Abstract
The relationship between enzyme architecture and substrate specificity among archaeal pre-tRNA splicing endonucleases has been investigated more deeply, by using biochemical assays and model building. The enzyme from Archeoglobus fulgidus (AF) is particularly interesting: it cleaves the bulge–helix–bulge target without requiring the mature tRNA domain, but, when the target is a bulge–helix–loop, the mature domain is required. A model of AF based on its electrostatic potential shows three polar patches interacting with the pre-tRNA substrate. A simple deletion mutant of the AF endonuclease lacking two of the three polar patches no longer cleaves the bulge–helix–loop substrate with or without the mature domain. This single deletion shows a possible path for the evolution of eukaryal splicing endonucleases from the archaeal enzyme.
Keywords: molecular evolution, RNA–protein interactions, tRNA endonucleases
Accuracy in tRNA splicing is essential for the formation of functional tRNAs and therefore for gene expression. In Bacteria, pre-tRNA introns are self-splicing group I introns, and the splicing mechanism is autocatalytic. In Eukarya, tRNA introns are small and invariably interrupt the anticodon loop one base 3′ to the anticodon. In Archaea, the introns are also small and often reside in the same location as eukaryal tRNA introns, but not always (1, 2). In both Eukarya and Archaea, the specificity for recognition of the pre-tRNA resides in the endonucleases (3–5). These enzymes remove the intron by making two independent endonucleolytic cleavages. It is generally accepted that the archaeal enzymes act without any reference to the mature domain but instead recognize specific structures that define the intron–exon boundaries (6–8). By contrast, the eukaryal enzyme normally acts in a mature-domain-dependent mode (3, 4).
The exact way in which the eukaryal endonucleases recognize the precursors has yet to be determined, but many RNA–protein interactions are presumably required. Previous studies have shown the role of the three-dimensional structure and of specific invariant bases of the mature domain in recognition and binding by the enzyme (3, 4). The importance of structural regularities in the precursors suggested an anchor-and-measure mechanism in which the endonuclease binds to one or more reference sites in the mature domain, which are common to all pre-tRNAs, and measures the distance to the equivalently positioned intron–exon junctions. This hypothesis was supported by experiments that involved the engineering of changes in the distance that separate the mature domain from the splice sites. These alterations changed the size of the intron in a predictable way: the insertion of 1 bp in the anticodon stem increased the size of the intron by two bases, one at each end, and the insertion of 2 bp in the anticodon stem increased the size of the intron by 4 nt (3, 4). These striking results, which had been observed in specific instances, were generalized and taken to mean that there are no important recognition elements at the intron–exon boundaries and that the enzyme binds only to the mature domain. Using a large number of constructs and in vitro selection techniques, we later found evidence that pre-tRNA introns also contribute to the specificity of splice site recognition (9, 10). There are also positions in the mature domain that participate in the splicing reaction, although they are not occupied by the same bases in all pre-tRNAs. We called these positions “cardinal positions” (CPs) (9). The eukaryal endonuclease is able to cut a precursor at the 3′ site if a base in the pre-tRNA single-stranded loop of the intron is allowed to form an anticodon-intron (A-I) base pair with the base of the 5′ exon situated at the position immediately following the anticodon stem (position 32 in yeast pre-tRNAPhe). A recently observed “A-minor” interaction between the first 3′ exon nucleotide and the A-I base pair provides the structural basis for their interdependence (11). These observations suggested that important recognition elements also lie at the intron–exon boundaries.
The mature domain dominates the scenario of eukaryal tRNA splicing. There is, however, a single striking exception: a specific structure consisting of two 3-nt bulges separated by a 4-bp helix, the so called bulge–helix–bulge (BHB) motif, has been shown to be recognized and cleaved in vitro by the eukaryal enzyme without any reference to the mature domain (12, 13). This finding presents interesting evolutionary implications. The BHB pre-tRNA is a universal substrate and is also cleaved by the archaeal enzymes. As a matter of fact, many intron-containing archaeal pre-tRNAs are characterized by a BHB constructed from the intron and from exonic sequences. The archaeal endonucleases, in general, remove the intron in a mature-domain-independent mode (14). It is because of this independence that the intron in archaeal pre-tRNAs can be located in different positions relative to the mature domain. Several archaeal pre-tRNAs contain relaxed forms of the BHB motif, one being the bulge–helix–loop (BHL), that consists of a single 3-nt bulge and an internal loop, separated by a 4-bp-helix (15). Some archaeal enzymes, such as the enzyme from Sulfolobus solfataricus (SS), can cleave the BHL substrate in a mature-domain-independent way. We reasoned, however, that, because the relaxed forms like the BHL resemble the motif found in most yeast pre-tRNAs (presenting, as a rule, introns located one base 3′ to the anticodon), they could represent a preliminary stage in the advent of the dominance of the tRNA splicing by the mature domain (15).
