Abstract
Cytochrome P450 aromatase (CYP19) plays an important role in steroid homoeostasis by converting androgens to estrogens. To evaluate the effects of benzo(a)pyrene (BaP), a model carcinogenic PAH and AhR ligand, on aromatase mRNA expression and enzyme activity, adult Fundulus were exposed to water-borne BaP (1 and 10 μg/L) for 15 days, and embryos were exposed to 10 μg/L for 10 days. Effects of BaP were examined by tissue, gender, and season in adults. Constitutively, the sexes did not have significantly different CYP19A2 mRNA levels, however females had higher brain aromatase activity. Female control killifish had more than 700-fold more CYP19A1 mRNA in their gonads compared to males. Within brain tissue of both sexes, there was 100-fold more CYP19A2 mRNA compared to CYP19A1. In ovary, CYP19A1 predominated by approximately 30-fold over the CYP19A2, but in testis there was relatively more CYP19A2. In embryos there was ~5-fold higher CYP19A2 expression. Due to high inter-individual variability, a significant effect of BaP treatment by gender, season or age was not observed for either aromatase mRNA. However, ovarian aromatase activity was significantly decreased by 10 μg/L BaP, while female brain activity was increased following winter exposure. These findings suggest that the aromatase enzyme is a potential target for disruption of fish developmental and reproductive physiology by BaP.
Keywords: Aromatase, Fundulus, Benzo(a)pyrene
1. Introduction
Estrogens are critical to the function and maintenance of a diverse array of tissues and physiological systems. Estrogens play important roles in the development, growth, sexual differentiation and reproduction in all vertebrates studied to date. In nonmammalian, egg-laying vertebrates like fish, estrogen influences a number of developmental and reproductive events including, hepatic vitellogenin, egg shell protein synthesis, oocyte growth, germ cell development, gonadal sex differentiation, and reproductive behavior (Callard et al., 2001; Fukada et al., 1996; Wallace, 1985). Therefore, estrogen biosynthesis is very crucial for normal reproductive physiology in fish.
Cytochrome P450 aromatase (CYP19) is the key steroidogenic enzyme responsible for conversion of androgens to estrogens. In mammals, it is expressed in a wide variety of tissues including brain, gonad, placenta, carcinoma, bone, skin, liver, adipose, and breast (Conley and Hinshelwood, 2001). The expression in lower vertebrates is typically restricted to the gonad, neural tissue (brain and retina), and the pituitary (Callard et al., 1981; Callard, 1983; Callard and Tchoudakova, 1997; Gelinas and Callard, 1993; Goto-Kazeto et al., 2004; Melo and Rand-Weaver, 2001). In mammals, with the exception of pig (Choi et al., 1997), there is a single CYP19 gene with multiple promoters (Simpson et al., 1997; Toda and Shizuta, 1994). Furthermore, in mammals, control of tissue-specific expression is obtained by the use of alternatively spliced 5′-untranslated exons associated with tissue-specific promoters. As exon I is not translated, the same protein is ultimately produced in all tissues. Interestingly, teleosts diverge from the typical vertebrate strategy in that they possess two separate and distinct CYP19 gene loci, CYP19A1 (predominantly gonad form) and CYP1A2 (predominantly brain form), each of which are regulated differently and encode proteins with different enzyme characteristics. Two CYP19 genes have been identified in several species of teleosts, including goldfish (Tchoudakova and Callard, 1998), medaka (Fukada et al., 1996), zebrafish (Kishida et al., 2001; Trant et al., 2001), Nile tilapia (Chang et al., 1997; Kwon et al., 2001), Mozambique tilapia, rainbow trout (Dalla Valle et al., 2002b; Tanaka et al., 1992), fathead minnow (Halm et al., 2001), sea bass (Dalla Valle et al., 2002a), black porgy (Liu et al., 2004), channel catfish (Trant, 1994), and killifish (Greytak et al., 2005). CYP19A2 is constitutively expressed at high levels in the brain, retina, and pituitary, where it is hypothesized to be involved in the neuronal differentiation, survival, morphology, synaptogenesis and sexual behavior, whereas CYP19A1 is expressed in the ovary and involved in sexual differentiation and oocyte growth (Kishida and Callard, 2001; Kishida et al., 2001; Kwon et al., 2001; Melo and Rand-Weaver, 2001; Trant et al., 2001).
Various endocrine disrupting chemicals are known to mimic sexual hormones and/or disrupt steroidogenic enzyme function, including aromatase, and as a consequence they are capable of altering normal reproductive function in wildlife. For example, sex steroids can influence phenotypic sex in all vertebrates, but this is especially true for lower vertebrates where complete sex reversal can be induced irrespective of the genetic background (Crisp et al., 1998). Polycyclic aromatic hydrocarbons (PAHs) are ubiquitously present in the environment as byproducts of incomplete combustion and have the potential to bioaccumulate in lipid-rich tissues. These chemicals enter the aquatic and marine environment in industrial effluent, and urban and agricultural run off. Thus, fish are potentially impacted by the endocrine disrupting effects of such chemicals.
There is in vivo evidence that PAH exposure results in reproductive and developmental deficits in fish from polluted environments (reviewed in Nicolas, 1999). Altered reproductive endpoints have been reported in both killifish and sole collected from PAH-impacted sites (Collier et al., 1998; Pait, 2001). Adult female Atlantic croaker that received BaP had significantly lower estradiol and testosterone concentrations, and ovarian growth was decreased compared to control fish (Thomas, 1990). 20-Methylcholanthrene and β-naphthoflavone, significantly decreased estradiol secretion by coho salmon ovarian follicles (Afonso et al., 1997). Likewise, estradiol production from fully vitellogenic flounder ovarian tissue was inhibited compared to control by 40, 25, and 10% by phenanthrene, BaP, and chrysene (Monteiro et al., 2000). To date, there has been limited understanding of PAH effects on fish aromatase (Kazeto et al., 2004), which is essential for testing the hypothesis that PAH-mediated alterations of aromatase could be responsible for decreased reproductive/developmental success.
In contrast, to the endocrine disrupting potential of PAHs, the carcinogenic potential of the higher molecular weight PAHs is well documented. Biotransformation of PAHs and metabolic activation of the carcinogenic PAHs, such as benzo(a)pyrene (BaP) and dimethylbenzanthracene (DMBA), occurs primarily through the aryl hydrocarbon receptor (AhR)-mediated induction of the CYP1 family of P450 monooxygenases. Bay region diol epoxide metabolites are specifically associated with stable DNA adduct formation and cancer initiation. In most species and tissues there are relatively low levels of CYP1 constitutively expressed, however they can be highly induced by exposure to AhR ligands. In fish, including killifish, exposure to BaP causes time and dose-related induction of EROD, CYP1A mRNA, biliary BaP metabolites, and DNA adduct formation (Willett et al., 1995). CYP1A induction can increase the metabolism of estrogens suggesting another potential mechanism for endocrine disruption. For example, liver microsomes from 10 mg/kg BaPexposed channel catfish converted estrogen to 1.3- and 6.3-fold more 2-hydroxyestradiol and 4-hydroxyestradiol, respectively compared to control microsomes (Butala et al., 2004).
In this study, Fundulus heteroclitus (Pisces: Cyprinodontidae), also known as killifish or mummichog, were used. Killifish are a common marine teleost found along the coastal eastern North America (Yozzo et al., 1994). Because they have a small home range (Lotrich, 1975), populations of killifish exist that have been exposed to and have tolerated a wide variety of contaminants including PAHs (Meyer et al., 2003; Vogelbein et al., 1990). Greytak et al. (2005) have reported CYP19A2 mRNA levels two-fold higher in killifish collected from a PCB-contaminated site compared to fish from a reference site. However, the in vivo consequences of PAH exposure on killifish aromatase (mRNA and protein) have not been previously documented.
The aim of this work was to confirm and extend the work of Greytak and coworkers by cloning and sequencing the killifish CYP19A1 and CYP19A2 cDNAs. Using the sequence information, we in turn measured the in vivo effects of waterborne BaP exposure on mRNA levels and ovarian and brain aromatase activity in adult and embryonic killifish.
2. Materials and methods
2.1. Fish source, care and handling
A parental population of F. heteroclitus collected from an uncontaminated site at the New River inlet near Beaufort, North Carolina was raised under the University Institutional Animal Care and Use Committee (IACUC) approved conditions. Sexually mature fish were bred and kept in salt water (20–25 ppt). The fish were maintained at a 14:10 light–dark cycle in summer and 10:14 light–dark cycle in winter. Adult fish were fed twice daily with tropical flake fish food (Tetramin, Tetra Werke, Germany) and live brine shrimp. First generation offspring, from wild parents, were used for the studies described here.
2.2. Cloning of Fundulus CYP19 cDNAs
Total RNA was isolated from fresh female gonad and brain tissues. Tissues were disrupted and homogenized in 1 mL of TRIzol ® Reagent (Invitrogen, Carlsbad, CA) and extracted by the manufacturer’s protocol. The total RNA was further cleaned by using an RNeasy® Kit (Qiagen, Valencia, CA) with a DNAse I digestion. Poly (A)+ RNA was isolated from total RNA using the Oligotex™ mRNA purification system (Qiagen) according to the supplier’s protocol.
cDNA was synthesized from 1 μg mRNA or total RNA from adult Fundulus brain and ovary using oligo (dT) primers and Superscript II reverse transcriptase (Invitrogen). Degenerate PCR primers were designed using Primer3 software and were purchased from Invitrogen. An aliquot (10%) of the first-strand reaction was amplified with Primers 1 and 2 for CYP19A2 and Primers 14 and 19 for CYP19A1 (Table 1) and PCR Master mix (Roche Diagnostics Corporation, Indianapolis, IN) using a Stratagene Robocycler Gradient 96 Temperature Cycler. The amplification procedure consisted of 10 min at 95 °C followed by 35 cycles at 95 °C (30 s), 40 °C (30 s), and 72 °C (2 min) and a final extension at 72 °C (10 min). A 509 bp fragment of CYP19A2 and a 755 bp fragment of CYP19A1 were detected.
Table 1.
