Abstract
The ribosomal protein S15 from Escherichia coli binds to a pseudoknot in its own messenger. This interaction is an essential step in the mechanism of S15 translational autoregulation. In a previous study, a recognition determinant for S15 autoregulation, involving a U⋅G wobble pair, was located in the center of stem I of the pseudoknot. In this study, an extensive mutagenesis analysis has been conducted in and around this U⋅G pair by comparing the effects of these mutations on the expression level of S15. The results show that the U⋅G wobble pair cannot be substituted by A⋅G, C⋅A, A⋅C, G⋅U, or C⋅G without loss of the autocontrol. In addition, the base pair C⋅G, adjacent to the 5′ side of U, cannot be flipped or changed to another complementary base pair without also inducing derepression of translation. A unique motif, made of only two adjacent base pairs, U⋅G/C⋅G, is essential for S15 autoregulation and is presumably involved in direct recognition by the S15 protein.
Keywords: pseudoknot, U⋅G base pair, RNA binding
Although RNA pseudoknots have been implicated in many different regulatory functions (1–8), very little is known about how proteins specifically recognize these structures. In one of the most studied cases, ribosomal frameshifting, protein–RNA recognition studies are difficult because the pseudoknot interacts with a complex protein-synthesizing machinery. A more favorable case is that of the autoregulation of ribosomal protein S15 from Escherichia coli. When S15 (10 kDa) is in excess of that required for ribosome assembly, it binds specifically to a pseudoknot structure that forms transiently around the ribosome loading site of its own messenger, thus repressing its own synthesis (9–11). The secondary structure of the pseudoknot was determined in vitro by enzymatic and chemical probing of S15 mRNA fragments (12, 13). These studies showed that the pseudoknot is unstable and is in equilibrium with another form consisting of two stem-loops. In vitro, S15 binds only to the pseudoknot, as shown by the tight correlation between S15-specific mRNA binding and the ability of the bound RNA to form a pseudoknot. Protection experiments with chemical probes identified two regions shielded by S15 in the pseudoknot (black dots in Fig. 1): one located at the distal end of stem 2 and covering loop 1 and the other in stem 1 near the junction of the two stems (13), suggesting, at first glance, that two contact areas in the minor groove might be involved. Protection by S15 against hydroxyl radical attack is restricted to the interaction essentially around the coaxial stack of the two stems (13). In all cases, the bases protected include the U of a U(−49)⋅G(−36) wobble base pair present in stem 1. In vivo, evidence for pseudoknot formation was obtained by analyzing the effects of compensatory mutations on the regulatory properties of a translational fusion between rpsO, the S15 gene, and lacZ (10). An extensive mutagenesis of stem 2 failed to reveal any base involved in autoregulation or S15 binding, suggesting that putative contacts in this region identified by the chemical protection experiments might be unspecific or associated to functional groups belonging to the backbone. On the other hand, mutational analysis of stem 1 showed that a recognition determinant is located in this stem and involves a U⋅G wobble pair (10). Several examples are known where G⋅U pairs are involved in RNA recognition (14–17). However, direct recognition of this pair has not been clearly established because it was generally possible (at least in vivo) to substitute the wobble pair by some other mispairs, suggesting that access to the functional groups is controlled by the helix geometry. Here it is shown that, in vivo, U⋅G cannot be substituted by other canonical or noncanonical base pairs and that U⋅G is included in a motif that probably determines a unique configuration of the wobble pair.
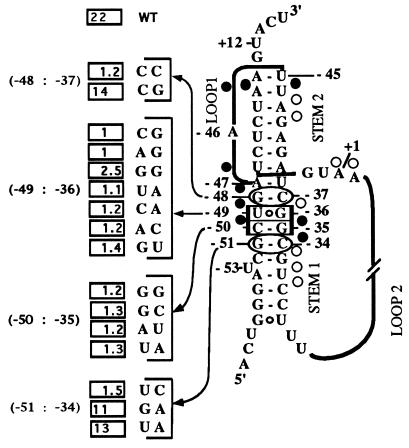
Figure 1.