At a certain stage, an enzyme could have evolved the capability of cleaving the BHL substrate only in mature-domain-dependent mode, while still capable of cleaving the BHB substrate in a mature-domain-independent mode. Such an enzyme would be characterized by surfaces providing specific interaction with the mature domain. In this article, we describe results of in vitro cleavage assays performed using a variety of substrate variants and the endonuclease from the archaeon Archeoglobus fulgidus (AF). This enzyme cleaves in a mature-domain-independent mode the BHB substrate but not the BHL substrate. In this respect, the AF enzyme resembles the eukaryal enzymes. An electrostatic potential calculation (16) applied to the AF enzyme structure permits the visualization, on the face of the protein that interacts with pre-tRNA, of positively charged patches, presumably involved in the interaction with specific parts of the substrate, including the mature domain.
Results
The AF Enzyme Cleaves the BHL Motif Substrate in a Mature-Domain-Dependent Mode.
We reasoned that, if in the archaeal world tRNA endonucleases exist that interact with the mature domain following a strategy similar to that adopted by the eukaryal enzymes, then in the corresponding organism there should exist pre-tRNAs presenting the intron located only at the canonical position in the anticodon loop region between nucleotides 37 and 38. Interestingly, two independent bioinformatic analyses have reached the same conclusion: the Euryarchaea encoding a homodimeric endonuclease present pre-tRNAs with the intron located exclusively at this canonical position (14). Do homodimeric enzymes, like the AF endonuclease, occupy a critical position in the evolutionary route from the archaeal to the eukaryal system?
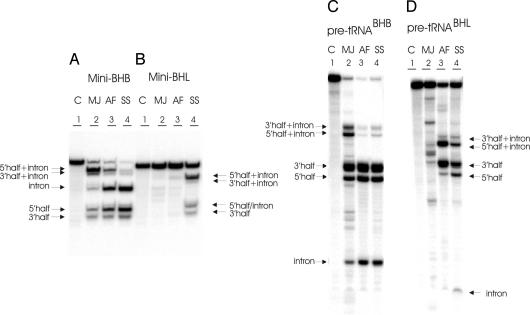
To investigate this possibility, we used two different pre-tRNA substrates for the in vitro cleavage assay. The first substrate pre-tRNABHB presents a BHB motif whereas the second pre-tRNABHL presents a BHL (Fig. 1 A and C). Each substrate was incubated with three different pre-tRNA endonucleases from Archaea, representing each of the three different architectures: the homotetramer (α4) of Methanocaldococcus jannaschii (MJ), the homodimer (α2) of AF, and the heterotetramer (α2 β2) of SS. All of the enzymes cleaved the BHB substrate (Fig. 2C). On the contrary, there are differences in the processing of the BHL substrate. The homotetrameric enzyme from MJ did not cleave the BHL. The heterotetrameric enzyme from SS and the homodimeric enzyme from AF cleaved the BHL substrate, which is, therefore, stable at the assay conditions (65°C) (Fig. 2D). These findings confirm results previously obtained by us (15). It seems, therefore, that the enzyme specificity for the BHB and BHL substrates is the result of adaptation of similar active sites, because the enzymes capable of processing the BHL pre-tRNA are also capable of processing the BHB. The BHB is a universal substrate. The question we address next is whether the ability to cleave the relaxed motifs is a prelude, in the archaeal world, to the advent of the dominance by the mature domain in the Eukarya.
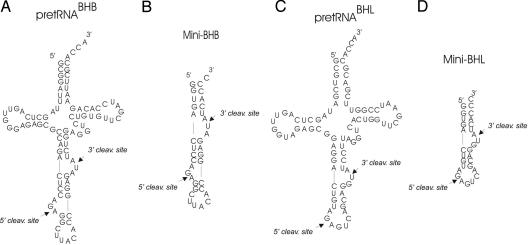
Fig. 1.