Primers used in RT-PCR and 5′and 3′-RACE analyses for sequencing CYP19A1 and CYP19A2 and for quantitative RT-RT PCR
| Primer name | Sequence 5′ → 3′ | Type and genea |
|---|---|---|
| Primer 1 | CTGATATTTGCTCAGAACCA | F-A2 |
| Primer 2 | AGGATGGCCTTCGTCATCACCAT | R-A2 |
| Primer 3 | CGCCGCATAGTGAAGTCAACCAC | R-A2 |
| Primer 4 | CTTCCCCAAGCCCAGAGAGTTCA | F-A2 |
| Primer 5 | ACTCCACGTTCCCTGGTCAGGAGAG | F-A2 |
| Primer 6 | ACAACAACAGATATGGCAGCACCGTC | F-A2 |
| Primer 7 | AGAGGCTGATGGAGAGCGTGTCA | R-A2 |
| Primer 8 | ATGGTGATGACGAAGGCCATCCT | F-A2 |
| Primer 9 | TTCGAGTTTAAATGAAAGGCCCACTT | R-A2 |
| Primer 10 | GAAGGTCCCCGTTCTGCAGCTCTCT | R-A1 |
| Primer 11 | GGTTTTGAGGAGCTACGCTG | F-A1 |
| Primer 12 | ACCTTAGTGGATGGAGAGCA | R-A1 |
| Primer 13 | CATGGATTTGATCTCTTCCTGC | F-A1 |
| Primer 14 | GTGGTCGACGTCTCCAACAGGCTCT | F-A1 |
| Primer 15 | AGAGAGCTGCAGAACGGGGACCTTC | F-A1 |
| Primer 16 | GAGTGTGGATCAATGGAGAGGAGAC | R-A1 |
| Primer 17 | AAGAGGAGGCAAATGGAGCAG | R-A1 |
| Primer 18 | TGTCATAGACGGCTACAGGGTCCCG | R-A1 |
| Primer 19 | GCAAGCACATCGCCATGGTGGATGAT | R-A1 |
| Primer 20 | AGAGCCTGTTGGAGACGTCGACCAC | R-A1 |
| Primer 21 | TATGAAAATTGACATGTAGCAAAGGG | F-A1 Realtime |
| Primer 22 | AACAGTGGCATGAATGGTGC | R-A1 Realtime |
| Primer 23 | CGAGGAGACCCAGATAACAACAC | F-A2 Realtime |
| Primer 24 | GCACTTCGCACACTACAGCATTAG | R-A2 Realtime |
| Primer 25 | TGGTTAATTCCGATAACGAACGA | F-18S Realtime |
| Primer 26 | CGCCACTTGTCCCTCTAAGAA | R-18S Realtime |
a F: forward primer, R: reverse primer, A1: CYP19A1, A2: CYP19A2, and 18S: 18S rRNA.
2.3. 5′-RACE-PCR
The 5′- and 3′-ends of the CYP19A2 cDNA and the 3′ end of CYP19A1 were obtained by rapid amplification of cDNA ends (RACE) using the SMART™ RACE cDNA Amplification kit (Clontech Laboratory, Inc., Palo Alto, CA) as per the supplier’s protocol. The amplification of double-stranded 5′-CYP19A2 cDNA was done with 10 μM Primer 3 (gene specific primer) (Table 1), using touchdown PCR with annealing temperatures of 72, 70, and 65 °C, respectively. Five microliters of the RACE product was amplified using 10 μM Nested Universal Primer and 25 cycles at 94 °C (3 s), 65 °C (10 s), and 72 °C (3 min). In order to isolate the 5′ end of the CYP19A1 cDNA the GeneRacer kit (Invitrogen) was used. mRNA was isolated from killifish ovary and converted to cDNA as described in the kit. The PCR reaction was performed with the GeneRacer 5′ primer and Primer 10 with touchdown PCR annealing temperatures of 72, 70 and 68 °C.
2.4. 3′-RACE-PCR
3′-RACE cDNA was synthesized from 1 μg total RNA using Superscript II (Invitrogen) and incubation for 1.5 h at 42 °C. The amplification of double-stranded 3′-CYP19A1 and CYP19A2 cDNA was done in the same way as 5′-reaction except that Primers 15 and 4 (Table 1) were used as the respective gene specific primers.
To get the full-length sequences of killifish CYP19s, 1 μg cDNA was amplified with Platinum high fidelity Taq polymerase (Invitrogen) and 10 μM Primers 5 and 9 (CYP19A2) or Primers 11 and 12 (CYP19A1) flanking the ORFs. PCR products were cloned and two to four independent clones were sequenced in both directions using SP6 and T7 primers, followed by internal Primers 2, 4, 6, 7, and 8 (CYP19A2) or Primers 13–20 (CYP19A1).
For cloning, DNA bands were excised from the gel and extracted using QIAquick gel extraction kit (Qiagen). pGEMT Easy Vector System I (Promega, Madison, WI) was used for ligation. Ligated DNA was transformed into DH5α E. coli competent cells (Life Technologies) and then plated on the LB/Amp/IPTG/X-gal plates. White colonies were picked and minipreped with a Qiaprep Spin miniprep kit (Qiagen). Sequencing was done by using the BigDye 3.1 sequencing kit from Applied Biosystems (Foster City, CA), except the reaction conditions were reduced to 5 μL using 0.25 μL of the BigDye mix. Sequencing reactions were analyzed on an ABI 3730Xl sequencer.
Sequence homology searches were carried out using the BLAST (Basic Local Alignment Search Tool) program at http://www.ncbi.nlm.nih.gov/BLAST/, whereas sequence alignment was performed using the ClustalW program at http://workbench.sdsc.edu/ or with MegAlign in DNAStar. The final contiguous sequence of the clone was arrived at using Lasergene sequence analysis software (DNAStar). Phylogenetic relationships of aromatase genes were derived by aligning the deduced amino acid sequences of killifish CYP19A1 and CYP19A2 forms together with aromatase sequences reported for other vertebrate species using the ClustalW program.
2.5. BaP exposure adults
Adult killifish were exposed to water-borne BaP (Sigma) during two seasons; winter and summer. For the winter exposure, both male and female adult killifish (n = 12) were exposed to the following treatment: control (180 μL ethanol), BaP 1.0 μg/L and BaP 10 μg/L for 15 days (complete reproductive cycle). Exposure conditions were 15–22 °C, 10:14 light–dark period, and 20–25 ppt. For summer exposure, only adult female killifish were exposed to the following treatment: control (ethanol), and BaP 10 μg/L for 15 days (complete reproductive cycle). Exposure conditions were 22–24 °C, 14:10 light–dark period, and 20–25 ppt. For both the exposure experiments, there was 1.5 L of water per fish and thus 18 L per tank. Fish were kept in the tanks for at least 2 days for acclimatization prior to the start of the exposure. The tanks were checked and water was changed (100% static water renewal) every 24 h. BaP stock solutions were made (0.1 and 1 μg/μL) in 100% ethanol. Once the exposure had started, water was changed and the fish were dosed every day at the same time. In order to confirm nominal concentrations, 200 mL of water samples were taken 20 min and 24 h after dosing and put in clean amber bottles.
The fish were anaesthetized with 3-aminobenzoic acid ethyl ester (MS-222, Sigma), and their weight and length recorded. From half of the animals in each exposure, brains, gonads, and livers were homogenized in phosphate buffer (10 mM potassium phosphate (dibasic), 100 mM potassium chloride, 1 mM EDTA, pH 7.4) using a motorized homogenizer and centrifuged at 10,000 × g/10 min/4 °C. The supernatant was collected and samples were frozen immediately on dry ice. The protein concentration of the supernatant was determined in 96-well plates by the Bio-Rad method (Bio-Rad Laboratories, Hercules, CA) using BSA as the standard. Brains and gonads were processed for measuring aromatase activity. Livers were used to confirm a dose-response of CYP1A by Western blot. The brains, gonads, and livers from the other half of the exposed animals were preserved in RNAlater (Ambion Inc., Austin, TX) for expression studies. Samples were stored at −80 °C until further processing.
2.6. BaP exposure embryos
Pooled oocytes and sperm stripped from parental killifish were mixed together for in vitro fertilization. Fertilized eggs were randomly sorted into three treatment groups namely, untreated, DMSO control (1 μL/mL) or BaP (10 μg/L). Each egg was place in 1 mL of water (~21 ppt) in an autosampler vial (Fisher C4013-1) with a Teflon lined cap (Fisher B7815-8) and exposure began at approximately 4 h post-fertilization (hpf). Each egg was inspected daily for normal development. Every other day water was changed and eggs were redosed. Exposure lasted 10 days at which time the eggs were removed and placed in a 96-well plate on a circle of damp filter paper. After 7 days, the eggs remaining from each treatment were pooled and hatched. Simultaneously hatched fish were raised an additional 2 weeks post-hatch in small finger bowls. Ten embryos were pooled per time point and were collected for CYP19 mRNA analysis at 120 and 240 hpf, immediately following hatch, and at 2 weeks post-hatch. Embryos were stored in RNAlater at −80 °C until they were extracted with Trizol and RNeasy® kits as described above. The experiment was repeated four times. Water samples (200 mL) were collected to confirm water control and BaP concentrations at least twice during each exposure.
2.7. Analysis of water samples from exposure
Water samples were passed over Waters Sep-Pak Vac RC C18, 500 mg (#Wat 036945, Waters Corporation, Milford, MA) columns pre-rinsed with methanol:water ratio of 3:1. Columns were extracted with 15 mL of methylene chloride and samples were allowed to dry under a gentle stream of nitrogen. Samples were suspended in 1 mL of iso-octane and analyzed for BaP concentrations using gas chromatography/mass spectrometry (GC/MS) in selected ion monitoring mode (SIM). BaP standards (0.1, 0.2, 0.5, 1 and 2 ppm) were prepared in iso-octane and used to develop a calibration curve. BaP was detected as an ion with molecular weight 252.
