Abstract
Recent studies of developmental biology have shown that the genes controlling phenotypic characters expressed in the early stage of development are highly conserved and that recent evolutionary changes have occurred primarily in the characters expressed in later stages of development. Even the genes controlling the latter characters are generally conserved, but there is a large component of neutral or nearly neutral genetic variation within and between closely related species. Phenotypic evolution occurs primarily by mutation of genes that interact with one another in the developmental process. The enormous amount of phenotypic diversity among different phyla or classes of organisms is a product of accumulation of novel mutations and their conservation that have facilitated adaptation to different environments. Novel mutations may be incorporated into the genome by natural selection (elimination of preexisting genotypes) or by random processes such as genetic and genomic drift. However, once the mutations are incorporated into the genome, they may generate developmental constraints that will affect the future direction of phenotypic evolution. It appears that the driving force of phenotypic evolution is mutation, and natural selection is of secondary importance.
For the last six decades, the dominant theory of evolution has been neo-Darwinism, which was developed by the three founders of theoretical population genetics, Fisher (1), Wright (2), and Haldane (3), and was later supported by various evolutionists (4–9). Neo-Darwinism asserts that natural selection is the driving force of evolution, and mutation merely provides raw genetic materials with which natural selection produces novel characters. This view is based on the argument that natural selection enhances the frequencies of advantageous alleles at many loci and makes it easy to recombine them into a single individual and produce a novel character, especially in the presence of gene interaction (1–3). By following this principle, evolutionary biologists have developed various theories of natural selection to explain the evolution of sex (9), formation of new species (10), development of social life in insects (11), evolution of altruism (12), etc. In these studies, it is customary to assume that there is a sufficient amount of genetic variation within populations, and therefore what is necessary is to study how natural selection produces complex characters or complex ways of life.
In the last four decades, the study of molecular evolution has shown that a majority of amino acid substitutions in proteins are neutral or nearly neutral and that only a minority of the substitutions change protein function (13–18). It has also been shown that the major factor of evolution at the molecular level is mutation, including gene duplication and other genetic changes (15–17). However, most evolutionists still believe in neo-Darwinism with respect to phenotypic evolution and are not interested in neutral evolution (19–22). Mayr (23) stated that neutral mutations apparently occur at the molecular level, but because they do not affect phenotypic characters, they are of little interest to evolutionists. In this respect, it is interesting to note that even Kimura (15), protagonist of the neutral theory of molecular evolution, believed in neo-Darwinism with respect to phenotypic evolution. By contrast, Nei (17, 24, 25) argued that because phenotypic characters are ultimately controlled by DNA sequences, both molecular and phenotypic evolution must occur in similar ways. He also suggested that a considerable portion of morphological evolution is caused by neutral or nearly neutral mutations, and the driving force of evolution is mutation at both molecular and phenotypic levels. However, the evidence for supporting this argument was rather weak.
In recent years substantial progress has occurred in the study of the molecular basis of phenotypic evolution, so that we can examine the relative importance of mutation and selection in detail. In this article, I will first consider phenotypic evolution controlled by multigene families, because there is a large amount of interesting data, and the interpretation of new findings in this area is relatively simple. I will then discuss the evolutionary changes of protein-coding and regulatory regions of genes in relation to phenotypic evolution and their implications for the general theory of evolution.
Multigene Families and Phenotypic Evolution
Conservative and Divergent Evolution.
Recent genomic studies of model organisms have made it clear that the genomes of eukaryotes contain a large number of multigene families and that most physiological and morphological characters are controlled by multigene families (26, 27). Many multigene families are of ancient origin and are shared by animals, plants, and fungi. Good examples are homeobox genes that encode transcription factors controlling various aspects of morphogenesis. The genomes of animals and plants contain a large superfamily of homeobox genes, with >200 genes in the human and ≈80 genes in the flowering plant Arabidopsis thaliana. Animal homeobox genes can be divided into at least 49 families (28, 29). The most well studied is the HOX gene family that controls the anterior–posterior segmentation of the animal body. The homeodomains encoded by orthologous and paralogous HOX genes from different animals are known to have the same or very similar amino acid sequences (28, 30, 31). In general, the transcription factor genes involved in the early stages of development are highly conserved (26). This suggests that the early stages of development are controlled by the same or similar sets of genes in many different phyla or classes of organisms.
The highly conserved genes stay in the genome not because of a low mutation rate but because of a high degree of purifying selection. The degree of purifying selection can be measured by comparing the number of synonymous nucleotide substitutions per synonymous site (dS) and the number of nonsynonymous substitutions per nonsynonymous site (dN) under the assumption that dS represents the number of neutral mutations. In the presence of purifying selection, nonsynonymous nucleotide substitutions resulting in amino acid changes may be eliminated. We therefore expect that dN is smaller than dS, and the extent of purifying selection can be measured by 1 − dN/dS. When I applied this equation to the concatenated nucleotide sequences (2,340 codons) of the homeoboxes of the 39 pairs of human and mouse HOX genes, I obtained 1 − 0.001/0.313 = 0.997. This suggests that 99.7% of nonsynonymous mutations are eliminated by purifying selection in homeobox regions.
However, most proteins are not as conserved as HOX homeodomains, and the average dN/dS ratio obtained from 1,000 randomly chosen human and mouse genes is ≈0.15 (18). This means that, on average, ≈85% of nonsynonymous nucleotide mutations are deleterious, and only 15% are fixed in the population. Many genes that are involved in various physiological functions of adult individuals usually evolve with a higher rate of nonsynonymous substitution than HOX genes. Examples are immune systems genes such as Ig and MHC genes, which are for protecting the host from parasites (viruses, bacteria, etc.). These genes tend to evolve faster to avoid the attack from ever changing parasites. However, the rate of nucleotide substitution in these genes is still much lower than that of pseudogenes, which is often regarded as the neutral substitution rate (17). Despite this conservative nature of amino acid substitution, multigene families may evolve relatively fast because of the rapid change of the number of member genes.
Evolutionary Change of the Number of Gene Copies.
The number of genes contained in a genome is not necessarily correlated with the complexity of the organism in eukaryotes (32). However, the number of gene copies in a gene family tends to increase with increasing complexity of the organism or the character involved. For example, the number of gene copies in the HOX gene family is only 8 in fruitflies but 39 in mammals (Table 1). This increase is understandable, because vertebrates need more homeobox genes to develop complex morphological characters. A large-scale study of this problem was conducted for 1,219 superfamilies of genes from 38 eukaryotic species, and it was shown that the number of genes within each superfamily is generally correlated with the number of cell types of the organism (26).
Table 1.