Substrates for the in vitro cleavage assays. (A) PretRNABHB consists of two regions derived from yeast pre-tRNAPHE (nucleotides 1–31 and 38–76), joined by a 25-nt insert that corresponds to the BHB motif of archaeal tRNATrp. (B) Mini-BHB is the BHB motif of pre-tRNABHB. (C) PretRNABHL corresponds to pre-tRNATyr from Caenorhabditis elegans. The synthetic substrate presents a residue change at the 5′ terminus (C to G) required for T7 transcription and a corresponding change (G to C) in the complementary strand. (D) Mini-BHL is the BHL motif of pre-tRNABHL. The base substitutions in the mini substrates were introduced to optimize T7 RNA polymerase transcription (5).
Fig. 2.
In vitro cleavage of BHB- and BHL-containing substrates. (A) The mini–BHB substrate was incubated with three different enzymes. (B) The mini–BHL substrate was incubated with three different enzymes. (C) Pre-tRNABHB was incubated with three different enzymes. (D) PretRNABHL was incubated with three different enzymes. The conditions of the reactions have been reported (15). The cleavage products were analyzed by electrophoresis on 10% polyacrylamide gel containing 29:1 monomer to bis and 8 M urea, followed by autoradiography. The identification of the reaction products is indicated. Lane 1 contains the control (C, no enzyme added). Lanes 2–4 show the products after incubation with the endonucleases from MJ, AF, and SS, respectively. The 2/3 molecules are produced by single cleavage.
We asked first whether each of the three archaeal enzymes, representing the different architectures (17), can operate independently of the mature domain on minisubstrates that correspond respectively to the BHB (mini-BHB) (Fig. 1B) or to the BHL (mini-BHL) (Fig. 1D) and lack the mature domain. Fig. 2A shows that the intron was correctly removed from the mini–BHB by all three enzymes whereas the mini-BHL was effectively cleaved only by the heterotetrameric SS endonuclease (Fig. 2B). The active sites of the enzyme from SS therefore allow the BHL to perform autonomously. We expected that the homotetrameric enzyme from MJ would not cleave this substrate. But to our surprise, the homodimeric AF enzyme also failed to cleave the mini-BHL (Fig. 2B). Inefficient cleavage of the 3′ site could occasionally be observed (see Fig. 5B). This striking observation is in contrast to our observation for the full-length pre-tRNABHL (Fig. 2D). These results suggest that the homodimeric enzyme of AF, like the eukaryal enzymes, requires the mature domain for cleavage. These observations led us to attempt to visualize a footprint of the full-length pre-tRNA, on the surface of the AF enzyme, a footprint of the full-length pre-tRNA.
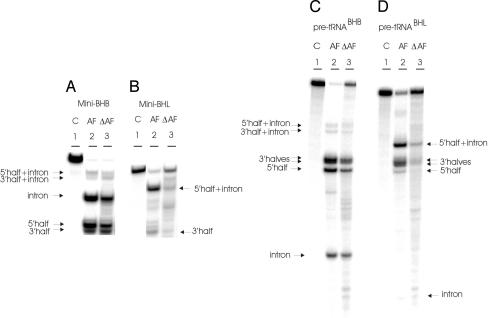
Fig. 5.
In vitro cleavage of BHB and BHL containing substrates by the ΔAF enzyme. (A) The mini-BHB substrate was incubated with the AF and ΔAF enzymes. (B) The mini-BHL substrate was incubated with the AF and ΔAF enzymes. (C) The pre-tRNABHB substrate was incubated with the AF and ΔAF enzymes. (D) The pre-tRNABHL substrate was incubated with the AF and ΔAF enzymes. The conditions of the reactions have been reported (12). The cleavage products were analyzed by electrophoresis on 10% polyacrylamide gel containing 29:1 monomer to bis and 8 M urea, followed by autoradiography. The identification of the reaction products is indicated. Lane 1 contains the control (C, no enzyme added). Lanes 2 and 3 show the products after incubation with the endonucleases from AF and ΔAF, respectively. The 2/3 molecules are produced by single cleavage.
The AF Enzyme Presents RNA Binding Sites Involved in Specific Interactions with Different tRNA Domains.