Pseudoknot structure of the S15 messenger site and regulatory properties of mutations in stem 1. Base pair changes were introduced in a rpsO–lacZ translational fusion carried in a λ phage. For each mutation, the effect of overexpression of S15 on the level of β-galactosidase was measured and compared with the value of a strain carrying a wild-type fusion (10). The mutated base pairs indicated are described in Table 1. Repression ratios correspond to the ratio of the β-galactosidase units in the control divided by the value in the mutant and are given in the squares. This ratio is 1 when all control is lost and is 22 for the wild type (WT). The recognition determinant, G⋅U/G⋅C, is boxed, and the adjacent base pairs are ovalized. Ribose protection of the messenger binding site against ethylnitrosourea (ENU) in the presence of S15 are taken from ref. 13 and are shown by circles: •, strong protection; ○, weak protection. +1 corresponds to the translational initiation codon.
MATERIALS AND METHODS
Mutagenesis.
Mutations were introduced into the pseudoknot by site-directed mutagenesis (18, 19) of M13 mp8 derivatives carrying a translational fusion between rpsO and lacZ (10). After mutagenesis, an EcoRI–HindIII fragment carrying all the regulatory region was sequenced and fused in frame to the distal part of lacZ, which is carried by a λ derivative phage (9). Encapsidation, infection of the E. coli strain AB5321 (argG, argE, his, rpsL, ΔlacX74), and plaque screening were carried out as previously described (9, 10). Monolysogens were isolated, and their translational regulatory properties were analyzed by measuring the repression of the β-galactosidase level in the presence of a plasmid overproducing S15 in trans (see Table 1).
Table 1.
In vivo effects of mutations on the autoregulation of a translational fusion rpsO-lacZ
| Pseudoknot
|
β-galactosidase units in the presence of plasmids
|
Repression ratio | ||
|---|---|---|---|---|
| Base pair | Mutation | pBR322 (control) | pBP111 (+ rpsO) | |
| Wild type | 869 ± 50 | 39 ± 3 | 22 | |
| G-48 C-37 | C·C | 3,297 ± 264 | 2,768 ± 166 | 1.2 |
| G-48 C-37 | C·G | 2,069 ± 69 | 184 ± 14 | 14 |
| U-49 G-36 | C·G | 3,873 ± 160 | 3,460 ± 132 | 1.1 |
| U-49 G-36 | A·G | 1,927 ± 199 | 1,894 ± 193 | 1.0 |
| U-49 G-36 | G·G | 3,399 ± 198 | 1,418 ± 84 | 2.5 |
| U-49 G-36 | U·A | 2,467 ± 56 | 2,322 ± 155 | 1.1 |
| U-49 G-36 | C·A | 3,188 ± 81 | 2,641 ± 197 | 1.2 |
| U-49 G-36 | A·C | 3,967 ± 565 | 3,293 ± 528 | 1.2 |
| U-49 G-36 | G·U | 4,387 ± 119 | 3,131 ± 80 | 1.4 |
| C-50 G-35 | G·G | 1,421 ± 94 | 1,392 ± 56 | 1.2 |
| C-50 G-35 | G·C | 3,963 ± 66 | 3,131 ± 49 | 1.4 |
| C-50 G-35 | A·U | 3,466 ± 281 | 2,994 ± 143 | 1.2 |
| C-50 G-35 | U·A | 3,769 ± 468 | 2,947 ± 341 | 1.3 |
| G-51 C-34 | G·A | 2,137 ± 68 | 188 ± 39 | 11 |
| G-51 C-34 | U·C | 2,621 ± 88 | 1,689 ± 245 | 1.5 |
| G-51 C-34 | U·A | 1,295 ± 42 | 98 ± 39 | 13 |
To analyze the expression of the ribosomal protein S15, a translational in frame fusion between rpsO, the gene encoding S15, and lacZ was used and the level of β-galactosidase (Miller units) measured in the presence of an overproduction of S15 in trans (pBP111) and in the absence (pBR322). The ratio between the two values (repression ratio) measures the efficiency of the regulation. The higher this value, the stronger the regulation. Conversely, when no control is present, this ratio decreases to 1.