Numbers of member genes of several homeobox gene families in animal species
| Gene family | Invertebrates |
Vertebrates |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Caenorhabditis elegans | Fruitfly | Tunicate | Puffer fish | Zebrafish | Frog | Mouse | Rat | Human | |
| NKX5 | 1 | 1 | 1 | 4 | 3 | 3 | 2 | 2 | 2 |
| DLX | 1 | 1 | 3 | 8 | 5 | 6 | 7 | 6 | 6 |
| CDX | 1 | 1 | 2 | 2 | 3 | 3 | 3 | 2 | 3 |
| HOX | 6 | 8 | 10 | 45 | 40 | 35 | 39 | 39 | 39 |
| PAX | 2 | 8 | 4 | 9 | 7 | 3 | 4 | 4 | 4 |
| POU | 3 | 5 | 2 | 16 | 13 | 15 | 14 | 13 | 16 |
| LIM | 7 | 7 | 7 | 21 | 14 | 12 | 12 | 11 | 12 |
Table is modified from Nam and Nei (29).
The increase of gene number is, of course, generally caused by gene duplication, but gene number sometimes decreases by gene deletion. Therefore, multigene families are generally subject to birth-and-death evolution (27, 33). In multigene families controlling physiological characters, variation in the number of gene copies among different species can be enormous. One of the most conspicuous is the variation of olfactory receptor (OR) genes among vertebrate species (Table 2). In this gene family, the number of functional OR genes is >1,000 in mice but 396 in humans. Interestingly, humans have more pseudogenes than mice, the proportion of pseudogenes being ≈55% in humans and 24% in mice. Dogs, which are supposed to have a good sense of smell, have 811 functional genes and 289 pseudogenes. However, the most notable organism in this respect is the chicken, which has only 82 functional genes but 478 pseudogenes.
Table 2.
Numbers of functional genes and pseudogenes in the sensory receptor and other multigene families of vertebrates
| Vertebrate | Olfactory receptor* |
Phermone receptor† |
Taste receptor‡ |
Ig§ |
||
|---|---|---|---|---|---|---|
| OR | V1R | T2R | VH | Vλ | Vκ | |
| Human | 396 (425)¶ | 5 (115)‖ | 25 (11) | 44 (60) | 31 (38) | 35 (43) |
| Mouse | 1,035 (356) | 187 (121) | 34 (7) | 97 (65) | 3 (0) | 80 (78) |
| Dog | 811 (289) | 8 (33) | 14 (4) | 43 (37) | 52 (64) | 16 (9) |
| Cow | 970 (1,159) | 40 (45) | 11 (13) | 11 (6) | 28 (30) | 9 (13) |
| Opossum | 1,188 (304) | 98 (30) | 25 (8) | 26 (6) | 48 (6) | 66 (49) |
| Chicken | 82 (476) | 0 (0) | 3 (0) | 1 (58) | 1 (25) | 0 (0) |
| Xenopus | 410 (478) | 21 (2) | 48 (5) | 39 (41) | 6 (1) | 71 (18) |
| Zebrafish | 102 (35) | 2 (0) | 4 (0) | 37 (10) | 33 (9)** | |
Y. Niimura and M. Nei (ref. 113 and unpublished work).
Shi and Zhang (114).
Shi and Zhang (115).
S. Das and M. Nei, unpublished work.
The numbers in parentheses indicate pseudogenes.
The V1R intact genes in humans are likely to be nonfunctional (112).
It is unclear whether these genes belong to the Vλ or Vκ genes family. Here, VH, Vλ, and Vκ stand for Ig heavy-chain variable, λ-chain variable, and κ-chain variable region genes, respectively.
Why do the numbers of functional genes and pseudogenes vary so much among vertebrate species? The obvious factor would be the requirement for a species to adapt to a particular environmental condition. For most vertebrate species, detection of millions of different odorants is crucial for their survival. Yet, animals living in different environments require different types and numbers of olfactory receptors (34). In some animals such as birds and primates, olfaction appears to be less important than in other terrestrial vertebrates because they are equipped with trichromatic color vision (35). For this reason, they appear to have smaller numbers of OR genes. However, dogs and cows, which have large numbers of functional OR genes, also possess large numbers of pseudogenes. In rats, it is known that even if up to 80% of glomeruli in the olfactory bulb are removed (OR genes knocked out), the individual still can live a normal life in the laboratory condition. Furthermore, Shepherd (36) pointed out the importance of processing of odor distinction in the brain, stating that although humans have a smaller number of OR genes, the proportion of brain concerned with olfaction is apparently greater in humans than in mice. If we consider these factors, variation in the number of functional OR genes among different species may not be directly related to the ability of olfaction required. This is particularly so in the presence of a large number of pseudogenes.
Great variation in the number of gene copies among vertebrate species is also observed with pheromone receptor, taste receptor, and Ig genes (Table 2). The reason for this variation is not always clear. However, it appears that the number of gene copies in these gene families was originally determined by their functional requirement, but after the copy number reached a required level, the number has fluctuated by random duplication and deletion of genes. We may call this event random genomic drift, in analogy with random genetic drift of allele frequencies in population genetics. This random genomic drift is apparently an important factor for the evolution of phenotypic characters. If the number of gene copies increases or decreases by chance for a group of individuals, these individuals may be able to adapt to a new environment. Genomic drift is not just confined to sensory receptor or immune systems genes but appears to be an important evolutionary mechanism for many multigene families. It is known that human populations harbor extensive polymorphisms of copy number of multigene families (37–39) and that many of these polymorphisms do not seem to affect the fitness of individuals even when they are caused by duplication of a genomic region containing ≈30 genes, as in the case of Ig variable region (Vκ) genes in humans (40). In plants, there is evidence that the types and numbers of genes in a genome are reshuffled extensively when polyploidization followed by diploidization occurs (41).
Multiple Signal Pathways and Genetic Networks.
So far, we have considered only DNA sequence conservation and genomic drift of multigene families. In general, a large number of different genes are involved in the development of phenotypic characters, and changes in the coordination of temporal and spatial expression of these genes in the developmental process play important roles in evolution. There are usually several signaling pathways for producing the same end character, and complex gene interaction occurs as a form of gene regulatory networks (42–44). The number of genes involved in these signaling pathways or genetic networks generally increases as the phenotypic character involved becomes more complex, and this increase in gene number is ultimately caused by gene duplication (45). For this reason, gene duplication is the fundamental process of generating complex organisms (26, 46–48).
In the past, it has been customary to treat each gene as a unit of evolution in population genetics. In reality, however, a large number of genes interact with one another temporally or spatially in the developmental process, and therefore the evolution of phenotypic characters should be studied by taking into account this gene interaction. If a character is controlled by a large number of interacting genes, it is possible that the genetic networks involved are robust and resistant to the effects of deleterious mutations (49). At the same time, the effects of advantageous mutations also may not be manifested significantly in a genetic network with many different developmental pathways. If this is the case, a large proportion of mutations may evolve in a more or less neutral fashion.