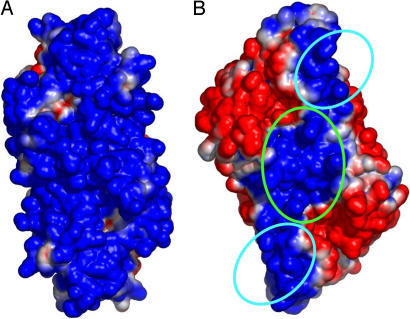
We first calculated an electrostatic potential using the MJ endonuclease structure determined by x-ray diffraction (18). This enzyme presents a face interacting with tRNA that is uniformly positively charged (Fig. 3A). Instead, the AF enzyme surface charge, calculated in the same way, omitting the RNA substrate, clearly shows three distinct positively charged polar patches (11) (Fig. 3B). The central patch is formed by residues belonging to the two catalytic repeats; it includes the two catalytic sites and is directly involved in the interaction with the BHB. The two flanking patches are identical and symmetrically placed with respect to one another. These two peripheral patches are formed by 11 basic residues located in the N-terminal domain of the AF protein. Four belong to helix 1 (Lys-12, Lys-14, Arg-18, and Arg-19); two belong to to helix 4 (Arg-74 and Arg-76); and two are on loop 7 (Lys-90 and Lys-91) (11).
Fig. 3.
Electrostatic surface potentials of MJ (A) and AF (B) endonucleases. Positively charged regions are shown in blue, the neutral regions are shown in white, and the negatively charged regions are shown in red. The green circle encompasses the catalytic site, whereas the cyan circles enclose the two putative sites recognizing the tRNA mature domain. The figure was generated with PyMOL (http://pymol.sourceforge.net), and the electrostatic properties were generated by using the APBS (Adaptive Poisson-Boltzman Solver) software package (16).
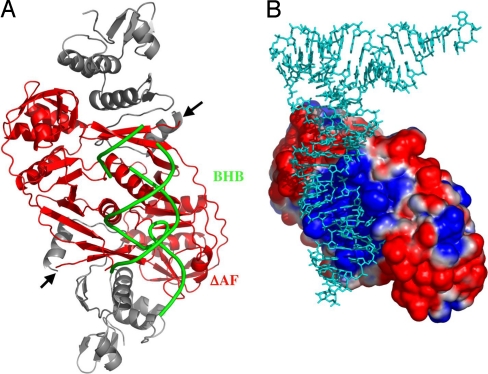
To confirm that we are dealing with three discrete patches and not a continuous elongated one, we considered the fact that each subunit of the homodimeric AF enzyme contains two similar repeating domains that are homologous to the subunit structure of the homotetrameric enzyme from MJ: the C-terminal repeat (CAF) and the N-terminal repeat (NAF). We designed a mutant enzyme that comprises a deletion of the NAF domain with the exception of the last two beta strands that comprise loop 10 at the C terminus, known to be important for the assembly of the enzyme (Fig. 4A). According to our expectations, the mutant enzyme (ΔAF) lacks the two peripheral patches, but it should still present all of the residues directly interacting with the BHB (Fig. 4B). To determine whether this truncated enzyme can assemble into an active form, we overexpressed and purified it as described in Materials and Methods. Fig. 5 A and C shows that the truncated enzyme is capable of cleaving the BHB in the presence or absence of a tRNA mature domain, but it is no longer capable of cleaving the BHL substrate, even in the presence of the mature domain (Fig. 5 B and C). We conclude that the positively charged peripheral patches interact with a set of recognition elements that are presented by the mature domain of AF tRNA precursors.
Fig. 4.
Structure of ΔAF enzyme and model of the interaction with its substrate. (A) Cartoon representation of the parts deleted from AF endonuclease (gray). The arrows indicate the new N termini of the subunits. The BHB substrate is shown in green. (B) Electrostatic surface potential of ΔAF. Positively charged regions are shown in blue, the neutral regions are shown in white, and the negatively charged regions are shown in red. The pre-tRNA model is shown in cyan.
Discussion
In Archaea, there are three distinct architectures of tRNA endonuclease, the enzyme responsible for the excision of the intron from pre-tRNA. The crystal structures of the enzymes from two Euryarcheota, AF and MJ, are known (18, 19). The AF enzyme is a homodimer (α2). Each subunit comprises two similar repeats, but the same repeat performs a different function in each of the two subunits. The NAF acts to stabilize the dimer, whereas the CAF is the catalytic domain. The MJ enzyme, a homotetramer (α4), is characterized by a similar functional subdivision. Two of the subunits play a catalytic role, whereas the other two have a structural role. We found that the Crenarcheote SS presents two genes homologous to the one coding for the MJ protein. One of the two genes is specialized for encoding the catalytic subunit and the other for encoding the subunit that maintains the structure of the heterotetramer (α2β2). Presumably, at the origin, in Archaea the situation was similar to that in MJ, with a single gene coding for a single protein able to serve both as catalytic or structural subunit. During evolution in AF and in SS, the single gene duplicated. In AF the two duplicated genes fused and subfunctionalization occurred between the two halves of the fused protein: the C-terminal half (CAF) retained the catalytic function and the N-terminal half (NAF) specialized as the structural subunit. In SS, one of the duplicated genes has specialized for encoding the catalytic and the other for encoding the structural subunit. Both subunits are necessary for substrate cleavage by the heterotetramer (15, 17, 20, 21).