2.8. Aromatase assay
Aromatase activity was quantified by the tritiated water release assay as described by Lephart and Simpson (1991) with minor modifications. The stereospecific loss of the 1β-3H of the substrate into the aqueous phase of the reaction mixture during aromatization is used as an indicator of aromatase activity. Tissue homogenates were incubated in a shaking water bath for 3 h at 28 °C/100 rpm with 3.0 nM 3H-androst-4-ene-3, 17-dione (specific activity 25.9 Ci/mmol (Perkin-Elmer Life Sciences, Boston, MA, USA)) in the presence of a reaction buffer (10 mM potassium phosphate (dibasic), 100 mM potassium chloride, 1 mM EDTA, 1 mM dithiothreitol, 1 mM NADPH, 10 mM glucose-6-phosphate and 1 U/mL glucose-6-phosphate dehydrogenase), in 200 μL total volume of reaction. Reactions were terminated by immersing the tubes in ice water and adding 100 μL of 30% trichloroacetic acid (TCA). The tubes were then centrifuged to remove the precipitated protein and unconverted substrate was then extracted with five volumes of chloroform by vortexing and centrifugation. The aqueous phase was increased to 1.5 mL with distilled water and centrifuged for 1 min at 800 × g. An aliquot of the 3H2O (1 mL) was placed into tubes containing 1 mL of 5% charcoal and 0.5% dextran T-70 (Pharmacia, Piscataway, NJ) in an aqueous solution, vortexed, and centrifuged. After centrifugation, 1 mL of the 3H2O was quantified by counting in 4 mL of premixed scintillation cocktail (Ultima Gold TMXR, Packard Biosciences, Boston, MA) in a liquid scintillation analyzer (TRI-CARB 2900 TR, Packard Biosciences). The specificity of the assay was determined by using 20 μM 4-hydroxyandrostenedione (Sigma), an irreversible inhibitor of the catalytic activity of aromatase, to block the formation of tritiated water. Aromatase activity (pmol/h/mg) was calculated by correcting for the tritium estimated in the blank tubes (containing substrate but no protein, dpm range 77–101), the dilution factor = 3, the 3H distribution of the [1β - 3H] androstenedione (75% at the 1β-position and 25% at the 1α-position) (Lephart and Simpson, 1991), protein concentration (150 μg for brain and 500 μg for gonad) and the specific activity of the substrate (dpm/mass; 66,020 ± 6000). Statistical differences across groups were determined by one-way analysis of variance (ANOVA) of log transformed data followed by Student–Newman–Keuls post hoc test, or by Student’s t-test for the summer exposures.
2.9. Quantitative real time RT-PCR
Primers for quantitative real time PCR were designed according to the requirements of Primer Express® v2.0 software for the ABI GeneAmp 5700 sequence detection system. Aromatase primers were designed in the gene specific 3′-UTR, and transcript abundance of the CYP19A1 or CYP19A2 was standardized against 18S rRNA response (CYP19A1, Primers 21 and 22 = 57 nt product; CYP19A2, Primers 23 and 24 = 51 nt product; 18S rRNA, Primers 25 and 26 = 97 nt product). Amplification efficiency of all three primer pairs was determined and was not statistically different (slopes = −3.63, −3.99, and −3.52 for CYP19A1, CYP19A2 and 18S rRNA, respectively, data not shown). Brain, gonad, or 10 pooled whole embryo RNA was isolated as described above. Quality and quantity of RNA was measured with an Agilent Bioanalyzer. The synthesis of cDNA was conducted with 250 ng total RNA using Taqman® Reverse Transcription Reagents (Applied Biosystems, Foster City, CA) in a 25 μL reaction. The cDNA corresponding to 10–20 ng for CYP19A1 or CYP19A2 and 10 pg for 18S of reverse-transcribed RNA served as templates for each of duplicate 25 μL PCR reactions using SYBR Green PCR Master Mix (Applied Biosystems). The PCR amplifications and fluorescence detection were performed with the GeneAmp® 5700 Sequence Detection System. Universal thermal cycling conditions according to the manufacturer were used. For quality control no template controls were done as well as melt curve analyses. CYP19 and 18S rRNA were amplified in duplicate and in separate reactions on the same plate. The amount of CYP19 mRNA, normalized to 18S, was given by the formula = 2−ΔΔCT, where CT is the threshold cycle indicating the fractional cycle number at which the amount of amplified CYP19 reaches a user selected threshold (fixed at 0.5 in these experiments). The ΔCT value is determined by subtracting the average 18S CT value from the average CYP19 CT value (plotted in Figs. 1 and 3). Then, the calculation of ΔΔCT involves subtraction of the ΔCT value of the calibrator (in our case the calibrator was average ΔCT value of control fish response in the BaP studies, or average female responses in the gender comparisons, or specific CYP19 form in the tissue comparisons) from the ΔCT value of each sample (ABI PRISM 7700 Sequence Detection System User’s Manual). Accordingly, CYP19 mRNA levels were reported as fold change in abundance relative to the average calibrator response. In the embryo experiments, CYP19 levels were reported relative to the untreated controls.
Fig. 1.
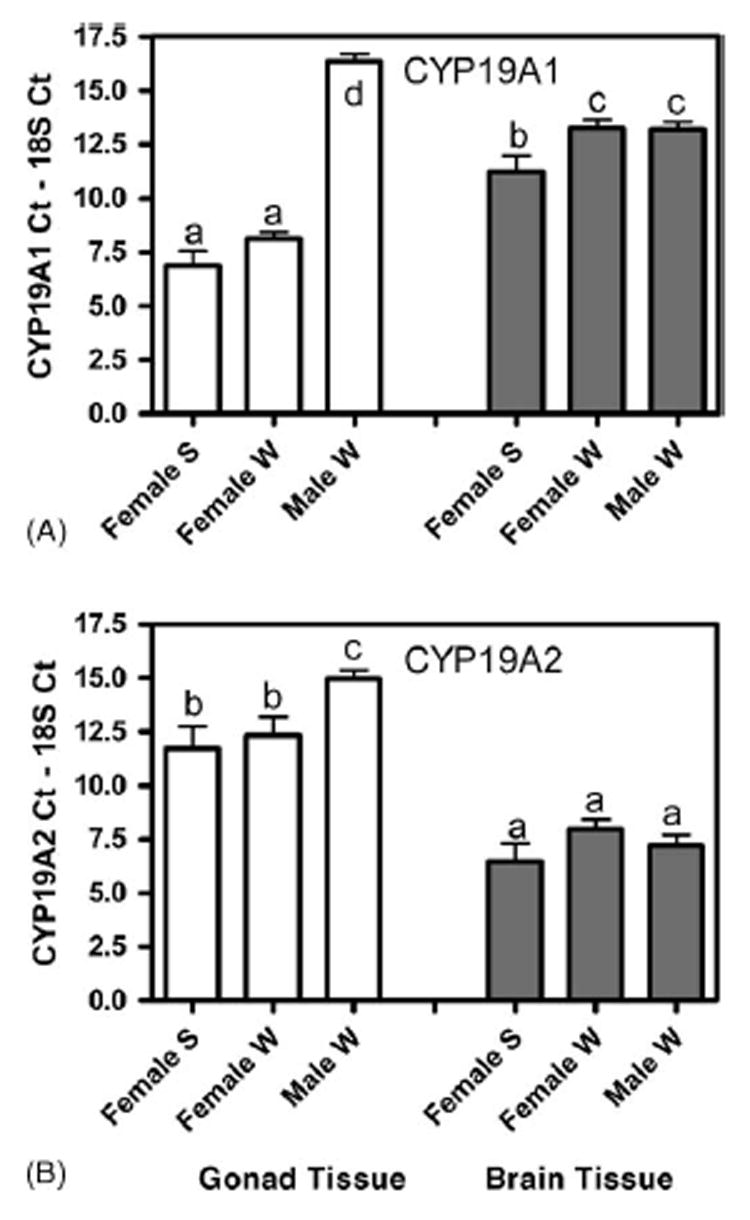
Constitutive CYP19A1 (A) and CYP19A2 (B) mRNA expression in control adult killifish gonad (white bars) and brain tissue (grey bars). S = summer exposure conditions while W = winter exposure conditions. Bar height represents the threshold cycle number for the CYP19 minus the threshold cycle number for the 18S for each sample. Note that the shorter the bar, the higher the mRNA expression (e.g. the sample crossed the threshold more quickly). Bars with the same letter are not statistically significant from each other (p > 0.05, ANOVA, n = 3–6). Data indicate there was significantly less CYP19A1 in testis compared to ovary, but there was not a sex difference in CYP19A2 in the brain. Female fish have statistically more CYP19A2 in the gonad and more CYP19A1 in brain during the summer exposure.
Fig. 3.
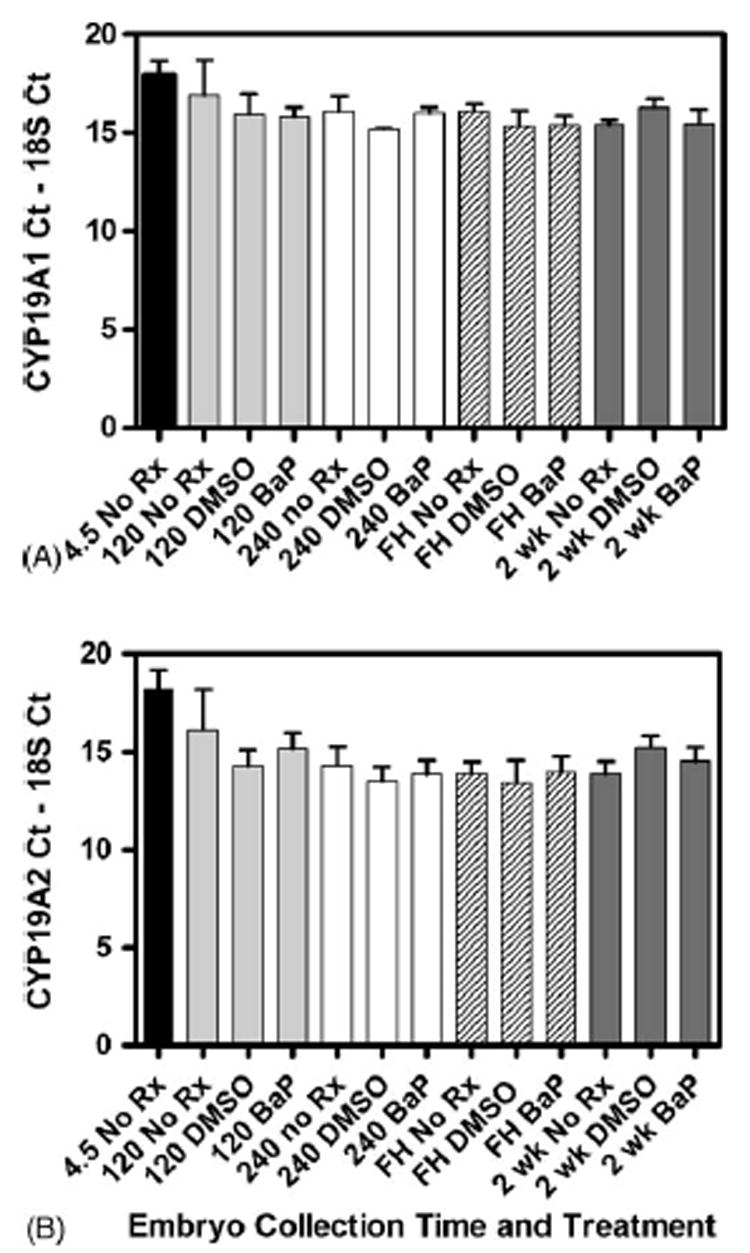
BaP effects on CYP19A1 (A) and CYP19A2 (B) mRNA levels in whole embryos. Killifish were untreated (No Rx) or exposed to 1 μL/mL DMSO or 10 μg/L BaP starting at ~4 to 240 hpf. Water was changed and embryos reexposed every other day. Ten embryos were pooled (n = 3 pools per time point) and collected at 120 and 240 hpf, at hatch (FH) and 2 weeks post-hatch (2 wk). RNA was isolated and amplified by qRT-RT PCR. Bar height represents the threshold cycle number for the CYP19 minus the threshold cycle number for the 18S for each sample. CYP19 mRNA expression was not statistically changed over the time course, however expression of CYP19A2 was ~5-fold higher than CYP19A1. BaP did not have a statistically significant effect on CYP19A1 or CYP19A2 expression.