Evolution of Physiological Characters
Strictly speaking, the principle of evolution of physiological characters cannot be distinguished from that of morphological characters, because the formation of morphological characters depends on various physiological processes in development, and the function of physiological characters depends on the anatomy or morphology of the organism. However, it is convenient to treat the evolution of physiological and morphological characters separately, because the former characters are concerned primarily with adult life, and the latter are products of morphogenesis in the developmental stage. For example, the transportation of oxygen from the lungs to various tissues in vertebrates is carried out primarily by hemoglobin and myoglobin. Therefore, by examining the molecular structures and expression patterns of these proteins from different organisms, one can study the mechanism of evolution of oxygen transportation. By contrast, to understand the evolution of morphological characters, one must study the evolutionary change of morphogenesis, which depends on complicated molecular and cellular processes carried out by a large number of genes. In this section, we consider the roles of mutation and selection in the evolution of physiological characters.
Changes in the Protein-Coding Regions of Genes.
The study of molecular evolution started with interspecific comparison of protein molecules concerned with various physiological functions (e.g., hemoglobin, cytochrome c, and insulin). This type of study soon revealed that most amino acid substitutions occurring in structural proteins are more or less neutral (16), and the functional change of proteins is caused primarily by amino acid substitutions occurring in the active sites of proteins [supporting information (SI) Table 3]. This is a general principle of evolution of proteins controlling physiological characters (15, 17, 18). Because recent papers on this subject have been reviewed by Nei (18), I shall not repeat the review here. The only comment I would like to make is that Kimura's (14) definition of neutral mutations (2Ns < 1, where N is the effective population size, and s is the selection coefficient for the mutant allele) is too strict to deal with long-term evolution, and, therefore, a more realistic definition based on functional change of genes by Nei (18) will be used in this paper. (He also proposed a more reasonable form of statistical definition of neutrality, which is given by for a reasonably large N or approximately −0.001 ≤ s ≤ 0.001 for N ≈ 106.)
Changes in the Regulatory Regions of Genes.
However, the evolution of physiological characters is also affected by mutational changes of the regulatory regions of genes that include promoters and enhancers surrounding the coding regions of genes. The β globin gene family in humans is known to consist of a cluster of duplicate genes ε, γA, γG, δ, and β (50). The gene ε is expressed in the early embryonic stage, γA and γG are expressed in fetal liver, and δ and β are expressed in adult individuals. The expression of these genes is controlled by the locus control region (LCR) that exists in an upstream region of the gene cluster. The molecular components of this LCR interact with the regulatory region of each globin gene and determine the successive activation and suppression of expression of β-family genes in development (50). A similar LCR is believed to control the expression of HOX genes (51) and the expression of olfactory receptor genes (52).
How these complex systems of gene expression evolved is unclear. However, the regulatory region of each gene must have changed gradually as the number of duplicate genes in the cluster increased. In fact, the nucleotide sequence of a cis-regulatory element is not fixed but changes in the evolutionary process, although it is generally quite conserved. The amino acid sequence of the DNA-binding region of a transcription factor also appears to change with time (53). These changes in the regulatory elements and the DNA-binding regions of transcription factors must be responsible for the evolutionary change of gene expression pattern and, consequently, the evolutionary change of physiological characters.
If this is the case, one would expect that physiological characters are generally conserved in the evolutionary process. Theoretically, when changes in internal or external environments occur, they may change relatively quickly because of the mutations occurring at the cis-regulatory elements and the DNA-binding regions of transcription factors. However, for cis-elements to bind transcription factors properly, they must coevolve with a delicate balance. Therefore, the evolution of physiological characters is expected to be a slow process. Of course, it is possible that the nucleotide sequences of the regulatory region outside the cis-elements evolve in a neutral fashion. However, because the DNA sequences in cis-elements are generally conserved, the average rate of nucleotide substitution of the entire regulatory region is expected to be lower than the rate of synonymous substitution in the coding regions but higher than the rate of nonsynonymous substitution. This expectation has been borne out by actual data for a large number of genes, and the sequence variations within and between species in the regulatory regions are generally in conformity with the pattern of neutral evolution (54–58).
In this connection, I want to emphasize that any mutation would never be strictly neutral, because its function depends on other genes and environmental conditions. In this sense, any mutation can only be nearly neutral as was conceived by early molecular evolutionists (18).
Evolution of Morphological Characters
Any specific morphological characters or organs such as animal eyes, hearts, and limbs and plant flowers, etc. are products of complex processes of temporal and spatial expression of many interacting genes in development. Developmental biologists often study and compare the developmental processes of distantly related organisms such as humans, zebrafish, sea urchins, and fruitflies. These studies show that each organism is uniquely adapted to its environmental condition or lifestyle, and, therefore, natural selection appears to have played important roles in producing morphological characters (32, 42). However, to understand the mechanism of evolution of morphological characters, one should study the differences in morphogenesis of closely related species or polymorphic individuals within species. In this case, the number of genes involved is likely to be small, so that it would be easier to understand the evolutionary process of morphological characters.
Changes in the Protein-Coding Regions of Genes.
To explain the conspicuous morphological difference between humans and chimpanzees despite a small degree of amino acid differences, King and Wilson (59) suggested that morphological evolution occurs by mutations of regulatory genes rather than structural genes. This view has been accepted by many developmental biologists (44, 60). By contrast, Nei (ref. 17, chapter 14) proposed that morphological evolution occurs by a small proportion of major effect mutations whether they are structural or regulatory (major gene effect hypothesis). It is still premature to conclude which hypothesis is right, but there are increasing data indicating the importance of structural gene mutations in morphological evolution.
One of the commonly observed morphological variations within and between related species is that of pigmentation of the hair, skin, and eyes of mammals and birds (SI Table 3). Many mammalian polymorphisms of black coat color (caused by the pigment eumelanin) and reddish or yellowish color (caused by the pigment phaeomelanin) are controlled by proteins called melanocortin-1 receptor (MC1R) and Agouti (61, 62). The wild-type coat color of jaguars of the cat family is reddish or yellowish and is determined by phaeomelanin. However, there are mutant genotypes with black coat color. This color is dominant to the wild type and is caused by deletion of several nucleotides as well as amino acid substitutions in the MC1R and Agouti genes (63). Jaguars live in the jungles of Central and South America, and the selective advantage or disadvantage of the black form over the wild type is unclear (62). It is possible that the mutant black form has spread through the population largely by genetic drift. Note that small selective advantage or disadvantage is easily swamped by the fluctuation of progeny size (18).
