10.1038/nmeth707. [DOI] [PubMed] [Google Scholar]
- 31.Cassidy PB, Edes K, Nelson CC, Parsawar K, Fitzpatrick FA, Moos PJ. Carcinogenesis. 2006;27:2538–2549. doi: 10.1093/carcin/bgl111. [DOI] [PubMed] [Google Scholar]
- 32.Baj G, Arnulfo A, Deaglio S, Mallone R, Vigone A, De Cesaris MG, Surico N, Malavasi F, Ferrero E. Breast Cancer Res Treat. 2002;73:61–73. doi: 10.1023/a:1015272401822. [DOI] [PubMed] [Google Scholar]
- 33.Diaz Z, Colombo M, Mann KK, Su H, Smith KN, Bohle DS, Schipper HM, Miller WH., Jr Blood. 2005;105:1237–1245. doi: 10.1182/blood-2004-05-1772. [DOI] [PubMed] [Google Scholar]
- 34.Slaby I, Holmgren A. J Biol Chem. 1989;264:16502–16506. [PubMed] [Google Scholar]
- 35.Chew EH, Matthews CS, Zhang J, McCarroll AJ, Hagen T, Stevens MF, Westwell AD, Bradshaw TD. Biochem Biophys Res Commun. 2006;346:242–251. doi: 10.1016/j.bbrc.2006.05.106. [DOI] [PubMed] [Google Scholar]
- 36.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Zhang J, Xu T. Toxicol Appl Pharmacol. 2007;218:88–95. doi: 10.1016/j.taap.2006.10.029. [DOI] [PubMed] [Google Scholar]
- 38.Witte AB, Anestal K, Jerremalm E, Ehrsson H, Arnér ES. Free Radic Biol Med. 2005;39:696–703. doi: 10.1016/j.freeradbiomed.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 39.Arnér ES, Nakamura H, Sasada T, Yodoi J, Holmgren A, Spyrou G. Free Radic Biol Med. 2001;31:1170–1178. doi: 10.1016/s0891-5849(01)00698-0. [DOI] [PubMed] [Google Scholar]
- 40.Mau BL, Powis G. Biochem Pharmacol. 1992;43:1621–1626. [PubMed] [Google Scholar]
- 41.Hashemy SI, Ungerstedt JS, Zahedi Avval F, Holmgren A. J Biol Chem. 2006;281:10691–10697. doi: 10.1074/jbc.M511373200. [DOI] [PubMed] [Google Scholar]
- 42.Holmgren A. Annu Rev Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- 43.Holmgren A. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- 44.Rhee SG, Chae HZ, Kim K. Free Radic Biol Med. 2005;38:1543–1552. doi: 10.1016/j.freeradbiomed.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 45.Kang SW, Rhee SG, Chang TS, Jeong W, Choi MH. Trends Mol Med. 2005;11:571–578. doi: 10.1016/j.molmed.2005.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoshi T, Heinemann S. J Physiol (London) 2001;531:1–11. doi: 10.1111/j.1469-7793.2001.0001j.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hirota K, Matsui M, Iwata S, Nishiyama A, Mori K, Yodoi J. Proc Natl Acad Sci USA. 1997;94:3633–3638. doi: 10.1073/pnas.94.8.3633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kondo N, Nakamura H, Masutani H, Yodoi J. Antioxid Redox Signal. 2006;8:1881–1890. doi: 10.1089/ars.2006.8.1881. [DOI] [PubMed] [Google Scholar]
- 49.Saitoh M, Nishitoh H, Fujii M, Takeda K, Tobiume K, Sawada Y, Kawabata M, Miyazono K, Ichijo H. EMBO J. 1998;17:2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hansen JM, Zhang H, Jones DP. Free Radic Biol Med. 2006;40:138–145. doi: 10.1016/j.freeradbiomed.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 51.Davis W, Jr, Ronai Z, Tew KD. J Pharmacol Exp Ther. 2001;296:1–6. [PubMed] [Google Scholar]
- 52.Shelton MD, Chock PB, Mieyal JJ. Antioxid Redox Signal. 2005;7:348–366. doi: 10.1089/ars.2005.7.348. [DOI] [PubMed] [Google Scholar]
- 53.Murata H, Ihara Y, Nakamura H, Yodoi J, Sumikawa K, Kondo T. J Biol Chem. 2003;278:50226–50233. doi: 10.1074/jbc.M310171200. [DOI] [PubMed] [Google Scholar]
- 54.Arthur JR. Cell Mol Life Sci. 2000;57:1825–1835. doi: 10.1007/PL00000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hayes JD, Flanagan JU, Jowsey IR. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 56.Yeh JY, Cheng LC, Ou BR, Whanger DP, Chang LW. Cell Mol Life Sci. 2002;59:1972–1982. doi: 10.1007/PL00012519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bahlis NJ, McCafferty-Grad J, Jordan-McMurry I, Neil J, Reis I, Kharfan-Dabaja M, Eckman J, Goodman M, Fernandez HF, Boise LH, Lee KP. Clin Cancer Res. 2002;8:3658–3668. [PubMed] [Google Scholar]
- 58.Jing Y, Wang L, Xia L, Chen GQ, Chen Z, Miller WH, Waxman S. Blood. 2001;97:264–269. doi: 10.1182/blood.v97.1.264. [DOI] [PubMed] [Google Scholar]
- 59.Akay C, Thomas C, III, Gazitt Y. Cell Cycle. 2004;3:324–334. [PubMed] [Google Scholar]
- 60.Seemann S, Hainaut P. Oncogene. 2005;24:3853–3863. doi: 10.1038/sj.onc.1208549. [DOI] [PubMed] [Google Scholar]
- 61.Tanaka H, Arakawa H, Yamaguchi T, Shiraishi K, Fukuda S, Matsui K, Takei Y, Nakamura Y. Nature. 2000;404:42–49. doi: 10.1038/35003506. [DOI] [PubMed] [Google Scholar]
- 62.Tapio S, Grosche B. Mutat Res. 2006;612:215–246. doi: 10.1016/j.mrrev.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 63.Lin S, Cullen WR, Thomas DJ. Chem Res Toxicol. 1999;12:924–930. doi: 10.1021/tx9900775. [DOI] [PubMed] [Google Scholar]
- 64.Thomas DJ, Styblo M, Lin S. Toxicol Appl Pharmacol. 2001;176:127–144. doi: 10.1006/taap.2001.9258. [DOI] [PubMed] [Google Scholar]
- 65.Schumacker PT. Cancer Cell. 2006;10:175–176. doi: 10.1016/j.ccr.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 66.Engel RH, Evens AM. Front Biosci. 2006;11:300–312. doi: 10.2741/1798. [DOI] [PubMed] [Google Scholar]
- 67.Ungerstedt JS, Sowa Y, Xu WS, Shao Y, Dokmanovic M, Perez G, Ngo L, Holmgren A, Jiang X, Marks PA. Proc Natl Acad Sci USA. 2005;102:673–678. doi: 10.1073/pnas.0408732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pelicano H, Carney D, Huang P. Drug Resist Update. 2004;7:97–110. doi: 10.1016/j.drup.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 69.Arnér ES, Sarioglu H, Lottspeich F, Holmgren A, Bock A. J Mol Biol. 1999;292:1003–1016. doi: 10.1006/jmbi.1999.3085. [DOI] [PubMed] [Google Scholar]
- 70.Holmgren A, Bjornstedt M. Methods Enzymol. 1995;252:199–208. doi: 10.1016/0076-6879(95)52023-6. [DOI] [PubMed] [Google Scholar]