RESULTS
Previous studies (10) suggested that recognition by S15 requires specific features given by a U⋅G wobble pair. Recognition may be either direct if S15 interacts precisely with functional groups of the U⋅G pair or indirect if S15 recognition depends only on a local helical distortion promoted by U⋅G. Direct recognition may rely on the presence of different accessible functional groups, like the 2-amino group of guanine, which is in an unusual orientation because of the deviation from standard geometry in the helical groove of U and G relative to the bases in C⋅G or U⋅A Watson–Crick pairs (20). To distinguish between these hypotheses, autoregulation of S15 was analyzed in vivo in the presence of different mutations affecting the U⋅G pair. If direct recognition is involved, no substitution will be tolerated, whereas some substitutions might be allowed by an indirect recognition mechanism. Several mutations were introduced into a rpsO–lacZ translational fusion carried on the chromosome, and their effect on autoregulation was estimated from their ability to deregulate the β-galactosidase level measured in the presence of a plasmid expressing S15 in trans.
Watson–Crick Pairs Cannot Substitute for the U⋅G Wobble Pair.
When shifting U⋅G for C⋅G, the repression ratio, which is 22 in the wild type, drops to 1 (Table 1 and Fig. 1). It has been previously shown that changing U⋅G for U⋅A also decreases the repression ratio to 1 (10). These observations clearly indicate that no Watson–Crick pair is tolerated in this position without inducing a total loss of autocontrol. These results do not distinguish between a mechanism involving direct recognition or indirect interaction. The formation of Watson–Crick base pairs presumably does not change the pseudoknot stability but rather removes or modifies the position of some specific functional group or, in another way, induces a change in the helix geometry by removing some local helix distortion.
The U⋅G Pair Cannot be Replaced by Other Noncanonical or Wobble Pairs.
If it is just the presence of a wobble base pair that is needed to properly position the S15 recognition determinants, it should be possible to keep autocontrol in vivo when U⋅G is shifted to C⋅A. This mispair, which can adopt a wobble configuration when protonated, has often been shown to be able to replace U⋅G. This is not the case here (Fig. 1). Moreover, other changes like flipping C⋅A to A⋅C, U⋅G to G⋅U have the same negative effect on regulation (Fig. 1 and Table 1). These observations might mean that the exocyclic amino group of guanosine, which is missing in C⋅A, is involved in S15 binding. However, replacing U⋅G by the noncanonical base pair A:G does not restore autocontrol. These experiments demonstrate that the polarity of U⋅G wobble pair is critical and that no change is tolerated at this position. The only substitution that allows just a very low level of regulation is G:G (repression ratio: 2.5, Table 1), suggesting that some limited specific interaction of S15 is still possible in this case.
U⋅G Is Part of a Recognition Determinant.
If direct recognition is involved, specific functional groups carried by the U⋅G pair should interact with residues on the protein. The exocyclic amino group of guanosine, which, in a wobble pair, projects in the minor groove, might be involved. However, the proper configuration of bases in the minor groove may also be dependent on neighboring bases and/or include several other functional groups like the 2′OH of ribose residues (21). If this is the case, flipping the base pair adjacent to the 5′ side of the U in the U⋅G pair is predicted to modify the position of the bases in the helix (22) and hence the position of their functional groups. A total loss of autocontrol was induced when the pair C(50)⋅G(−35), 5′ to the U side, was flipped or replaced by A⋅U or U⋅A (Table 1 and Fig. 1). These observations demonstrate that the two stacked base pairs U⋅G/C⋅G constitute a motif that seems necessary for the proper configuration of the recognition determinants.
The RNA Determinant Is Constituted by Only Two Base Pairs.
To see if the other adjacent base pairs are also involved, the pair G(−48)⋅C(−37) adjacent to the 3′ side of the U was flipped. Only a limited effect in autoregulation was observed as shown by the repression ratio, which decreases to 14 (Table 1). Practically the same value is observed when the pair G(−51)⋅C(−34) is changed for U⋅A. Single changes at positions (−48) or (−51) create the mispairs C(−48)⋅C or U(−51)⋅C. These changes, which are known to disrupt base pairing, abolish the autoregulation. Interestingly, another single change (A(−34)), induces only a limited effect on autoregulation. In contrast to the other single changes, a noncanonical base pair (G⋅A) is probably formed in this case, which would induce only limited structural changes and would not interfer with the access or the position of determinants. These results clearly show that the recognition determinant is constituted by only two base pairs.