However, there are cases in which coat color is clearly related to the adaptation of organisms. In the Pinacate region of southwest Arizona, the rock pocket mouse, Chaetodipus intermedius, inhabits both dark and sandy rocky areas of the region. Dark areas have been formed by laval flow from a volcanic eruption that occurred >1 million years ago (Mya). Rock pocket mice are generally light-colored, but in the laval areas dark-colored individuals are observed. Nachman et al. (64) showed that there are four amino acid differences in MC1R between dark-colored and light-colored individuals in this region. Because dark-colored mice were derived from light-colored mice by mutation, the former were apparently adapted to the dark environment to avoid the attack from predators such as birds and large mammals. Similar adaptation to new environments caused by a single amino acid substitution in MC1R has been reported in the beach mouse, Peromyscus polionatus, in Florida (65). These examples suggest that new mutations are responsible for adaptation (preadaptation) to new environments and they have spread through the population by natural selection (elimination of previous genotypes). The importance of changes of protein sequences has also been reported for the HOX genes determining body segmentation of insects (66, 67), the Vrs1 transcription factor controlling the six-rowed spike in barley (68), and others (SI Table 3).
These examples show that morphological characters can be changed by a few amino acid substitutions, but it should be noted that, as in the case of physiological characters, most amino acid substitutions do not affect them appreciably. In the case of MC1R, there are 63 aa differences (of 315 shared sites compared) between wild-type mice and wild-type rock pocket mice, but the two species have essentially the same coat color, indicating that only a few specific mutations can change coat color. This observation supports the major gene effect hypothesis (17).
Changes in the Regulatory Regions of Genes.
Generally speaking, the genetic basis of morphological evolution is more complicated than that of physiological characters. Darwin's finches, consisting of 14 species, in the Galapagos Islands are often used as a textbook example of adaptive radiation of morphological characters. One character that has been studied well is the beak shape of the birds living on different islands. Several species of the finches eat insects and flowers of cactuses, whereas some others feed on seeds dropped on the ground. Cactus finches generally have long and pointed beaks, whereas ground finches have broad and thick beaks used for crushing seeds. Abzhanov et al. (69) found that there is a high correlation between the extent of beak breadth and the expression level of bone morphogenic protein, BMP4, in the frontal part of beak in the embryonic stage. Later, they searched for other genes affecting the beak shape and showed that calmodulin (CaM), a protein involved in mediating calcium signaling, is expressed at higher levels in the long and pointed beak of cactus finches than in the more broad beaks of ground finches (70). Therefore, it appears that the breadth and length of finch's beaks are controlled primarily by the expression levels of genes Bmp4 and CaM, respectively. Darwin's finches are believed to have originated from the finches in South or Central America ≈2 Mya (71) through a bottleneck of population size. It is therefore likely that the beak shape of the finches evolved by new regulatory mutations and natural selection that occurred during the last 2 million years. In this case, many different regulatory mutations appear to have occurred, because there is continuous variation in the beak shape and the expression levels of BMP4 and calmodulin among different species of Darwin's finches.
Another example of rapid morphological evolution by regulatory mutations is that of freshwater stickleback fish living in lakes near the northern Atlantic and Pacific. They were apparently derived from the oceanic marine sticklebacks ≈12,000 years ago when glaciation started to retreat. Marine sticklebacks have relatively long pelvic (rear) fins, but the fins are almost absent or substantially reduced in freshwater sticklebacks. It has been shown that the presence of pelvic fins is associated with a high level of expression of transcription factor gene, Pitx1, in the pelvic region of the embryo (72). By contrast, freshwater sticklebacks showed no or low levels of expression of the gene. Study of the PITX1 proteins from marine and freshwater sticklebacks showed that there were no amino acid differences between them. From these observations, it was concluded that the formation of pelvic fins is initiated by the expression of Pitx1, and the evolutionary change of the regulatory region of the Pitx1 gene is responsible for the reduction of pelvic fins. There are many other examples of cis-regulatory mutations that have generated morphological changes (73). Therefore, this form of mutation seems to play important roles in phenotypic evolution. Many developmental biologists seem to believe that cis-regulatory mutations are more important than the mutations in coding regions of genes, because new morphological characters are often associated with changes in the expression level of genes rather than changes in the amino acid sequences encoded.
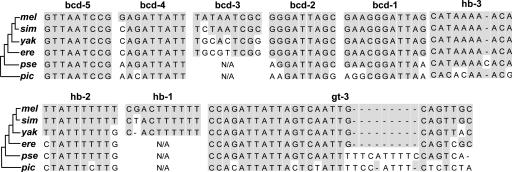
A number of authors have studied the evolutionary change of gene regulatory systems using relatively closely related species. The Drosophila homeotic gene even-skipped (eve) is known to produce seven transverse stripes along the anterior–posterior axis of the early embryo. Expression of each of these stripes is regulated by >12 cis-elements in the enhancer (activator and repressor) region. Ludwig, Patel, and Kreitman (74) studied the tripe 2 enhancers of the eve gene from six different Drosophila species and showed that the cis-elements of this gene are generally highly conserved, but a few of them were absent in some species (Fig. 1). Furthermore, the number of nucleotide differences in each element increased as the genomic divergence between species increased. Nevertheless, when the genetic constructs of enhancers and coding regions from different species were examined, all of them showed essentially the same tripe 2 expression. A similar but more complicated evolutionary change of the regulatory region of a gene resulting in the same phenotype has been reported with respect to the mating type MATa and MATα genes in the ascomycete yeast lineages (75).
Fig. 1.
Nucleotide sequences of the cis-elements for five bicoid (bcd), three hunchback (hb), and one giant (gt) transcription factors in the regulatory region of the even skipped (eve) enhancer 2 gene in six Drosophila species. N/A, no homologous sequence identified. –, nucleotide deletion. mel, D. melanogaster; sim, D. simulans; yak, D. yakuba; ere, D. erecta; pse, D. pseudoobscura; pic, D. picticornis. Adapted from Ludwig et al. (74). The phylogenetic relationships of these species are shown at the left-hand side of the diagram.
These results indicate that most nucleotide substitutions in the regulatory region evolve in a more or less neutral fashion, similar to those in the protein-coding region. It is therefore possible that the evolutionary change of gene regulation is also controlled by major gene mutations.
Polymorphism in cis-Regulatory Regions and Gene Expression Level.
Mendelian geneticists have established that quantitative characters are controlled by a large number of genes (76, 77). This can be explained partly by the presence of a high degree of protein polymorphism observed by electrophoresis at many loci (24, 78). Recent studies have also shown that there is a large amount of variation in the level of gene expression among different alleles at a locus (79–83). In humans, it has been reported that, in 63% of the genes examined, the expression level of one allele is 2-fold or more greater than that of another allele (81).
These allelic differences of gene expression level as well as the amino acid differences detected by protein electrophoresis are likely to influence phenotypic characters. Probably for this reason, most quantitative characters in outbreeding populations contain a large amount of genetic variation, and artificial selection is almost always effective (5). Previously, many neo-Darwinians claimed that the genetic variation within species is maintained primarily by balancing selection such as overdominant selection (5, 8). If this is the case, the extent of nonsynonymous nucleotide diversity is expected to be higher than that of synonymous diversity as in the case of MHC loci (84). In reality, however, this type of genetic variation has been observed in only a small proportion of genes in diploid organisms (85).