The protagonist of this paper is the AF enzyme. The homodimer cleaves pre-tRNABHB, pre-tRNABHL, and mini-BHB, but not mini-BHL. The AF enzyme, therefore, can cleave the BHL only in a mature-domain-dependent mode. Because mature-domain dependence for cleavage of non-BHB substrates is a main feature of the eukaryotic enzymes, we observe in AF the dawn of dominance of the mature domain in tRNA splicing. In AF and in all other Euryarchaea encoding homodimeric endonucleases, as in Eukarya, the intron is located at the canonical position in the anticodon loop region between nucleotides 37 and 38. The latter feature is a necessary requirement for mature-domain dependence, because it assures a fixed geometry relative to the mature domain for all pre-tRNAs. Three RNA-binding sites, one central and two peripheral, characterize the surface of the AF enzyme that interacts with the substrate (Fig. 3B). The central site contains the two symmetric catalytic triads and presumably interacts with the BHB and its relaxed forms, like the BHL. The two flanking patches are identical and symmetrically placed and are presumably involved in the interaction with the mature domain. They function one at a time, because the enzyme can accommodate only one tRNA molecule. In an experiment that attempted to reverse the proposed evolutionary process, we deleted a portion of the N-terminal domain of the AF enzyme; the resulting ΔAF mutant should lack the two peripheral RNA binding regions (Fig. 4 A and B). As predicted, ΔAF is unable to perform in a mature-domain-dependent mode. It can cleave only pre-tRNABHB and mini-BHB; it does not cleave pre-tRNABHL or, of course, the mini-BHL (Fig. 5).
Interaction of a Pre-tRNABHB with the AF Enzyme.
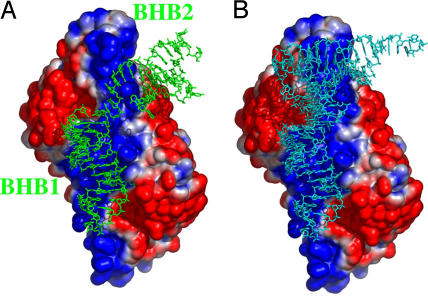
We built a model using the recently solved structure of the AF enzyme in complex with a minimal BHB substrate, generously provided by H. Li (11). Interestingly, the asymmetric unit of the co-crystal contains two molecules of RNA–enzyme complex and the analysis of the crystal packing made it possible to see that each enzyme, besides interacting with its substrate through the BHB structure, also interacts with the A-helix belonging to the substrate of a symmetry-related enzyme molecule (Fig. 6A). These interactions involve the side chains of Lys-90 and Lys-91, which belong to the group of residues forming the above described peripheral positive patch (Fig. 3B). Almost certainly, the observed RNA–protein contacts are the result of different packing constraints in the crystal lattice and only mimic the interaction with a cognate mature-domain substrate in solution, but they clearly suggest that a site can be available to host the tRNA mature body as we propose in our model. Because of the symmetry of the catalytic site, the enzyme can accommodate one tRNA molecule at a time, interacting through one or another patch. We superimposed, using the program O, the 6-bp stem of the anticodon arm of the tRNAPhe to the corresponding 6-bp stem of the BHB minimal substrate. The superimposed pre-tRNA mature domain faces one of the peripheral patches of the enzyme with the variable pocket and T-arm-acceptor stem helix (Fig. 6B). The enzyme can approach the tRNA only by using this strategy; introns are also present in class II tRNAs (tRNACAALeu in AF). These tRNAs present a long extra arm that could otherwise impede, by steric hindrance, interaction with an enzyme approaching from the opposite side. Moreover, the T-arm-acceptor coaxial A-helix presents its deep and narrow major groove to the enzyme so that the latter can interact only with the phosphate backbone, thereby permitting recognition of the tRNA substrate that is not sequence specific.
Fig. 6.
Model of the interactions between the AF enzyme and the substrates. Electrostatic surface potential of AF bound either to two BHB RNA molecules (shown in green) obtained from the crystallographic structure (11) (A) or to a model of pre-tRNA (shown in cyan) (B).