DISCUSSION
All of these observations show that the recognition determinant for S15 binding is located in a motif comprising the two stacked base pairs U⋅G/C⋅G. The function of this motif, located inside stem 1 of the pseudoknot, is to properly configure the local functional groups interacting with S15. It exhibits specific structural properties linked to the presence of the wobble pair U⋅G. This pair is a key element that cannot be substituted by other noncanonical pairs. This observation is in contrast with other studies showing that conserved U⋅G pairs involved in recognition of RNA sites can be successfully substituted by other unusual base pairs. For example, a G⋅U pair, which constitutes a specific protein determinant in the transcript of yeast ribosomal protein L32 (17), can be shifted to G⋅A, G⋅G, U⋅C, and C⋅C (but not to C⋅A) and still give appreciable protein binding in vitro. It should be noted though that the G⋅U pair of the L32 messenger closes an internal loop, which is not the case for the pseudoknot studied here. In the group I intron of Tetrahymena, a U⋅G wobble pair, located near the center of helix P1, is involved in splicing, and the only change giving efficient cleavage in vitro (about 50% of the wild-type activity) is a change to C⋅A (14). Although in this example, only RNA–RNA and not an RNA–protein interaction is involved, there seems to be some similarity in the mode of ligand binding (21, 23). In another case, C⋅A or G⋅A can replace G⋅U in tRNAAla and still allow efficient tRNA aminoacylation in vivo (24) but not in vitro (25), suggesting that, in vivo, aminoacylation efficiency might be affected by other extrinsic factors. In any case, the high specificity of S15 recognition observed in vivo for only U⋅G stresses the selectivity of the recognition process and suggests that a direct contact might be involved between this base pair and the protein S15.
Another possibility is that the exact U⋅G configuration is determined by the adjacent base pair located at the 5′ side of the U or that S15 interacts with determinants carried by the two base pairs of the motif. It has previously been shown that a U⋅G pair has a unique stacking configuration, whereby U⋅G bases stack on the bases to the 3′ side of the guanine and unstack from the bases to the 5′ side of the guanine (22, 23, 26, 27). In addition, NMR data on the U⋅G pair embedded in the P1 helix from group I self-splicing introns show that not only the positions of functional groups of the bases but also the local strand conformations are affected by U⋅G pairs and that these new structural features might be involved in recognition (23). Thus, in addition to the stacking discussed above, an undertwist of the U strand has been shown to be associated with an overwinding of the G strand to the 3′ side of guanine. This means that functional groups like the 2′OH of the U⋅G and groups from adjacent pairs might then adopt a specific configuration dictated by the sequence context. This hypothesis is best illustrated by the comparison between tRNAAla and the 5′ splice site of group I intron. In tRNAAla, purine nucleotides are located at either side of the guanine, instead of pyrimidines, as in the P1 helix of self-splicing introns. Several structural differences are observed by NMR between the U⋅G sites of these two RNAs. In tRNAAla, base stacking is limited (28), whereas it is extensive in the P1 helix so that variations in the helical twist across the base pair are small (23, 28), and backbone angles, α and ζ, instead of being in the canonical gauche−/gauche+ conformation, are trans/trans.
The U(111)⋅G(9) wobble pair of helix I from the 5 S RNA of E. coli exhibits neighboring adjacent base pairs identical to those of the S15 pseudoknot (27). If the sequence context is a primary determinant of double helical geometry, it should be possible to deduce some information on the local structure of the S15 binding site from the 5 S structure. NMR analysis has shown that, in 5 S RNA, the wobble pair stacks with the pair to the 5′ side of U, whereas the bases of the pair 3′ to the U are completely unstacked. This suggests that the local structure might be similar to what is observed in the P1 helix of the self-splicing intron. However, local detailed RNA structure sufficient to reveal the precise conformation of the bases and of the sugar-phosphate backbone in this region is still lacking, and so the degree of similarity between helix 1 from 5 S RNA and the P1 helix has not been determined. Thus, the extent of local helix distortions induced by the G⋅U wobble pair in this base pair context is not known. Only the presence of a stacking between the U⋅G/C⋅G pairs in the pseudoknot can be deduced from these studies.