It is generally more difficult to study the roles of mutation and natural selection in phenotypic evolution than in protein evolution, because phenotypic characters are usually controlled by many genes and affected by environmental factors. Khaitovich et al. (86–88) studied the evolutionary divergence of gene expression levels of >10,000 genes in several primate species using the microarray technique and showed that the extent of evolutionary divergence increased roughly in proportion to the time of divergence between species. They also showed that the extent of gene expression divergence is generally higher for the genes whose intraspecific variation is high than for the genes whose intraspecific variation is low. From these observations, Khaitovich et al. concluded that the evolutionary divergence in gene expression level has occurred in a more or less neutral fashion. In genes from tissues such as the testes, there were some deviations from the above general pattern, possibly because of positive selection, but they were rare. Similar results have been observed in fruitflies (88, 89), yeasts (90), fish (91), and others. Although the authors of these studies did not necessarily support neutral evolution, the results indicate that mutation is the driving force in gene expression evolution. Essentially the same evolutionary pattern has been observed with protein variation detected by electrophoresis (92, 93).
Evolution of Phenotypic Diversity.
If we compare different phyla or classes of organisms, we are deeply impressed with the enormous amount of phenotypic diversity. For example, sea urchin and starfish, which belong to different classes of the phylum Echinodermata and diverged >540 Mya, show strikingly different morphologies, and they are apparently well adapted to different lifestyles in different environments. However, studies of the early stage of embryonic development have shown that sea urchin and starfish have similar morphologies and developmental patterns, and there is a common form of gene regulatory network (GRN) consisting of approximately six transcription factor genes (94). This basic core of GRN is specific for the early development of echinoderms and has not changed for the last 540 million years. However, as the development proceeds, the GRN in each of the two species expands into a more complex form including a large number of genes for transcription factors, signaling proteins, and structural proteins. In this process of expansion of GRN, different genes are added in the two species so that their GRNs are gradually differentiated. This gradual differentiation of GRNs is responsible for the formation of the very different morphologies of sea urchin and starfish. The basic core of GRN is highly conserved, and any significant change of the core results in deformation of the organism. This is also true with the GRNs operating in successive developmental stages, but the extent of developmental constraint gradually becomes weaker as the development proceeds.
This property appears to apply to many different animal phyla, and new species in each phylum are generated by mutational change of GRNs in the final or near-final stages of development (94, 95). In fact, the evolution of eye spots in the wings of some butterflies or the evolutionary changes of the number and form of body segments in insects and vertebrates have occurred by modification of GRNs in late stages of development (43, 44, 96). In this view, the evolution of phenotypic diversity of different phyla has occurred by a continuous process of novel mutations and elimination of preexisting less-fit genotypes. Evolutionists have proposed various mechanisms by which evolution can occur so fast that enormous amounts of phenotypic diversity among organisms can be explained (1, 2, 97). In reality, evolution is an intrinsically slow process, and the current phenotypic diversity has been generated only because there has been a long evolutionary time, >3 billion years.
Theoretically, the evolutionary change of phenotypic characters can be generated by neutral or nearly neutral mutations that may be fixed in the population by chance. The observation that the pattern of variation of gene expression levels within and between closely related species is consistent with that of neutral evolution supports this idea. However, once these mutations are incorporated into the genome, they may generate new developmental constraints that affect the future direction of phenotypic evolution. This is particularly so when the environmental condition changes. Large random phenotypic evolution may also be generated when geological changes such as mass extinction and continental drift occur.
Prospective and Retrospective Views of Evolution
The teleological view of evolution has been out of fashion for more than a century. Yet, human minds appear to be susceptible to this view consciously or unconsciously. In the evolutionary literature, it is not uncommon to see such phrases as “making of Homo sapiens” and “faster evolution of humans than other primate species.” Using these phrases, investigators often discuss the evolution of complex organisms without considering the fact that many closely related species have become extinct in the past. For example, humans appear to have evolved a higher level of phenotypic complexity than chimpanzees after their divergence ≈6 Mya. We are therefore tempted to believe that the genes controlling phenotypic characters have been subjected to positive Darwinian selection more often in the human lineage than in the chimpanzee (98).
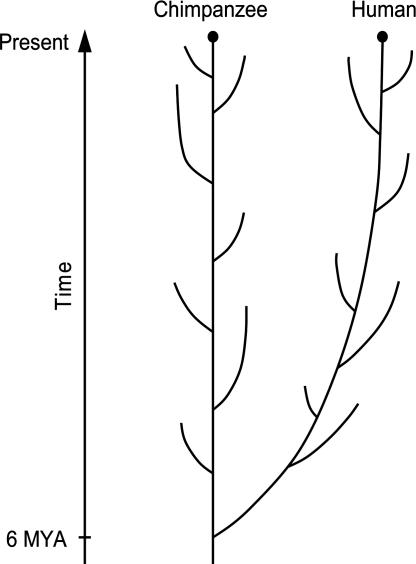
However, if we consider evolution as a forward process, this view becomes dubious. Fig. 2 shows a schematic representation of the evolution of humans and chimpanzees. Let us imagine that we can go back to the time when the two populations leading to humans and chimpanzees diverged ≈6 Mya and are asked whether one can predict which of the two populations is destined to produce humans later. Most evolutionists would say “it is impossible.” This would be true even if the two populations are genetically differentiated to a considerable extent. In other words, although we know that evolution occurs by mutation and natural selection, we cannot predict the outcome of evolution. Evolution occurs without purpose, and therefore it is intrinsically unpredictable.
Fig. 2.
Schematic representation of human and chimpanzee evolution. The branches of the human and chimpanzee lineages are the species or subspecies that have become extinct in the past. The retrospective view of evolution is depicted by the smooth lines aiming at the current morphologies of the two species. It is assumed that the morphology of chimpanzees is similar to that of the common ancestor of the two species, whereas the human morphology has changed substantially. In the prospective view, the future evolution is unpredictable, and therefore the evolutionary process might have been deviated considerably from the smooth lines.
By contrast, if we study human evolution retrospectively by using the knowledge of current humans and chimpanzees, we can always make a sensible story of evolution, although the story will be somewhat teleological because it is based on the final products of evolution. One such story would be that the population leading to the human lineage moved to a new habitat, whereas the chimpanzee lineage stayed in the original place. For this reason, many new adaptive mutations may have been fixed in the human lineage and these mutations led to the evolution of current H. sapiens. However, there is no reason to believe that a smaller number of adaptive mutations have been fixed in the chimpanzee lineage than in the human. The chimpanzee lineage also may have enhanced the adaptability to its own habitat, of which the climate and ecological community surely changed over geological time. In fact, this view is supported by microarray studies of gene expression levels between humans and chimpanzees (88). In this case, the types of mutations fixed in the two lineages would be different, but if the morphological differences are generated by a relatively small number of “major effect mutations” (17), it would be difficult to detect them by standard statistical methods. Experimental studies would be necessary.