From Archaea to Eukarya.
The salient features that characterize the AF enzyme are the result of subfunctionalization and creation of the binding sites for elements of the mature domain of the pre-tRNA. As far as subfunctionalization is concerned, presumably, the extreme environments that characterize the habitat of Archaea is not ideal for α4 architectures, where a single polypeptide plays two very different roles. It is not clear whether drift alone or positive selection or both had a role in subfunctionalization.
We should not forget, however, that subfunctionalization did not occur only in homodimeric (αβ) enzymes, like AF, but also in heterotetrameric (α2β2) enzymes like SS. The fact, therefore, that it occurred twice independently, and by different routes in the same gene, argues in favor of positive selection. The acquisition of the three RNA-binding sites points to the extraordinary and spectacular series of events that saw evolution engraving on the surface of the AF enzyme the overlapping portraits of two pre-tRNAs. The ΔAF mutant tells us that we can reverse the evolutionary process by deleting a sequence of amino acids from the N-terminal part of the NAF. The truncated enzyme loses the ability to act in a mature-domain-dependent mode. If we compare Fig. 4B with Fig. 6B, we see that ΔAF completely lacks the peripheral sites that interact, in the wild-type enzyme, with the mature domain.
In the intact AF enzyme, the two active sites are completely interchangeable. Depending on which peripheral binding site is used by the pre-tRNA substrate, a given active site will cleave at the 5′ or at the 3′ junction. In the case of the yeast tRNA splicing endonuclease, the situation is radically different. The eukaryal enzyme is a heterotetramer (α, β, γ, δ) (22). The four subunits (Sen15, Sen2, Sen34, and Sen54 in yeast) contribute to the formation of two composite active sites that, because of specific interactions with the mature domain, interact rigidly and specifically either with the 5′ or the 3′ cleavage sites (23). If we wanted, therefore, to modify the AF enzyme to make it more similar to its eukaryal counterpart, we should think of ways to eliminate one of the two peripheral patches. In this way, the active sites would be rigidly assigned to one or the other of the two cleavage sites.
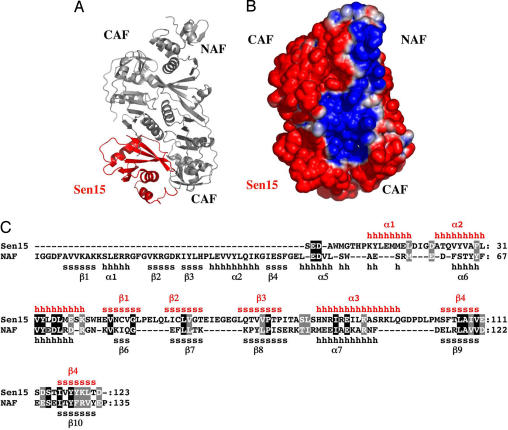
The ΔAF result teaches us that the peripheral sites comprise a sequence of amino acids located in the N-terminal part of the NAF. We observed that the sequence of the eukaryal subunit Sen15 does not contain the catalytic triad (22) and that therefore it must be a structural subunit. Alignment of the Sen15 and NAF sequences shows that the eukaryal peptide lacks a segment corresponding to the N-terminal part of NAF, which constitute the peripheral site, but is homologous to the C-terminal sequence of the AF enzyme (Fig. 7C). Because the NMR structure of the human Sen15 (24) is available, we aligned the atomic structures of the Sen15 monomer with NAF using the program VMD (25). The alignment clearly shows that Sen15 and the C-terminal portion of NAF share a common fold (Fig. 7A). On the basis of these observations, we constructed a model of a chimeric enzyme comprising two CAFs, one NAF, and the human Sen15 substituting for the other NAF. Electrostatic potential calculations show that this model chimeric enzyme has lost one of the peripheral patches and, like a eukaryal enzyme, is characterized by active sites rigidly assigned to either the 5′ or the 3′ cleavage sites of the substrate (Fig. 7B).
Fig. 7.
Model of a chimeric enzyme presenting two CAFs, one NAF, and the human Sen15. (A) Representation of the model chimeric enzyme. The archaeal parts are shown in gray, and the human part is shown in red. (B) Electrostatic surface potential of the model chimeric enzyme. Positively charged regions are shown in blue, neutral regions are shown in white, and negatively charged regions are shown in red. (C) Primary sequences and secondary structures of the NAF and of the human Sen15. Secondary structure elements, as determined by the crystallographic structure of AF (19) and the NMR structure of Sen15 (24), are shown in black and red, respectively. The “s” and β indicate β-strands, and “h” and α indicate α-helices. Black columns indicate residues that are conserved; gray columns indicate residues that are similar.