2.10. Western blot
Liver tissue homogenates were analyzed for CYP1A protein content by Western blot. Tissue homogenate aliquots containing 20 μg protein were resolved by SDS-polyacrylamide gel electrophoresis (10% separating gel and 4% stacking gel) and transferred electrophoretically onto a PVDF membrane (Bio-Rad Laboratories). Following transfer, the membrane was blocked (1% casein, 20 mM Tris, 0.5 M NaCl pH 7.4, Bio-Rad Laboratories), and the membrane was incubated with casein buffer containing a CYP1A primary antibody (1:500, mouse anti-fish monoclonal antibody CYP1A, IgG3, Cayman Chemicals, Ann Arbor, MI) (Lee et al., 2000). The membrane was washed with 0.02 M Tris–base containing 10% Triton-X, and incubated with buffer containing the alkaline phosphatase-conjugated antibody (goat anti-mouse IgG, 1:5000, Sigma Chemicals). The membrane was developed with alkaline phosphatase buffer containing BCIP (5-bromo-4-chloro-3-indoylphosphate p-toluidine salt) and NBT (p-nitro blue tetrazolium chloride) color developing reagents. A positive control of human supersomes (Gentest Corporation, Woburn, MA) was used to estimate the approximate size of the immunopositive CYP1A1 band. Killifish CYP1A was identified as a single band between 48 and 91-kDa markers. Relative band intensities were quantified by determining the optical density (OD) by image analysis of the scanned Western blots using the VersaDoc imaging system (Bio-Rad Laboratories). Statistical differences between control and dosed samples of each tissue were determined with one-way ANOVA of log transformed data followed by Student–Newman–Keuls post hoc test (p < 0.05) using Graph-Pad Prism 3.0 version.
3. Results
3.1. Isolation of Fundulus CYP19 cDNA
CYP19A1 (GenBank Accession AY713118) and CYP19A2 (GenBank Accession AY494837) were obtained by RT-PCR and 5′- and 3′-RACE analyses. Killifish CYP19A1 cDNA (gonad form) contained an ORF of 1551 bp encoding a putative protein of 517 amino acids which spanned 9 exons. The 5′- and 3′-UTRs of CYP19A1 cDNA determined by RACE were 70 and 344-bp long, respectively. The deduced 500 amino acid sequence derived from the killifish CYP19A2 cDNA was based on an ORF of 1500-bp. The 5′- and 3′-UTRs of CYP19A2 cDNA were 66 and 628-bp long, respectively.
Killifish CYP19A1 and CYP19A2 were 60% identical to each other at the amino acid level. Greytak et al. (2005) have also recently cloned the killifish aromatase genes (GenBank Accession AY428665 and AY428666). There were only two amino acid differences T163S and P169L between the CYP19A1 forms, however in CYP19A2 there were 14 amino acid differences including an additional leucine in the membrane binding domain. It should be noted that Greytak et al. (2005) also found six amino acid differences among the CYP19A2 clones that they sequenced, suggesting that there is a potentially significant amount of sequence variability in Fundulus CYP19A2.
Compared to other teleosts, killifish CYP19A2 was 62–80% identical to CYP19A2 of other teleost brain aromatases and was most similar to Mozambique tilapia. Killifish CYP19A1 was 62–84% identical to CYP19A1 of other teleost ovarian aromatases with the highest similarity to medaka, sea bass and sea bream. Killifish CYP19A1 and CYP19A2 showed 50 and 51% identity, respectively, to human placental aromatase.
3.2. Quantitative real time RT-PCR
In order to quantitate CYP19A1 and CYP19A2 expression in killifish brain, gonad and embryos, primers were designed to the unique regions of the 3′-UTR. Both primer sets were used for each adult tissue type. In this way, we could address which aromatase mRNA was more prevalent in each tissue for each sex. Fig. 1A shows that there was no difference in ovarian CYP19A1 mRNA between the summer and winter fish (p > 0.05, ANOVA). However, there was significantly less (760 ± 230-fold) CYP19A1 in testis compared to ovarian tissue. Note that in the figure the data is presented as CYP19 CT –the 18S CT. Therefore, shorter bars represent higher mRNA expression. Relative fold induction is calculated by the equation 2−ΔΔCT as described in the methods section. The amount of CYP19A1 was also investigated in brain tissue and was 6.7-fold (±4.2) higher in females during the summer conditions compared to females or males in the winter conditions. In the brain, male and female CYP19A2 mRNA expression did not differ (Fig. 1B). In the ovary, there was 13-fold (±4.5) more CYP19A2 than in testis. Within tissues (comparing panel A and panel B, in Fig. 1), there was 29-fold (±8.7) more CYP19A1 in ovary compared to CYP19A2, and in brain there was 100-fold (±44) more CYP19A2 than CYP19A1.
3.3. Water concentrations of BaP
Exposure to BaP at a concentration of 1 μg/L (4 nM) and 10 μg/L (40 nM) had no effect on survivorship (100% survival in 15 days exposure) in either of the two adult exposure experiments. Similarly, there were no developmental delays or significant defects noted in DMSO or 10 μg/L BaP exposed embryos. Actual BaP water concentrations in tanks averaged 1.02 ± 0.08 and 6.88 ± 1.61 μg/L, as determined by GC/MS for the nominal concentrations of 1 and 10 μg/L, respectively. Water concentrations after 24 h in the adult tanks were only about 20% of the initial, and therefore, water was changed and redosed daily. For the embryo exposures, the DMSO samples all had nondetectable BaP while the nominal 10 μg/L concentration was actually 18.6 ± 1.19 μg/L. All figures indicate the nominal concentrations.
3.4. CYP19 mRNA in BaP exposed animals
When comparing the BaP exposed fish to controls, either 1 or 10 μg/L from the winter exposure or just 10 μg/L BaP exposed females from the summer exposure, there was no statistically significant effect on either CYP19A1 or CYP19A2 mRNA levels (Fig. 2). Furthermore, there was a high degree of inter-individual variability between fish.
Fig. 2.
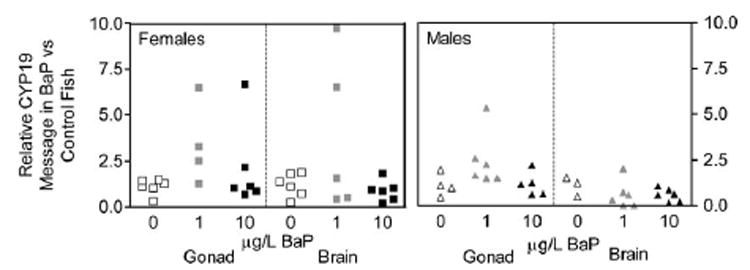
BaP effects on CYP19A1 and CYP19A2 mRNA levels in gonad and brain, respectively following the winter exposure. Adult killifish were exposed 15 days to 0, 1 or 10 μg/L and RNA was isolated from fish brains and gonads and amplified by qRT-RT PCR. Relative fold induction of BaP exposed animals was calculated by 2−ΔΔCT compared to the average ethanol treated animals (n = 3–6). BaP did not have a statistically significant effect on CYP19 expression in these or the summer studies (data not shown).
Similar to the adult exposure, there was not a significant effect on either CYP19A1 or CYP19A2 at the four developmental stages investigated following a 10 day, 10 μg/L BaP exposure (Fig. 3). These data are plotted as CYP CT –18S CT and show that there was on average 4.9-fold (±0.8) more CYP19A2 (shorter bars) compared to CYP19A1 in the embryos. There was less CYP19 mRNA at 4.5 hpf compared to the older embryos.
3.5. Aromatase enzyme activity
Aromatase activity in tissues of exposed killifish was measured by a tritiated water release assay. Ovarian (1000 μg) protein was incubated with 3 nM 3H-androstenedione for 0.5, 1.5, 2.5 and 3.5 h. Aromatase activity increased linearly with time (data not shown). Three hour incubations were used for all the further assays. Ovarian (500 μg) and brain (150 μg) protein, respectively, were incubated with 0.5–5 nM 3H-androstenedione concentrations [S] for 3 h at 28 °C. Aromatase activity [V] increased linearly obeying Michaelis–Menten kinetics.