The fact that S15 can apparently distinguish between U⋅G and G⋅U in an identical base pair context might be interpreted as a functional discrimination between two different angular orientations of the amino exocyclic group of guanine (29). This observation would mean that S15 recognizes specifically a single atomic group carried by the wobble pair, suggesting that the recognition of the operator site by S15 is more likely dependent upon the presence of specific functional groups than upon local helix distortions induced by the G⋅U base pair in its base pair context. However, other explanations can be proposed. The importance of the C(−50)⋅G(−35) pair adjacent to the U⋅G pair indicates that either other functional groups carried by this pair are recognized or that the determinant carried by U⋅G must be properly positioned to be recognized. In fact, both aspects might be intermingled if modifications to the helix geometry reorientate the RNA determinants, facilitating contacts with specific groups of the S15 protein. Whether these determinants correspond only to the amino exocyclic group of guanine or also to 2′ hydroxyl groups (30, 31) or even to another factor (like a divalent metal ion bound near U⋅G) (32) remains to be demonstrated.
The other base pairs adjacent to the motif, as long as they are engaged in Watson-Crick pairing, have a very limited influence on this motif, as shown by the tolerance of complementary base substitutions in these positions. Even small structural perturbations, presumably induced by the presence of a noncanonical base pair, have practically no influence on S15 binding. Moreover, a deletion of the bulge base U(−53) has been shown to have no drastic effect (10). Thus, the size of the motif seems to be limited to only two base pairs. They form a quite specific structure, embedded in a stable helix, which cannot be modified without losing S15 binding. Consequently, a direct S15–RNA interaction at the U⋅G motif is strongly suggested and is fully supported by probing experiments with ethylnitrosourea, which showed that this region is shielded by S15 (13).
If the U⋅G motif is sufficient for S15 binding, a mutant unable to form the pseudoknot but still carrying the U⋅G motif should nevertheless be able to bind S15 and to induce autocontrol. As this is not the case (10, 13), one can imagine that another contact located in another part of the pseudoknot is necessary for S15 binding. In fact, backbone probing experiments show that, in addition to the U⋅G region, the upper edge of stem 2 and loop 1 are also shielded by S15. Although bases do not seem to be involved in this putative contact(s) as shown by mutational analysis (10), recognition of specific functional groups in the backbone might be involved. Another possibility would be that the protections observed correspond to nonspecific backbone interactions stabilizing the complex formed. This would imply that direct and indirect effects are intermingled in recognition and binding. In vitro chemical mutagenesis experiments should help to resolve this problem (21, 25).
In conclusion, the U⋅G motif is a major, if not unique, recognition determinant located in stem 1 of the pseudoknot. Because no change is tolerated at the U⋅G motif without a complete loss of autoregulation, a direct recognition is probably located in this area. The exocyclic amino group of guanosine, known to be projected in the minor groove, is very likely involved as well as contacts with ribose in the adjacent C(−50)⋅G(−35) base pair.
Acknowledgments
We are very grateful to C. Cachia for preparing the S15 protein and to J. Plumbridge for constructive comments on the manuscript. This work was supported by grants from Centre National de la Recherche Scientifique. L.B. was supported by a doctoral grant from the Ministère de l’Enseignement Supérieur et de la Recherche.
References
- 1.Wang C, Le S-Y, Ali N, Siddiqui A. RNA. 1995;1:526–537. [PMC free article] [PubMed] [Google Scholar]