We expect that, if the route that we hypothesize was really followed in evolution, the active sites would drift, obviously conserving the catalytic triad and the ability to cleave the BHB substrate, adapting to the specific intron–exon boundaries. Clearly, much remains to be done in terms of defining the parts of the mature domain that are required for correct positioning of the pre-tRNA on the enzyme surface. What we have established is that at least one current archeon, AF, retains the memory of the first step on the path to the eukaryal system.
Materials and Methods
Expression and Purification of the Protein Constructs.
The construction of the vectors coding for the genes of the endonucleases from AF, SS, and MJ and the purification of the enzymes are described elsewhere (15, 17). The gene coding for the ΔAF construct was obtain by PCR-amplification by using as a template the previously cloned AF gene and the following DNA primers: (i) 5′GGAATTCCATATGGACGAGTTAAGGCTTGCTGTCG and (ii) 5′CGCGGATCCTCAAACCTTAACCCTCTCAAAGC. The two primers were designed to obtain an amplified fragment presenting an NdeI site upstream of the gene and a BamHI site downstream. After digest, the fragment was cloned into pET28b (Novagen, Madison, WI). The correct clones were verified by DNA sequencing. The protein was overexpressed as hexa-histidine-tagged forms (pET28b) in Escherichia coli Rosetta (Novagen). Cells were grown in 1-liter cultures of Terrific Broth broth at 37°C in the presence of 30 μg/ml kanamycin (pET28) with the addition of 30 μg/ml chloramphenicol. The purification consisted of a metal affinity column as a first step, followed by gel filtration using an analogous strategy already described for the other enzymes (17). The purity of the enzyme was assessed by Coomassie blue staining of SDS polyacrylamide gels.
In Vitro RNA Synthesis and Pre-tRNA Splicing Reactions.
DNA templates were produced as described (17). T7 RNA polymerase transcription reactions were carried out following the conditions of the Ambion (Austin, TX) T7-Megashortscript kit. [α-32P]UTP (800 Ci/mmol; Amersham Pharmacia) (1 Ci = 37 GBq) was included to radiolabel the RNA transcripts. All reactions were purified by electrophoresis on a 10% denaturing polyacrylamide gel. RNA products corresponding to the correct size were eluted, phenol was extracted, and ethanol was precipitated.
tRNA splicing reactions were typically performed in 25 mM Tris·HCl (pH 7.5), 5 mM MgCl2, 100 mM NaCl, and 10% glycerol, and included 20 fmol of tRNA precursor substrates. Purified splicing endonucleases were added and incubated at 65°C for 1 h. The reactions were stopped by phenol extraction, and ethanol was precipitated and separated on 10% denaturing polyacrylamide gels. Cleavage products were visualized by analysis on a Molecular Dynamics model Storm 860 PhosphorImager by using ImageQuant software, version 4.
Acknowledgments
This article is dedicated to the memory of our beloved friend and colleague Domenica “Nica” Gandini-Attardi, prematurely deceased, who was very helpful for continuous encouragement and critical reading of the manuscript. We thank Hong Li for providing the coordinates of the AF splicing endonuclease bound with an RNA BHB substrate (Protein Data Bank ID code 2GJW), A. Ferrara and T. Cuccurullo for secretarial assistance, and G. Di Franco for technical assistance. This work was supported by Italian Ministry of Research FIRB (G. Armenise-Harvard Foundation, Italia-Canada and Idee Progett. 2005) and SVIFASTA grants and European FP6 contracts (MUGEN, EURASNET, and EUMODIC).
Abbreviations
- NAF
N-terminal repeat
- CAF
C-terminal repeat
- BHB
bulge–helix–bulge
- BHL
bulge–helix–loop
- SS
Sulfolobus solfataricus
- AF
Archeoglobus fulgidus
- MJ
Methanocaldococcus jannaschii.
Footnotes
The authors declare no conflict of interest.