The first exposure, done in winter, used two doses of BaP (1 and 10 μg/L). In killifish ovaries, 10 μg/L BaP inhibited aromatase activity by 2.8-fold (p < 0.05, ANOVA) (Fig. 4A). A dose-dependent inhibition in ovarian aromatase activity (0.0025, 0.0021, 0.0009 pmol/h/mg protein) with 0, 1, and 10 μg/L BaP, respectively was observed. The aromatase assay was not done with male gonads because the tissue preparations did not provide enough protein. Female brains showed a 1.9-fold (p < 0.05, ANOVA) induction in aromatase activity following BaP 10 μg/L treatment (Fig. 4B). Male brain aromatase activity was not statistically affected by BaP (Fig. 4C). For the second exposure experiment done in summer only one BaP dose (10 μg/L) was used because the lower BaP dose did not show any significant response in winter exposure. Only females were exposed in summer owing to the fact that in winter, BaP had no effect on male brain aromatase activity and male gonads did not provide sufficient tissue for aromatase assays. BaP significantly inhibited ovarian aromatase activity by 2.5-fold (p = 0.0192, t-test, Fig. 4A) but had no statistical effect on brain aromatase activity (p = 0.275, t-test, Fig. 4B). Aromatase activity was decreased by 99% in the presence of 4-hydroxyandrostenedione, an irreversible inhibitor, validating the activity to be due to aromatase enzyme (data not shown).
Fig. 4.
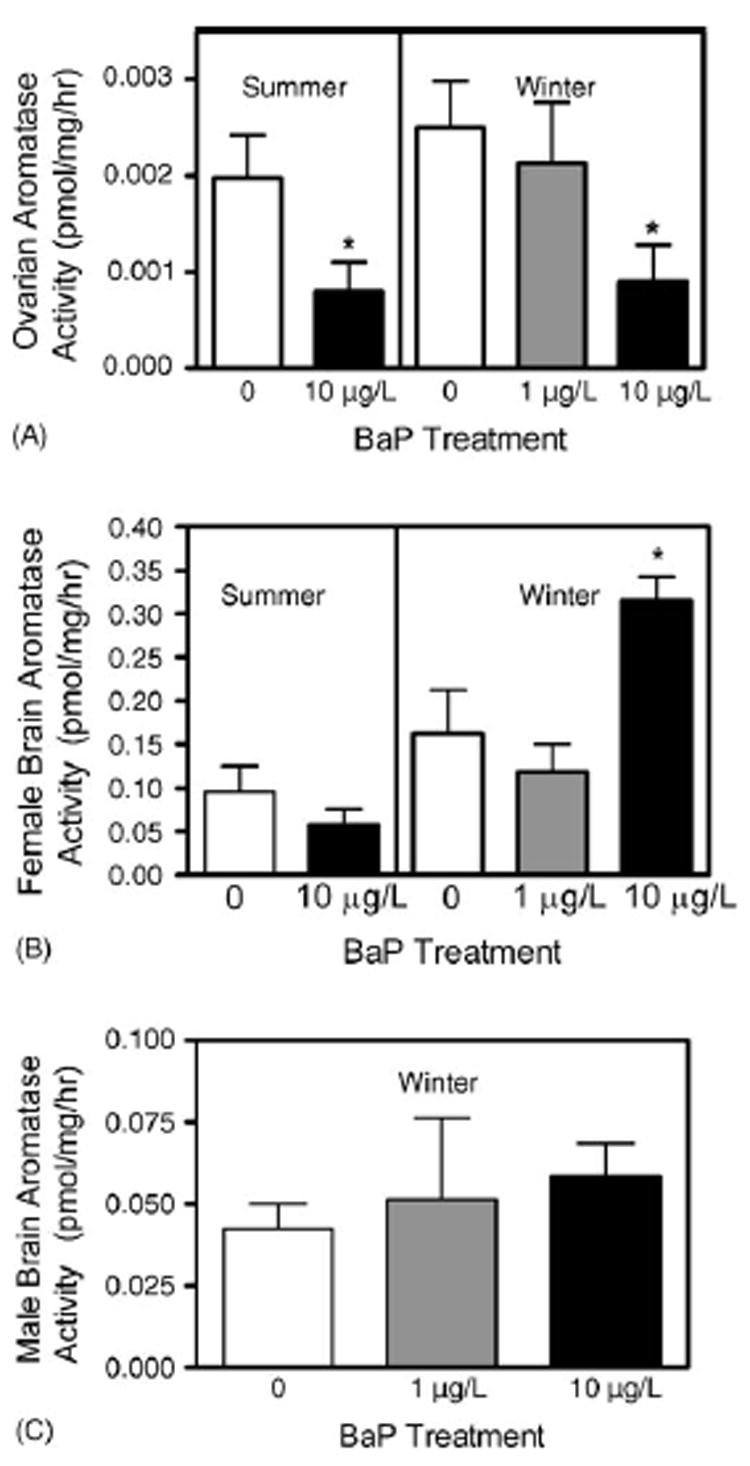
Aromatase activity in killifish exposed to water-borne BaP for 15 days. Ethanol (180 μL) was used as the solvent control. Ovary or brain homogenate (500 or 150 μg protein, respectively) was incubated with 3 nM 3H-androstenedione for 3 h at 28 °C. Aromatase activity was measured as the release of tritiated water and reported as pmol/mg/h. Each bar represents the mean ± S.E. of 5–7 fish per treatment. (A) Ovarian activities. Following the summer and winter exposure 10 μg/L BaP inhibited aromatase by 2.5- and 2.8-fold, respectively (p = 0.019, t-test and p = 0.027, ANOVA, respectively). (B) Female brain activities. Summer brain activities were not statistically different from controls (p = 0.275, t-test), however, in winter BaP significantly increased brain activity (p = 0.029, ANOVA). (C) Male brain activities (p = 0.531 ANOVA).
3.6. CYP1A Western blot
BaP caused a dose-dependent increasing trend in induction of CYP1A protein levels in both male and female liver from winter and female liver from summer exposures (Fig. 5). Average CYP1A protein levels were 0.005, 0.016, and 0.026 OD (5.2-fold, p = 0.054, ANOVA) for BaP 0, 1 and 10 μg/L in male liver and 0.003, 0.006, and 0.028 OD (9.3-fold, p = 0.013, ANOVA) for BaP 0, 1 and 10 μg/L in female liver from winter exposure. CYP1A protein levels were 0.005, and 0.014 OD for BaP 0 and 10 μg/L in female liver from summer exposure (p = 0.14, t-test). Induction of CYP1A protein by BaP confirms that water-borne BaP had its expected physiological effect in the fish at the low concentrations used.
Fig. 5.
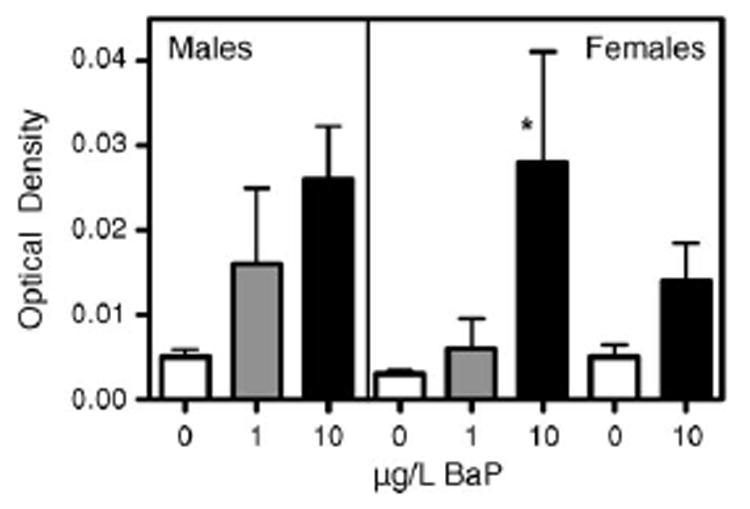
CYP1A protein measured by Western blot analysis in the liver of killifish exposed to aqueous BaP for 15 days. Males from winter exposure and females from both winter and summer exposures are shown. Ethanol (180 μL) was used as the solvent control. Results are expressed as the optical density (OD) ±S.E. for the 4–6 samples from each treatment group. OD was measured using VersaDoc software, *p < 0.05.
4. Discussion
CYP19 in humans is present as a single gene in the haploid genome. In contrast, several teleosts have shown the presence of two separate CYP19 genes. Regulation of CYP19 gene is complex and poorly understood (Goto-Kazeto et al., 2004; Kishida and Callard, 2001; Pellegrini et al., 2005). We cloned the full-length CYP19 cDNAs from brain and ovary of F. heteroclitus, with an anticipation of studying the gene structure and regulation. While some differences were found between the aromatase sequences previously reported (Greytak et al., 2005) especially for CYP19A2, these may be related to differences in the northern (Massachusetts) and more southern (North Carolina) populations of killifish. Differences in genetic makeup and gene expression between killifish from different locations have been established (Oleksiak et al., 2002; Segal et al., 1999). The physiological significance of the CYP19 sequence differences is currently unknown. Full-length cDNAs of CYP19A1 and CYP19A2 encoded proteins of 517 and 500 amino acids, respectively. The I-helix region, aromatase specific conserved region and the heme-binding region, were found in both Fundulus sequences. The amino acids in these regions were 81, 43 and 71% identical, respectively when both CYP19 forms of medaka, Mozambique tilapia, killifish, rainbow trout, zebrafish, and channel catfish were all compared. However, when killifish CYP19A2 was compared to Mozambique tilapia CYP19A2, these regions were 97, 83 and 100% conserved, respectively. Similarly, killifish and medaka CYP19A1s were 97, 96 and 93% identical in these aromatase specific areas.
With quantitative RT-RT PCR we investigated distribution of the CYP19 mRNA in the different tissues and sexes. Both CYP19 forms were found in brain and gonad which has previously been reported in adult zebrafish and killifish (Greytak et al., 2005; Trant et al., 2001). While the amount of CYP19A2 was similar in male and female control brains, there was significantly more CYP19A1 in ovary compared to testis. When investigating the CYP19 form present in each tissue, the ratio of CYP19A2 to CYP19A1 in the brain was consistent between sexes at about 100-fold (±44). This was less than the approximately 300-fold more relative CYP19A2:CYP19A1 reported in zebrafish (Trant et al., 2001). However in the testis, the CYP19A2 actually predominated over the CYP19A1. Largely due to high variability between fish, we did not detect a significant effect of BaP on CYP19A1 or CYP19A2 mRNA expression. Similarly, CYP19A1 mRNA levels were not different in killifish collected from a highly PCB-contaminated Superfund site compared to fish from a reference site (Greytak et al., 2005). However, recent work by Kazeto et al. (2004) showed that zebrafish juveniles exposed to 10 μM BaP from days 17 to 20 post-fertilization had increased CYP19A2 transcript while levels of CYP19A1 were unchanged relative to controls. The difference may be explained by the higher sensitivity of the juvenile zebrafish or the much higher concentration of BaP used (10 μM versus 40 nM in this study). The 10 μg/L BaP concentration used in these studies was based on the maximum solubility of BaP in water (~4 μg/L). CYP19A2 mRNA was also increased in killifish of both sexes from the PCB-impacted site (Greytak et al., 2005). Together these studies suggest that AhR ligands can increase the brain aromatase, however in our studies, we detected this increase at the level of enzyme activity not mRNA.