Note also that any extant species represents only one evolutionary lineage surviving among many that appeared but became extinct in the past (Fig. 2). This is well documented in the case of human evolution (62), but it should be true with the chimpanzee lineage as well. The cause of extinction of many lineages is not well understood, but part of the reason must be the random change of genetic materials (genomic drift) and extrinsic environmental changes. If the H. sapiens lineage had become extinct and another lineage of Homo erectus had survived, this world would have been quite different. This indicates that evolution is opportunistic at the species level too.
Discussion and Perspectives
In this article, I have examined various types of molecular data concerning the evolution of phenotypic characters from the point of view of the selectionism/mutationism controversy. The main conclusions are as follows. (i) A multigene family concerned with basic developmental processes (e.g., HOX genes) is generally highly conserved, but the number of gene copies involved tends to increase with increasing complexity of the organism or the character. (ii) When a physiological character is controlled by a gene family, the number of gene copies may vary extensively among different organisms, and there are many pseudogenes in the genome. There are also extensive polymorphisms of copy number within species. This high degree of copy number variation is caused by genomic drift as well as by environmental factors. (iii) The evolutionary change of physiological and morphological characters occurs by mutational changes of the protein-coding and regulatory regions of genes. The genes controlling the characters expressed in the early stage of development are highly conserved, and evolutionary changes occur primarily in the characters expressed in later stages of development. At the nucleotide level, the driving force of phenotypic evolution is mutation, and there is a significant component of neutral or nearly neutral changes. (iv) The prospective view of evolution suggests that evolution occurs without purpose by mutation and adaptation to new environmental conditions, and therefore it is intrinsically unpredictable.
As mentioned in the introduction, a majority of current evolutionists believe in neo-Darwinism. In one of the most popular textbooks on evolution, Futuyma (ref. 20, p. 10) states that evolutionary change is a population process in which one genotype replaces other ones, and for this process to occur, mutation is quite ineffective because of its low rate of occurrence, whereas even the slightest intensity of natural selection can bring about substantial change in a realistic amount of time. He also states “Natural selection can account for both slight and great differences among species, and adaptations are traits that have been shaped by natural selection.” Although this type of statement is quite common in the evolutionary literature, it is obvious that any advantageous genotype is produced by mutation including all kinds of genetic changes. Natural selection occurs as a consequence of mutational production of different genotypes, and therefore it is not the fundamental cause of evolution.
Most molecular evolutionists are well aware of the importance of mutation in protein evolution. Yet, many investigators are trying to identify even the slightest trace of natural selection using various statistical methods (99–102). Using these methods, a number of authors have reported that a substantial proportion of amino acid substitutions are caused by positive Darwinian selection (103–106). However, the statistical methods used are based on many assumptions, which are not necessarily satisfied with actual data (18, 107, 108). Furthermore, their estimates of selection coefficients are often of the order of 10−6 (100, 106) and are unlikely to affect gene function (18). Note also that although these authors emphasized natural selection, they are actually estimating the proportion of mutations that are adaptive.
Historically, the word mutationism was used to refer to William Bateson's saltationism or similar ideas, in which natural selection plays little role. Later Morgan (109) presented a more reasonable form of mutationism taking into account the role of natural selection. His view was abstract and based on a few lines of speculative arguments. However, recent molecular studies of phenotypic evolution support the basic ideas of his view and have extended it to a more comprehensive view presented in this article. If the new form of mutation theory described here is right, even in its crudest form, more emphasis should be given on the roles of mutation in the study of evolution. Neo-Darwinians developed an impressive set of selection theories concerning the evolution of sex (110), altruism (12), new species (10), and others, without considering mutations that affect the characters involved such as male and female reproductive organs. These theories should be reexamined by studying the molecular basis of physiological and morphological components of the characters involved. It is also important to clarify the mechanism of formation of novel characters by mutation whether selection is involved or not. A group of molecular biologists are already working in this direction (111), but participation of population biologists and genomic scientists in this enterprise would speed up our understanding of phenotypic evolution.
Supplementary Material
Acknowledgments
I thank E. Holmes, J. Klein, Z. Lin, K. Makova, J. Nam, N. Nikolaidis, Y. Suzuki, N. Takahata, and J. Zhang for valuable comments. This study was supported by National Institutes of Health Grant GM020293.
Abbreviations
- GRN
gene regulatory network
- MC1R
melanocortin-1 receptor
- OR
olfactory receptor.
Footnotes
The author declares no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703349104/DC1.
References
- 1.Fisher RA. The Genetical Theory of Natural Selection. Oxford: Clarendon; 1930. [Google Scholar]
- 2.Wright S. Proc 6th Int Cong Genet. 1932;1:356–366. [Google Scholar]
- 3.Haldane JBS. The Causes of Evolution. London: Longmans, Green & Co; 1932. [Google Scholar]
- 4.Dobzhansky T. Genetics and the Origin of Species. New York: Columbia Univ Press; 1937. [Google Scholar]
- 5.Dobzhansky T. Genetics of the Evolutionary Process. New York: Columbia Univ Press; 1970. [Google Scholar]