- 2.Wills N M, Gesteland R F, Atkins J F. EMBO J. 1994;13:4137–4144. doi: 10.1002/j.1460-2075.1994.tb06731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen X, Chamorro M, Lee S I, Shen L X, Vines J V, Tinoco I J, Varmus H E. EMBO J. 1995;14:842–852. doi: 10.1002/j.1460-2075.1995.tb07062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Takamatsu N, Watanabe Y, Meshi T, Okada Y. J Virol. 1990;64:3686–3693. doi: 10.1128/jvi.64.8.3686-3693.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shamoo Y, Tam A, Konigsberg W H, Williams K R. J Mol Biol. 1993;232:89–104. doi: 10.1006/jmbi.1993.1372. [DOI] [PubMed] [Google Scholar]
- 6.Tang C K, Draper D E. Cell. 1989;57:531–536. doi: 10.1016/0092-8674(89)90123-2. [DOI] [PubMed] [Google Scholar]
- 7.Ten Dam E B, Verlaan P W G, Pleij C W A. RNA. 1995;1:146–154. [PMC free article] [PubMed] [Google Scholar]
- 8.Asano K, Kato A, Moriwaki H, Hama C, Shiba K, Mizobuchi K. J Biol Chem. 1991;266:3774–3781. [PubMed] [Google Scholar]
- 9.Portier C, Dondon L, Grunberg-Manago M. J Mol Biol. 1990;211:407–414. doi: 10.1016/0022-2836(90)90361-O. [DOI] [PubMed] [Google Scholar]
- 10.Bénard L, Philippe C, Dondon L, Grunberg-Manago M, Ehresmann B, Ehresmann C, Portier C. Mol Microbiol. 1994;14:31–40. doi: 10.1111/j.1365-2958.1994.tb01264.x. [DOI] [PubMed] [Google Scholar]
- 11.Philippe C, Eyermann F, Bénard L, Portier C, Ehresmann B, Ehresmann C. Proc Natl Acad Sci USA. 1993;90:4394–4398. doi: 10.1073/pnas.90.10.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Philippe C, Portier C, Mougel M, Grunberg-Manago M, Ebel J P, Ehresmann B, Ehresmann C. J Mol Biol. 1990;211:415–426. doi: 10.1016/0022-2836(90)90362-P. [DOI] [PubMed] [Google Scholar]
- 13.Philippe C, Bénard L, Portier C, Westhof E, Ehresmann B, Ehresmann C. Nucleic Acids Res. 1995;23:18–28. doi: 10.1093/nar/23.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doudna J A, Cormack B P, Szostak J W. Proc Natl Acad Sci USA. 1989;86:7402–7406. doi: 10.1073/pnas.86.19.7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McClain W H, Foss K. Science. 1988;240:793–796. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- 16.Hou Y-M, Schimmel P. Nature (London) 1988;333:140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- 17.White S A, Li H. RNA. 1996;2:226–234. [PMC free article] [PubMed] [Google Scholar]
- 18.Nakamaye K L, Eckstein F. Nucleic Acids Res. 1986;78:9679–9698. doi: 10.1093/nar/14.24.9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunkel T A. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sänger W. In: Principles of Nucleic Acid Structure. Cantor C R, editor. Heidelberg: Springer; 1984. pp. 116–158. [Google Scholar]
- 21.Strobel S A, Cech T R. Science. 1995;267:675–678. doi: 10.1126/science.7839142. [DOI] [PubMed] [Google Scholar]
- 22.van Knippenberg P H, Formenoy L J, Heus H A. Biochim Biophys Acta. 1990;1050:14–17. doi: 10.1016/0167-4781(90)90134-n. [DOI] [PubMed] [Google Scholar]
- 23.Allain F H-T, Varani G. J Mol Biol. 1995;250:333–353. doi: 10.1006/jmbi.1995.0381. [DOI] [PubMed] [Google Scholar]
- 24.Gabriel K, Schneider J, McClain H. Science. 1996;271:195–197. doi: 10.1126/science.271.5246.195. [DOI] [PubMed] [Google Scholar]
- 25.Beuning P J, Yang F, Schimmel P, Musier-Forsyth K. Proc Natl Acad Sci USA. 1997;94:10150–10154. doi: 10.1073/pnas.94.19.10150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mizuno H, Sundaralingam M. Nucleic Acids Res. 1978;5:4451–4461. doi: 10.1093/nar/5.11.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White S A, Nilges M, Huang A, Brünger A T, Moore P B. Biochemistry. 1992;31:1610–1621. doi: 10.1021/bi00121a005. [DOI] [PubMed] [Google Scholar]
- 28.Ramos A, Varani G. Nucleic Acids Res. 1997;25:2083–2090. doi: 10.1093/nar/25.11.2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frugier M, Schimmel P. Proc Natl Acad Sci USA. 1997;94:11291–11294. doi: 10.1073/pnas.94.21.11291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holbrook S R, Cheong C, Tinoco I, Kim S-H. Nature (London) 1991;353:579–581. doi: 10.1038/353579a0. [DOI] [PubMed] [Google Scholar]
- 31.Musier-Forsyth K, Schimmel P. Nature (London) 1992;357:513–515. doi: 10.1038/357513a0. [DOI] [PubMed] [Google Scholar]
- 32.Allain F H-T, Varani G. Nucleic Acids Res. 1995;23:341–350. doi: 10.1093/nar/23.3.341. [DOI] [PMC free article] [PubMed] [Google Scholar]