The tritiated water release assay is a very sensitive measure of potential effects at the protein level allowing for detection of activities <10 fmol/mg protein/h (Gonzalez and Piferrer, 2002). Killifish female brain showed 65- and 49-fold higher rate of conversion of 3H-androst-4-ene-3, 17-dione to estrone compared to ovary in winter and summer, respectively; consistent with what has been reported in other fish. Of all the vertebrates, teleost fish are characterized by exceptionally high levels (100–1000-fold) of neural estrogen biosynthesis when compared with the brains of other vertebrates or to the ovaries of the same fish (Pasmanik and Callard, 1985). Brain aromatase activity in sea bass was 7.8 pmol/mg/h, or four times higher than that from ovarian tissue (2.1 pmol/mg/h) (Gonzalez and Piferrer, 2002). Sea bass brain aromatase activities were similar in both sexes as juveniles but approximately one-tenth of the activity measured in first time spawners (Gonzalez and Piferrer, 2003). Killifish activities (range ~2–300 fmol/mg/min) were lower than those reported in sea bass but similar to those reported in Japanese medaka (Contractor et al., 2004). Brain activities were higher in killifish females compared to males (by 3.8-fold, p = 0.041, unpaired t-test). The high activity levels of aromatase in the brain of teleosts suggest an important role in regulatory pathways and constitute a potential target for disruption of the endocrine system by xenobiotics.
Two to 10-fold seasonal variations have been reported in adult killifish, sea bass and goldfish brain aromatase, with peaks associated with spawning (Gelinas et al., 1998; Gonzalez and Piferrer, 2003; Greytak et al., 2005; Pasmanik and Callard, 1988). While there was no significant seasonal difference in control female brain or ovarian enzyme activities or CYP19 mRNA levels, our results showed a seasonal difference in the BaP effects on killifish female brain aromatase activity. Following the winter BaP exposure female brain aromatase activity was increased, but there was no effect following the summer exposure. However, in the same fish and both seasons, BaP did cause a decrease in ovarian aromatase activity. While it is unclear at this time, BaP-mediated induction in female brain aromatase activity in the winter may be attributed to positive feedback in a sexually immature fish. However, no difference was observed in the GSI in either season.
BaP exposure from 4.5 hpf to 10 days post-fertilization did not affect CYP19A1 or CYP19A2 mRNA levels in embryos collected at 120 and 240 hpf, at hatch or 2 weeks post-hatch. Developmental expression of the CYP19s and estrogen induction of CYP19A2 has been previously described in zebrafish (Kishida and Callard, 2001; Trant et al., 2001). In zebrafish, CYP19 genes are maternally transferred which is followed by embryonic transcription beginning 12–24 hpf (Kishida and Callard, 2001). Killifish development (Armstrong and Child, 1965) is considerably slower than zebrafish, but both CYP19 mRNAs were detectable in equal amounts at 4.5 hpf. By 120 hpf to 2 weeks post-hatch (744 hpf), CYP19A2 predominated ~5-fold over the CYP19A1. As described above, in juvenile zebrafish a high BaP exposure did cause an increase in CYP19A2 mRNA (Kazeto et al., 2004) which was not seen in the killifish exposures. Therefore, higher concentrations of BaP, which could potentially be achieved through maternal transfer, could also impact killifish CYP19 expression and possibly development. No significant developmental defects were observed throughout the 10-day BaP exposure which was consistent with work by Wassenberg and Di Giulio (2004) where waterborne concentrations up to 100 μg/L caused no increase in deformities.
An environmental endocrine disruptor can act through estrogenic or anti-estrogenic mechanisms. In these studies, BaP showed activities consistent with both mechanisms of disruption by inhibiting ovarian aromatase while increasing the brain activity. When considering the estrogenic pathways, estrogen can exert its organizational and activational effects in the central nervous system either directly, or indirectly after aromatization of circulating androgen to estrogen within the brain. Therefore, an endocrine disrupting chemical can potentially affect the estrogen-dependent process by either or both of these two pathways. Some examples include adult fathead minnows where environmentally relevant concentrations of 17β-estradiol upregulated CYP19A2 mRNA expression in the testis and ovary in a dose-response manner after 14-day exposure (Halm et al., 2002). Estrogen also induced the CYP19A2 mRNA in goldfish by eight-fold (Gelinas et al., 1998) and in zebrafish embryos by four-fold (Kishida et al., 2001). Japanese medaka exposed to 500 ng/L ethinylestradiol also had increased male brain and gonad and female brain aromatase activity (Contractor et al., 2004).
There are several reports of estrogen-like activity of BaP. Unlike BaP itself, its monohydroxy metabolites like 1, 2, 3, and 9-OH BaP are considered potentially estrogenic because they bind to hER α and β in vitro (Hirose et al., 2001), but they did not induce estrogenic effects in the mouse uterus (Fertuck et al., 2001). BaP at concentrations ≥1 μM produced responses comparable to 0.1 nM estrogen in MCF-7 human breast cancer cells transiently transfected with a chimeric ER, and this response was completely blocked by the ER antagonist ICI 182,780, indicating that the response was ER-mediated (Charles et al., 2000).
Alternatively, several studies have reported the anti-estrogenic activity of BaP-like compounds. In fish, flounder (Platichthys flesus L.) ovarian tissue treated in vitro with BaP (15 μM) showed inhibition of ovarian steroidogenesis and a 25% decrease in estradiol production due to an inhibitory effect on aromatase (Monteiro et al., 2000). In vitro inhibition of aromatase activity by PAHs has also been reported with coho salmon ovarian tissue (Afonso et al., 1997). BaP and some other PAHs weakly displaced [3H]estradiol-binding from the human ER expressed in yeast (Tran et al., 1996), and treatment of MCF-7 human breast cancer cells with BaP resulted in a decrease in nuclear ER (Chaloupka et al., 1992). In vivo, uterine ER density was significantly decreased in female rats treated with BaP (20 μg, subcutaneously) within the first week after birth (Csaba and Inczefi-Gonda, 1993), and in Atlantic croaker BaP caused a decrease in circulating steroid concentrations and ovarian growth (Thomas, 1990).
The mechanisms of PAH-mediated inhibition of estrogen-induced responses are complicated and unresolved, but two different pathways have been hypothesized to contribute to this anti-estrogenicity. One involves PAHs inducing aryl hydrocarbon receptor-mediated CYP1A family genes (Willett et al., 1995) which metabolize estrogen to catechol estrogens (Badawi et al., 2001). The decreased circulating estrogen levels, in turn, inhibit the transcription of estrogen responsive genes. This hypothesis could be further tested by measuring aromatase activity following co-exposures of BaP and a CYP1A inhibitor (Wassenberg and Di Giulio, 2004). The other possibility is that transcription is suppressed through the interaction of AhR/ARNT and estrogen response elements (ERE) in estrogen responsive genes (Safe, 2001). For example, TCDD (a potent AhR ligand) inhibited estrogen-induced gene expression of cathepsin D due to the disruption of ER/Sp1 complex by targeted interaction with an overlapping xenobiotic response element (XRE) by AhR complex (Krishnan et al., 1995). Identification of the potential responsive elements in the 5′-flanking regions of two teleosts, zebrafish and goldfish, CYP19A1 and CYP19A2 genes has provided new insight as to how the two genes are differentially regulated in fish. Consensus sequences of TATA box, cAMP-responsive elements (CRE), a steroidogenic factor 1 (SF-1) site, and an XRE were observed in the 5′-flanking region of CYP19A1. In contrast, CYP19A2 showed the presence of CRE, an ERE, and a TATA box (Callard et al., 2001; Kazeto et al., 2001; Tchoudakova et al., 2001). The presence of XRE that binds AhR/ARNT is consistent with reports that TCDD (AhR agonist) decreased the aromatase mRNA in rat granulosa cells (Dasmahapatra et al., 2000) and the decreased CYP19A2 expression in PCB-exposed killifish (Greytak et al., 2005). The killifish CYP19A1 promoter has not been cloned, so either of these hypotheses may contribute to the mechanism for BaP-mediated decrease in ovarian aromatase activity seen in these studies.
Our results imply that AhR ligands, like BaP, have the potential to cause endocrine disruption by modulating aromatase activity. We found an increase of 1.4- to 2-fold in male and female brain aromatase activity and a decrease of ovarian aromatase activity by >2.5-fold by water-borne BaP in killifish. Although the observed ~2-fold induction or inhibition is relatively weak and was not observed at the mRNA level, aromatase is the rate-limiting enzyme in the conversion of androgens to estrogens. Alteration of this physiologically important enzyme in vivo would be expected to lead to altered local production of estrogens with the potential to cause or contribute to estrogen-related disorders. Future work will further investigate this hypothesis by directly investigating the correlation between reproductive and developmental success, BaP exposure and aromatase activity.
In conclusion, killifish aromatases are similar to other teleost CYP19s. Expression of aromatase differed by tissue and sex. BaP inhibited ovarian aromatase activity whereas it induced female brain aromatase activity suggesting both estrogen and anti-estrogen-like mechanisms of action. By modulating the two CYP19 gene activities, BaP can potentially disrupt steroid homeostasis and lead to reproductive deficits in fish.
Acknowledgments
This project was funded by start-up funds from the University of Mississippi and from the National Institutes of Environmental Health Sciences, Grant R01 ES012710-01 A1. Parental killifish were collected and provided by Dr. Patricia McClellan-Green, Duke University. We appreciate the help of Dr. Scott Bearson (USDA) with real time PCR primer design and instrument use. Special thanks also to Rooha Contractor for training with the aromatase assay; Annette Ford, Christine Metzger and Zankhana Master for their help and guidance in the experiments, and Kate Argote for help in maintaining the laboratory killifish.