References
- 1.Abelson J, Trotta CR, Li H. J Biol Chem. 1998;273:12685–12688. doi: 10.1074/jbc.273.21.12685. [DOI] [PubMed] [Google Scholar]
- 2.Belfort M, Weiner A. Cell. 1997;89:1003–1006. doi: 10.1016/s0092-8674(00)80287-1. [DOI] [PubMed] [Google Scholar]
- 3.Reyes VM, Abelson J. Cell. 1988;55:719–730. doi: 10.1016/0092-8674(88)90230-9. [DOI] [PubMed] [Google Scholar]
- 4.Mattoccia E, Baldi IM, Gandini-Attardi D, Ciafre S, Tocchini-Valentini GP. Cell. 1988;55:731–738. doi: 10.1016/0092-8674(88)90231-0. [DOI] [PubMed] [Google Scholar]
- 5.Di Nicola Negri E, Fabbri S, Bufardeci E, Baldi MI, Gandini Attardi D, Mattoccia E, Tocchini-Valentini GP. Cell. 1997;89:859–866. doi: 10.1016/s0092-8674(00)80271-8. [DOI] [PubMed] [Google Scholar]
- 6.Palmer JR, Nieuwlandt DT, Daniels CJ. J Bacteriol. 1994;176:3820–3823. doi: 10.1128/jb.176.12.3820-3823.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kleman-Leyer K, Armbruster DW, Daniels CJ. Cell. 1997;89:839–847. doi: 10.1016/s0092-8674(00)80269-x. [DOI] [PubMed] [Google Scholar]
- 8.Lykke-Andersen J, Garrett RA. EMBO J. 1997;16:6290–6300. doi: 10.1093/emboj/16.20.6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baldi MI, Mattoccia E, Bufardeci E, Fabbri S, Tocchini-Valentini GP. Science. 1992;255:1404–1408. doi: 10.1126/science.1542788. [DOI] [PubMed] [Google Scholar]
- 10.Bufardeci E, Fabbri S, Baldi MI, Mattoccia E, Tocchini-Valentini GP. EMBO J. 1993;12:4697–4704. doi: 10.1002/j.1460-2075.1993.tb06158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xue S, Calvin K, Li H. Science. 2006;312:906–910. doi: 10.1126/science.1126629. [DOI] [PubMed] [Google Scholar]
- 12.Fruscoloni P, Baldi MI, Tocchini-Valentini GP. EMBO Rep. 2001;2:217–221. doi: 10.1093/embo-reports/kve040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabbri S, Fruscoloni P, Bufardeci E, Di Nicola Negri E, Baldi MI, Attardi DG, Mattoccia E, Tocchini-Valentini GP. Science. 1998;280:284–286. doi: 10.1126/science.280.5361.284. [DOI] [PubMed] [Google Scholar]
- 14.Marck C, Grosjean H. RNA. 2003;9:1516–1531. doi: 10.1261/rna.5132503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tocchini-Valentini GD, Fruscoloni P, Tocchini-Valentini GP. Proc Natl Acad Sci USA. 2005;102:15418–15422. doi: 10.1073/pnas.0506750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baker N, Holst M, Wang F. J Comput Chem. 2000;21:1343–1352. [Google Scholar]
- 17.Tocchini-Valentini GD, Fruscoloni P, Tocchini-Valentini GP. Proc Natl Acad Sci USA. 2005;102:8933–8938. doi: 10.1073/pnas.0502350102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Trotta CR, Abelson J. Science. 1998;280:279–284. doi: 10.1126/science.280.5361.279. [DOI] [PubMed] [Google Scholar]
- 19.Li H, Abelson J. J Mol Biol. 2000;302:639–648. doi: 10.1006/jmbi.2000.3941. [DOI] [PubMed] [Google Scholar]
- 20.Randau L, Calvin K, Hall M, Yuan J, Podar M, Li H, Soll D. Proc Natl Acad Sci USA. 2005;102:17934–17939. doi: 10.1073/pnas.0509197102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calvin K, Hall MD, Xu F, Xue S, Li H. J Mol Biol. 2005;353:952–960. doi: 10.1016/j.jmb.2005.09.035. [DOI] [PubMed] [Google Scholar]
- 22.Trotta CR, Miao F, Arn EA, Stevens SW, Ho CK, Rauhut R, Abelson JN. Cell. 1997;89:849–858. doi: 10.1016/s0092-8674(00)80270-6. [DOI] [PubMed] [Google Scholar]
- 23.Trotta CR, Paushkin SV, Patel M, Li H, Peltz SW. Nature. 2006;441:375–377. doi: 10.1038/nature04741. [DOI] [PubMed] [Google Scholar]
- 24.Song J, Markley JL. J Mol Biol. 2007;366:155–164. doi: 10.1016/j.jmb.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Humphrey W, Dalke A, Schulten K. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]