- 6.Mayr E. Animal Species and Evolution. Cambridge, MA: Harvard Univ Press; 1963. [Google Scholar]
- 7.Mayr E. The Growth of Biological Thought. Cambridge, MA: Harvard Univ Press; 1982. [Google Scholar]
- 8.Ford EB. Ecological Genetics. 4th Ed. London: Chapman & Hall; 1975. [Google Scholar]
- 9.Maynard Smith J. Evolutionary Genetics. Oxford: Oxford Univ Press; 1989. [Google Scholar]
- 10.Coyne JA, Orr HA. Speciation. Sunderland, MA: Sinauer; 2004. [Google Scholar]
- 11.Wilson EO. Sociobiology. Cambridge, MA: Harvard Univ Press; 1975. [Google Scholar]
- 12.Hamilton WD. J Theor Biol. 1964;7:1–52. doi: 10.1016/0022-5193(64)90038-4. [DOI] [PubMed] [Google Scholar]
- 13.Zuckerkandl E, Pauling L. In: Evolving Genes and Proteins. Bryson V, Vogel HJ, editors. New York: Academic; 1965. pp. 97–166. [DOI] [PubMed] [Google Scholar]
- 14.Kimura M. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- 15.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, UK: Cambridge Univ Press; 1983. [Google Scholar]
- 16.King JL, Jukes TH. Science. 1969;164:788–798. doi: 10.1126/science.164.3881.788. [DOI] [PubMed] [Google Scholar]
- 17.Nei M. Molecular Evolutionary Genetics. New York: Columbia Univ Press; 1987. [Google Scholar]
- 18.Nei M. Mol Biol Evol. 2005;22:2318–2342. doi: 10.1093/molbev/msi242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dawkins R. The Blind Watchmaker. New York: Norton; 1987. [Google Scholar]
- 20.Futuyma DJ. Evolution. Sunderland, MA: Sinauer; 2005. [Google Scholar]
- 21.Grant PR, Grant BR. Science. 2006;313:224–226. doi: 10.1126/science.1128374. [DOI] [PubMed] [Google Scholar]
- 22.Ayala FJ. Proc Natl Acad Sci USA. 2007;104:8567–8573. doi: 10.1073/pnas.0701072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mayr E. What Evolution Is. New York: Basic Books; 2001. [Google Scholar]
- 24.Nei M. Molecular Population Genetics and Evolution. Amsterdam: North–Holland; 1975. [PubMed] [Google Scholar]
- 25.Nei M. In: Evolution of Genes and Proteins. Nei M, Koehn RK, editors. Sunderland, MA: Sinauer; 1983. pp. 165–190. [Google Scholar]
- 26.Vogel C, Chothia C. PLoS Comput Biol. 2006;2:e48. doi: 10.1371/journal.pcbi.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nei M, Rooney AP. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burglin TR. In: Encyclopedia of Molecular Cell Biology and Molecular Medicine. Meyers RA, editor. Weinheim, Germany: Wiley–VCH; 2005. pp. 179–222. [Google Scholar]
- 29.Nam J, Nei M. Mol Biol Evol. 2005;22:2386–2394. doi: 10.1093/molbev/msi229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Duboule D. Guidebook to the Homeobox Genes. New York: Oxford Univ Press; 1994. [Google Scholar]
- 31.Zhang J, Nei M. Genetics. 1996;142:295–303. doi: 10.1093/genetics/142.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carroll SB. The Making of the Fittest. New York: Norton; 2006. [Google Scholar]
- 33.Ota T, Nei M. Mol Biol Evol. 1994;11:469–482. doi: 10.1093/oxfordjournals.molbev.a040127. [DOI] [PubMed] [Google Scholar]
- 34.Firestein S. Nature. 2001;413:211–218. doi: 10.1038/35093026. [DOI] [PubMed] [Google Scholar]
- 35.Gilad Y, Wiebe V, Przeworski M, Lancet D, Paabo S. PLoS Biol. 2004;2:e5. doi: 10.1371/journal.pbio.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shepherd GM. PLoS Biol. 2004;2:e146. doi: 10.1371/journal.pbio.0020146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, Maner S, Massa H, Walker M, Chi M, et al. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 38.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharp AJ, Cheng Z, Eichler EE. Annu Rev Genomics Hum Genet. 2006;7:407–442. doi: 10.1146/annurev.genom.7.080505.115618. [DOI] [PubMed] [Google Scholar]
- 40.Schable KF, Zachau HG. Biol Chem Hoppe Seyler. 1993;374:1001–1022. [PubMed] [Google Scholar]
- 41.Adams KL, Wendel JF. Trends Genet. 2005;21:539–543. doi: 10.1016/j.tig.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 42.Wilkens SA. The Evolution of Developmental Pathways. Sunderland, MA: Sinauer; 2002. [Google Scholar]
- 43.Carroll SB, Grenier JK, Weatherbee SD. From DNA to Diversity. Oxford: Blackwell; 2005. [Google Scholar]
- 44.Davidson EH. The Regulatory Genome. San Diego: Academic; 2006. [Google Scholar]
- 45.Pires-daSilva A, Sommer RJ. Nat Rev Genet. 2002;4:39–49. doi: 10.1038/nrg977. [DOI] [PubMed] [Google Scholar]
- 46.Lewis EB. Cold Spring Harb Symp. 1951;16:159–174. doi: 10.1101/sqb.1951.016.01.014. [DOI] [PubMed] [Google Scholar]
- 47.Nei M. Nature. 1969;221:40–42. doi: 10.1038/221040a0. [DOI] [PubMed] [Google Scholar]
- 48.Ohno S. Evolution by Gene Duplication. Berlin: Springer; 1970. [Google Scholar]
- 49.Wagner A. Nat Genet. 2000;24:355–361. doi: 10.1038/74174. [DOI] [PubMed] [Google Scholar]
- 50.Wolpert L. Principles of Development. 3rd Ed. New York: Oxford Univ Press; 2007. [Google Scholar]
- 51.Lee AP, Koh EG, Tay A, Brenner S, Venkatesh B. Proc Natl Acad Sci USA. 2006;103:6994–6999. doi: 10.1073/pnas.0601492103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- 53.Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA. Mol Biol Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- 54.Purugganan MD. Mol Ecol. 2000;9:1451–1461. doi: 10.1046/j.1365-294x.2000.01016.x. [DOI] [PubMed] [Google Scholar]
- 55.Miyashita NT. Mol Biol Evol. 2001;18:164–171. doi: 10.1093/oxfordjournals.molbev.a003790. [DOI] [PubMed] [Google Scholar]
- 56.de Meaux J, Goebel U, Pop A, Mitchell-Olds T. Plant Cell. 2005;17:676–690. doi: 10.1105/tpc.104.027839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Meaux J, Pop A, Mitchell-Olds T. Genetics. 2006;174:2181–2202. doi: 10.1534/genetics.106.064543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Keightley PD, Lercher MJ, Eyre-Walker A. PLoS Biol. 2005;3:e42. doi: 10.1371/journal.pbio.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.King MC, Wilson AC. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 60.Carroll SB. PLoS Biol. 2005b;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bennett DC, Lamoreux ML. Pigment Cell Res. 2003;16:333–344. doi: 10.1034/j.1600-0749.2003.00067.x. [DOI] [PubMed] [Google Scholar]
- 62.Carroll SB. Endless Forms Most Beautiful. New York: Norton; 2005. [Google Scholar]
- 63.Eizirik E, Yuhki N, Johnson WE, Menotti-Raymond M, Hannah SS, O'Brien SJ. Curr Biol. 2003;13:448–453. doi: 10.1016/s0960-9822(03)00128-3. [DOI] [PubMed] [Google Scholar]