References
- Afonso LO, Campbell PM, Iwama GK, Devlin RH, Donaldson EM. The effect of the aromatase inhibitor fadrozole and two polynuclear aromatic hydrocarbons on sex steroid secretion by ovarian follicles of coho salmon. Gen Comp Endocrinol. 1997;106(2):169–174. doi: 10.1006/gcen.1996.6855. [DOI] [PubMed] [Google Scholar]
- Armstrong PB, Child JS. Stages in normal development of Fundulus heteroclitus. Biol Bull. 1965;128:143–169. [Google Scholar]
- Badawi AF, Cavalieri E, Rogan EG. Role of human cytochrome P450 1A1, 1A2, 1B1 and 3A4 in the 2-, 4- and 16α-hydroxylation of 17β-estradiol. Metabolism. 2001;50(9):1001–1003. doi: 10.1053/meta.2001.25592. [DOI] [PubMed] [Google Scholar]
- Butala H, Metzger C, Rimoldi J, Willett KL. Microsomal estrogen metabolism in channel catfish. Mar Environ Res. 2004;58:489–494. doi: 10.1016/j.marenvres.2004.03.034. [DOI] [PubMed] [Google Scholar]
- Callard GV. Androgen and estrogen actions in the vertebrate brain. Am Zool. 1983;23:607–620. [Google Scholar]
- Callard GV, Petro Z, Ryan KJ. Biochemical evidence for aromatization of androgen to estrogen in the pituitary. Gen Comp Endocrinol. 1981;44(3):364–369. doi: 10.1016/0016-6480(81)90013-7. [DOI] [PubMed] [Google Scholar]
- Callard GV, Tchoudakova A. Evolutionary and functional significance of two CYP19 genes differentially expressed in brain and ovary of goldfish. J Steroid Biochem Mol Biol. 1997;61(3–6):387–392. doi: 10.1016/s0960-0760(97)80037-4. [DOI] [PubMed] [Google Scholar]
- Callard GV, Tchoudakova AV, Kishida M, Wood E. Differential tissue distribution, developmental programming, estrogen regulation and promoter characteristics of cyp19 genes in teleost fish. J Steroid Biochem Mol Biol. 2001;79(1–5):305–314. doi: 10.1016/s0960-0760(01)00147-9. [DOI] [PubMed] [Google Scholar]
- Chaloupka K, Krishnan V, Safe S. Polynuclear aromatic hydrocarbon carcinogens as antiestrogens in MCF-7 human breast cancer cells: role of the Ah receptor. Carcinogenesis. 1992;13(12):2233–2239. doi: 10.1093/carcin/13.12.2233. [DOI] [PubMed] [Google Scholar]
- Chang XT, Kobayashi T, Kajiura H, Nakamura M, Nagahama Y. Isolation and characterization of the cDNA encoding the tilapia (Oreochromis niloticus) cytochrome P450 aromatase (P450arom): changes in P450arom mRNA, protein and enzyme activity in ovarian follicles during oogenesis. J Mol Endocrinol. 1997;18(1):57–66. doi: 10.1677/jme.0.0180057. [DOI] [PubMed] [Google Scholar]
- Charles GD, Bartels MJ, Michael J, Zacharewski TR, Gollapudi BB, Freshour N, Carney EW. Activity of benzo[a]pyrene and its hydroxylated metabolites in an estrogen receptor-α reporter gene assay. Toxicol Sci. 2000;55(2):320–326. doi: 10.1093/toxsci/55.2.320. [DOI] [PubMed] [Google Scholar]
- Choi I, Troyer DL, Cornwell DL, Kirby-Dobbels KR, Collante WR, Simmen FA. Closely related genes encode developmental and tissue isoforms of porcine cytochrome P450 aromatase. DNA Cell Biol. 1997;16(6):769–777. doi: 10.1089/dna.1997.16.769. [DOI] [PubMed] [Google Scholar]
- Collier TK, Johnson LL, Stehr CM, Myers MS, Stein JE. A comprehensive assessment of the impacts of contaminants on fish from an urban waterway. Mar Environ Res. 1998;46(1–5):243–247. [Google Scholar]
- Conley A, Hinshelwood M. Mammalian aromatases. Reproduction. 2001;121(5):685–695. doi: 10.1530/rep.0.1210685. [DOI] [PubMed] [Google Scholar]
- Contractor RG, Foran CM, Li S, Willett KL. Evidence of gender- and tissue-specific promoter methylation and the potential for ethinylestradiol-induced changes in Japanese medaka (Oryzias latipes) estrogen receptor and aromatase genes. J Toxicol Environ Health. 2004;67:1–22. doi: 10.1080/15287390490253633. [DOI] [PubMed] [Google Scholar]
- Crisp TM, Clegg ED, Cooper RL, Wood WP, Anderson DG, Baetcke KP, Hoffmann JL, Morrow MS, Rodier DJ, Schaeffer JE, Touart LW, Zeeman MG, Patel YM. Environmental endocrine disruption: an effects assessment and analysis. Environ Health Perspect. 1998;106(Suppl 1):11–56. doi: 10.1289/ehp.98106s111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csaba G, Inczefi-Gonda A. Uterus estrogen receptors’ binding capacity is reduced in rat if exposed by benzpyrene neonatally. J Dev Physiol. 1993;19(5):217–219. [PubMed] [Google Scholar]
- Dalla Valle L, Lunardi L, Colombo L, Belvedere P. European sea bass (Dicentrarchus labrax L.) cytochrome P450arom: cDNA cloning, expression and genomic organization. J Steroid Biochem Mol Biol. 2002a;80(1):25–34. doi: 10.1016/s0960-0760(01)00170-4. [DOI] [PubMed] [Google Scholar]
- Dalla Valle L, Ramina A, Vianello S, Belvedere P, Colombo L. Cloning of two mRNA variants of brain aromatase cytochrome P450 in rainbow trout (Oncorhynchus mykiss Walbaum) J Steroid Biochem Mol Biol. 2002b;82(1):32. doi: 10.1016/s0960-0760(02)00143-7. [DOI] [PubMed] [Google Scholar]
- Dasmahapatra AK, Wimpee BA, Trewin AL, Wimpee CF, Ghorai JK, Hutz RJ. Demonstration of 2,3,7,8-tetrachlorodibenzo-p-dioxin attenuation of P450 steroidogenic enzyme mRNAs in rat granulosa cell in vitro by competitive reverse transcriptase-polymerase chain reaction assay. Mol Cell Endocrinol. 2000;164(12):5–18. doi: 10.1016/s0303-7207(00)00245-8. [DOI] [PubMed] [Google Scholar]
- Fertuck KC, Kumar S, Sikka HC, Matthews JB, Zacharewski TR. Interaction of PAH-related compounds with the α and β isoforms of the estrogen receptor. Toxicol Lett. 2001;121(3):167–177. doi: 10.1016/s0378-4274(01)00344-7. [DOI] [PubMed] [Google Scholar]
- Fukada S, Tanaka M, Matsuyama M, Kobayashi D, Nagahama Y. Isolation, characterization, and expression of cDNAs encoding the medaka (Oryzias latipes) ovarian follicle cytochrome P-450 aromatase. Mol Reprod Dev. 1996;45(3):285–290. doi: 10.1002/(SICI)1098-2795(199611)45:3<285::AID-MRD4>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Gelinas D, Callard GV. Immunocytochemical and biochemical evidence for aromatase in neurons of the retina, optic tectum and retinotectal pathways in goldfish. J Neuroendocrinol. 1993;5(6):635–641. doi: 10.1111/j.1365-2826.1993.tb00533.x. [DOI] [PubMed] [Google Scholar]
- Gelinas D, Pitoc GA, Callard GV. Isolation of a goldfish brain cytochrome P450 aromatase cDNA: mRNA expression during the seasonal cycle and after steroid treatment. Mol Cell Endocrinol. 1998;138(12):81–93. doi: 10.1016/s0303-7207(98)00015-x. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Piferrer F. Characterization of aromatase activity in the sea bass: effects of temperature and different catalytic properties of brain and ovarian homogenates and microsomes. J Exp Zool. 2002;293:500–510. doi: 10.1002/jez.90005. [DOI] [PubMed] [Google Scholar]
- Gonzalez A, Piferrer F. Aromatase activity in the European sea bass (Dicentrarchus labraz L.) brain. Distribution and changes in relation to age, sex, and the annual reproductive cycle. Gen Comp Endocrinol. 2003;132:223–230. doi: 10.1016/s0016-6480(03)00086-8. [DOI] [PubMed] [Google Scholar]
- Goto-Kazeto R, Kight KE, Zohar Y, Place AR, Trant JM. Localization and expression of aromatase mRNA in adult zebrafish. Gen Comp Endocrinol. 2004;139:72–84. doi: 10.1016/j.ygcen.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Greytak SR, Champlin D, Callard GV. Isolation and characterization of two cytochrome P450 aromatase forms in killifish (Fundulus heteroclitus): differential expression in fish from polluted and unpolluted environments. Aquat Toxicol. 2005;71:371–389. doi: 10.1016/j.aquatox.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Halm S, Pounds N, Maddix S, Rand-Weaver M, Sumpter JP, Hutchinson TH, Tyler CR. Exposure to exogenous 17beta-oestradiol disrupts p450aromB mRNA expression in the brain and gonad of adult fathead minnows (Pimephales promelas) Aquat Toxicol. 2002;60(34):285–299. doi: 10.1016/s0166-445x(02)00011-5. [DOI] [PubMed] [Google Scholar]
- Halm S, Rand-Weaver M, Sumpter JP, Tyler CR. Cloning and molecular characterization of an ovarian-derived (brain-like) P450 aromatase cDNA and development of a competitive RT-PCR assay to quantify its expression in the fathead minnow (Pimephalespromelas) Fish Physiol Biochem. 2001;24:49–62. [Google Scholar]
- Hirose T, Morito K, Kizu R, Toriba A, Hayakawa K, Ogawa S, Inoue S, Muramatsu M, Masamune Y. Estrogenic/antiestrogenic activities of benzo[a]pyrene monohydroxy derivatives. J Health Sci. 2001;47(6):552–558. [Google Scholar]
- Kazeto Y, Ijiri S, Place AR, Zohar Y, Trant JM. The 5′-flanking regions of CYP19A1 and CYP19A2 in zebrafish. Biochem Biophys Res Commun. 2001;288(3):503–508. doi: 10.1006/bbrc.2001.5796. [DOI] [PubMed] [Google Scholar]