- 64.Nachman MW, Hoekstra HE, D'Agostino SL. Proc Natl Acad Sci USA. 2003;100:5268–5273. doi: 10.1073/pnas.0431157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. Science. 2006;313:101–104. doi: 10.1126/science.1126121. [DOI] [PubMed] [Google Scholar]
- 66.Galant R, Carroll SB. Nature. 2002;415:910–913. doi: 10.1038/nature717. [DOI] [PubMed] [Google Scholar]
- 67.Ronshaugen M, McGinnis N, McGinnis W. Nature. 2002;415:914–917. doi: 10.1038/nature716. [DOI] [PubMed] [Google Scholar]
- 68.Komatsuda T, Pourkheirandish M, He C, Azhaguvel P, Kanamori H, Perovic D, Stein N, Graner A, Wicker T, Tagiri A, et al. Proc Natl Acad Sci USA. 2007;104:1424–1429. doi: 10.1073/pnas.0608580104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- 70.Abzhanov A, Kuo WP, Hartmann C, Grant BR, Grant PR, Tabin CJ. Nature. 2006;442:563–567. doi: 10.1038/nature04843. [DOI] [PubMed] [Google Scholar]
- 71.Sato A, Tichy H, O'Huigin C, Grant PR, Grant BR, Klein J. Mol Biol Evol. 2001;18:299–311. doi: 10.1093/oxfordjournals.molbev.a003806. [DOI] [PubMed] [Google Scholar]
- 72.Shapiro MD, Marks ME, Peichel CL, Blackman BK, Nereng KS, Jonsson B, Schluter D, Kingsley DM. Nature. 2004;428:717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- 73.Wray GA. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 74.Ludwig MZ, Patel NH, Kreitman M. Development (Cambridge, UK) 1998;125:949–958. doi: 10.1242/dev.125.5.949. [DOI] [PubMed] [Google Scholar]
- 75.Tsong AE, Tuch BB, Li H, Johnson AD. Nature. 2006;443:415–420. doi: 10.1038/nature05099. [DOI] [PubMed] [Google Scholar]
- 76.Mather K. Biometrical Genetics. London: Methuen; 1949. [Google Scholar]
- 77.Falconer DS. Introduction to Quantitative Genetics. 2nd Ed. London: Longman; 1981. [Google Scholar]
- 78.Lewontin RC. The Genetic Basis of Evolutionary Change. New York: Columbia Univ Press; 1974. [Google Scholar]
- 79.Cavalieri D, Townsend JP, Hartl DL. Proc Natl Acad Sci USA. 2000;97:12369–12374. doi: 10.1073/pnas.210395297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brem RB, Yvert G, Clinton R, Kruglyak L. Science. 2002;296:752–755. doi: 10.1126/science.1069516. [DOI] [PubMed] [Google Scholar]
- 81.Rockman MV, Wray GA. Mol Biol Evol. 2002;19:1991–2004. doi: 10.1093/oxfordjournals.molbev.a004023. [DOI] [PubMed] [Google Scholar]
- 82.Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, et al. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- 83.Balhoff JP, Wray GA. Proc Natl Acad Sci USA. 2005;102:8591–8596. doi: 10.1073/pnas.0409638102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hughes AL, Nei M. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 85.Hughes AL. Adaptive Evolution of Genes and Genomes. New York: Oxford Univ Press; 1999. [Google Scholar]
- 86.Khaitovich P, Weiss G, Lachmann M, Hellmann I, Enard W, Muetzel B, Wirkner U, Ansorge W, Paabo S. PLoS Biol. 2004;2:e132. doi: 10.1371/journal.pbio.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Khaitovich P, Paabo S, Weiss G. Genetics. 2005;170:929–939. doi: 10.1534/genetics.104.037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Khaitovich P, Enard W, Lachmann M, Paabo S. Nat Rev Genet. 2006;7:693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
- 89.Rifkin SA, Kim J, White KP. Nat Genet. 2003;33:138–144. doi: 10.1038/ng1086. [DOI] [PubMed] [Google Scholar]
- 90.Fay JC, McCullough HL, Sniegowski PD, Eisen MB. Genome Biol. 2004;5:R26. doi: 10.1186/gb-2004-5-4-r26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Whitehead A, Crawford DL. Proc Natl Acad Sci USA. 2006;103:5425–5430. doi: 10.1073/pnas.0507648103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Chakraborty R, Fuerst PA, Nei M. Genetics. 1978;88:367–390. doi: 10.1093/genetics/88.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Skibinski DOF, Ward RD. Nature. 1982;298:490–492. [Google Scholar]
- 94.Davidson EH, Erwin DH. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 95.Gehring WJ. J Hered. 2005;96:171–184. doi: 10.1093/jhered/esi027. [DOI] [PubMed] [Google Scholar]
- 96.Brakefield PM, Gates J, Keys D, Kesbeke F, Winjgaarden PJ, Monteiro A, French V, Carroll SB. Nature. 1996;384:236–242. doi: 10.1038/384236a0. [DOI] [PubMed] [Google Scholar]
- 97.Muller HJ. Am Nat. 1932;68:118–138. [Google Scholar]
- 98.Vallender EJ, Lahn BT. Hum Mol Genet. 2004;13:245–254. doi: 10.1093/hmg/ddh253. [DOI] [PubMed] [Google Scholar]
- 99.Smith NG, Eyre-Walker A. Nature. 2002;415:1022–1024. doi: 10.1038/4151022a. [DOI] [PubMed] [Google Scholar]
- 100.Sawyer SA, Kulathinal RJ, Bustamante CD, Hartl DL. J Mol Evol. 2003;57(Suppl 1):S154–S164. doi: 10.1007/s00239-003-0022-3. [DOI] [PubMed] [Google Scholar]
- 101.Zhang J, Nielsen R, Yang Z. Mol Biol Evol. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- 102.Tang H, Wu CI. Mol Biol Evol. 2006;23:372–379. doi: 10.1093/molbev/msj043. [DOI] [PubMed] [Google Scholar]
- 103.Clark NL, Swanson WJ. PLoS Genet. 2005;1:e35. doi: 10.1371/journal.pgen.0010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Eyre-Walker A. Trends Ecol Evol. 2006;21:569–575. doi: 10.1016/j.tree.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 105.Gojobori J, Tang H, Akey JM, Wu CI. Proc Natl Acad Sci USA. 2007;104:3907–3912. doi: 10.1073/pnas.0605565104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sawyer SA, Parsch J, Zhang J, Hartl DL. Proc Natl Acad Sci USA. 2007;104:6504–6510. doi: 10.1073/pnas.0701572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hughes AL, Friedman R, Glenn NL. Curr Genomics. 2006;7:227–234. [Google Scholar]
- 108.Subramanian S, Kumar S. Mol Biol Evol. 2006;23:2283–2287. doi: 10.1093/molbev/msl123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Morgan TH. The Scientific Basis of Evolution. New York: Norton; 1932. [Google Scholar]
- 110.Michod RE, Levin BR. The Evolution of Sex: An Examination of Current Ideas. Sunderland, MA: Sinauer; 1987. [Google Scholar]
- 111.The ENCODE Project. Nature. 2007;447:799–815. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang J, Webb DM. Proc Natl Acad Sci USA. 2003;100:8337–8341. doi: 10.1073/pnas.1331721100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Niimura Y, Nei M. J Hum Genet. 2006;51:505–517. doi: 10.1007/s10038-006-0391-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shi P, Zhang J. Genome Res. 2007;17:166–174. doi: 10.1101/gr.6040007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shi P, Zhang J. Mol Biol Evol. 2006;23:292–300. doi: 10.1093/molbev/msj028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.