- Kazeto Y, Place AR, Trant JM. Effects of endocrine disrupting chemicals on the expression of CYP19 genes in zebrafish (Danio rerio) juveniles. Aquat Toxicol. 2004;69(1):25–34. doi: 10.1016/j.aquatox.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Kishida M, Callard GV. Distinct cytochrome P450 aromatase isoforms in zebrafish (Danio rerio) brain and ovary are differentially programmed and estrogen regulated during early development. Endocrinology. 2001;142(2):740–750. doi: 10.1210/endo.142.2.7928. [DOI] [PubMed] [Google Scholar]
- Kishida M, McLellan M, Miranda JA, Callard GV. Estrogen and xenoestrogens upregulate the brain aromatase isoform (P450aromB) and perturb markers of early development in zebrafish (Danio rerio) Comp Biochem Physiol B Biochem Mol Biol. 2001;129(23):261–268. doi: 10.1016/s1096-4959(01)00319-0. [DOI] [PubMed] [Google Scholar]
- Krishnan V, Porter W, Santostefano M, Wang X, Safe S. Molecular mechanism of inhibition of estrogen-induced cathepsin D gene expression by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) in MCF-7 cells. Mol Cell Biol. 1995;15(12):6710–6719. doi: 10.1128/mcb.15.12.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon JY, McAndrew BJ, Penman DJ. Cloning of brain aromatase gene and expression of brain and ovarian aromatase genes during sexual differentiation in genetic male and female Nile tilapia Oreochromis niloticus. Mol Reprod Dev. 2001;59(4):359–370. doi: 10.1002/mrd.1042. [DOI] [PubMed] [Google Scholar]
- Lee YH, Lee FY, Yueh WS, Tacon P, Du JL, Chang CN, Jeng SR, Tanaka H, Chang CF. Profiles of gonadal development, sex steroids, aromatase activity, and gonadotropin II in the controlled sex change of protandrous black porgy, Acanthopagrus schlegeli Bleeker. Gen Comp Endocrinol. 2000;119(1):111–120. doi: 10.1006/gcen.2000.7499. [DOI] [PubMed] [Google Scholar]
- Lephart ED, Simpson ER. Assay of aromatase activity. Meth Enzymol. 1991;206:477–483. doi: 10.1016/0076-6879(91)06116-k. [DOI] [PubMed] [Google Scholar]
- Liu X, Liang B, Zhang S. Sequence and expression of cytochrome P450 aromatase and FTZ-F1 genes in the protandrous black porgy (Acanthopagrus schlegeli) Gen Comp Endocrinol. 2004;138(3):247–254. doi: 10.1016/j.ygcen.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Lotrich VA. Summer home range and movements of Fundulus heteroclitus (Pisces: Cyprinodontidae) in a tidal creek. Ecology. 1975;56(1):191–198. [Google Scholar]
- Melo CA, Rand-Weaver M. Sexual dimorphism of brain aromatase activity in medaka: induction of a female phenotype by estradiol. Environ Health Perspect. 2001;109(3):257–264. doi: 10.1289/ehp.01109257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JN, Wassenberg D, Karchner S, Hahn ME, Di Giulio RT. Expression and inducibility of aryl hydrocarbon receptor pathway genes in wild-caught killifish (Fundulus heteroclitus) with different contaminantexposure histories. Environ Toxicol Chem. 2003;22(10):2337–2343. doi: 10.1897/02-495. [DOI] [PubMed] [Google Scholar]
- Monteiro PR, Reis-Henriques MA, Coimbra J. Polycyclic aromatic hydrocarbons inhibit in vitro ovarian steroidogenesis in the flounder (Platichthys flesus L.) Aquat Toxicol. 2000;48(4):549–559. doi: 10.1016/s0166-445x(99)00055-7. [DOI] [PubMed] [Google Scholar]
- Nicolas J-M. Vitellogenesis in fish and the effects of polycyclic aromatic hydrocarbon contaminants. Aquat Toxicol. 1999;45:77–90. [Google Scholar]
- Oleksiak MF, Churchill GA, Crawford DL. Variation in gene expression within and among natural populations. Nat Genet. 2002;32:261–266. doi: 10.1038/ng983. [DOI] [PubMed] [Google Scholar]
- Pait AS. Reproductive Endocrine Disruption in the Killifish Fundulus heteroclitus in the Chesapeake Bay. University of Maryland College Park; 2001. pp. 1–271. (PhD dissertation) [Google Scholar]
- Pasmanik M, Callard GV. Aromatase and 5 alpha-reductase in the teleost brain, spinal cord, and pituitary gland. Gen Comp Endocrinol. 1985;60(2):244–251. doi: 10.1016/0016-6480(85)90320-x. [DOI] [PubMed] [Google Scholar]
- Pasmanik M, Callard GV. Changes in brain aromatase and 5 alpha-reductase activities correlate significantly with seasonal reproductive cycles in goldfish (Carassius auratus) Endocrinology. 1988;122(4):1349–1356. doi: 10.1210/endo-122-4-1349. [DOI] [PubMed] [Google Scholar]
- Pellegrini E, Menuet A, Lethimonier C, Adrio F, Gueguen M-M, Tascon C, Anglade I, Pakdel F, Kah O. Relationships between aromatase and estrogen receptors in the brain of teleost fish. Gen Comp Endocrinol. 2005;142:60–66. doi: 10.1016/j.ygcen.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Safe S. Molecular biology of the Ah receptor and its role in carcinogenesis. Toxicol Lett. 2001;120(1–3):1–7. doi: 10.1016/s0378-4274(01)00301-0. [DOI] [PubMed] [Google Scholar]
- Segal JA, Barnett JL, Crawford DL. Functional analyses of natural variation in Sp1 binding sites of a TATA-less promoter. J Mol Evol. 1999;49(6):736–749. doi: 10.1007/pl00006596. [DOI] [PubMed] [Google Scholar]
- Simpson ER, Michael MD, Agarwal VR, Hinshelwood MM, Bulun SE, Zhao Y. Expression of the CYP19 (aromatase) gene: an unusual case of alternative promoter usage. FASEB J. 1997;11(1):29–36. doi: 10.1096/fasebj.11.1.9034163. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Telecky TM, Fukada S, Adachi S, Chen S, Nagahama Y. Cloning and sequence analysis of the cDNA encoding P-450 aromatase (P450arom) from a rainbow trout (Oncorhynchus mykiss) ovary; relationship between the amount of P450arom mRNA and the production of oestradiol-17 beta in the ovary. J Mol Endocrinol. 1992;8(1):53–61. doi: 10.1677/jme.0.0080053. [DOI] [PubMed] [Google Scholar]
- Tchoudakova A, Callard GV. Identification of multiple CYP19 genes encoding different cytochrome P450 aromatase isozymes in brain and ovary. Endocrinology. 1998;139(4):2179–2189. doi: 10.1210/endo.139.4.5899. [DOI] [PubMed] [Google Scholar]
- Tchoudakova A, Kishida M, Wood E, Callard GV. Promoter characteristics of two CYP19 genes differentially expressed in the brain and ovary of teleost fish. J Steroid Biochem Mol Biol. 2001;78(5):427–439. doi: 10.1016/s0960-0760(01)00120-0. [DOI] [PubMed] [Google Scholar]
- Thomas P. Teleost model for studying the effects of chemicals on female reproductive endocrine function. J Exp Zool. 1990;(Suppl 4):126–128. doi: 10.1002/jez.1402560421. [DOI] [PubMed] [Google Scholar]
- Toda K, Shizuta Y. Identification and characterization of cisacting regulatory elements for the expression of the human aromatase cytochrome P-450 gene. J Biol Chem. 1994;269(11):8099–8107. [PubMed] [Google Scholar]
- Tran DQ, Ide CF, McLachlan JA, Arnold SF. The antiestrogenic activity of selected polynuclear aromatic hydrocarbons in yeast expressing human estrogen receptor. Biochem Biophys Res Commun. 1996;229(1):102–108. doi: 10.1006/bbrc.1996.1764. [DOI] [PubMed] [Google Scholar]
- Trant JM. Isolation and characterization of the cDNA encoding the channel catfish (Ictalurus punctatus) form of cytochrome P450arom. Gen Comp Endocrinol. 1994;95(2):155–168. doi: 10.1006/gcen.1994.1113. [DOI] [PubMed] [Google Scholar]
- Trant JM, Gavasso S, Ackers J, Chung C, Place RA. Developmental expression of cytochrome P450 aromatase genes (CYP19a and CYP19b) in zebrafish fry (Danio rerio) J Exp Zool. 2001;290(5):475–483. doi: 10.1002/jez.1090. [DOI] [PubMed] [Google Scholar]
- Vogelbein WK, Fournie JW, Van Veld PA, Huggett RJ. Hepatic neoplasms in the mummichog Fundulus heteroclitus from a creosote-contaminated site. Cancer Res. 1990;50(18):5978–5986. [PubMed] [Google Scholar]
- Wallace RA. Vitellogenesis and oocyte growth in nonmammalian vertebrates. Dev Biol (NY 1985) 1985;1:127–177. doi: 10.1007/978-1-4615-6814-8_3. [DOI] [PubMed] [Google Scholar]
- Wassenberg DM, Di Giulio RT. Synergistic embryotoxicity of polycyclic aromatic hydrocarbon aryl hydrocarbon receptor agonists with cytochrome P4501A inhibitors in Fundulus heteroclitus. Environ Health Perspect. 2004;112:1658–1664. doi: 10.1289/ehp.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett K, Steinberg MA, Thomsen J, Narasimhan TK, Safe SH, McDonald SJ, Beatty KB, Kennicutt MC. Exposure of killifish to benzo(a)pyrene: Comparative metabolism, DNA adduct formation and aryl hydrocarbon (Ah) receptor agonist activities. Comp Biochem Physiol. 1995;112B(1):93–103. [Google Scholar]
- Yozzo DJ, Hester KI, Smith DE. Abundance and spawning site utlilization of Fundulus heteroclitus at the Virginia Coast Reserve. Virginia J Sci. 1994;45:187–197. [Google Scholar]


