Abstract
Multiple protein arginine methyltransferases are involved in transcriptional activation of nuclear receptors. Coactivator-associated arginine methyltransferase 1 (CARM1)-mediated histone methylation has been shown to activate nuclear receptor-dependent transcription; however, little is known about the regulation of its enzymatic activity. Here, we report that the methyltransferase activity of CARM1 is negatively regulated through phosphorylation at a conserved serine residue. When the serine residue is mutated to glutamic acid, which mimics the phosphorylated serine residue, the mutant CARM1 exhibits diminished ability to bind the methyl donor adenosylmethionine and diminished histone methylation activity. Moreover, such mutation leads to the inhibition of CARM1 transactivation of estrogen receptor-dependent transcription. Our results provide an example for the regulation of protein arginine methyltransferase activity by phosphorylation. As CARM1 is a potent transcriptional coactivator of estrogen receptor, our results suggest that phosphorylation of CARM1 serves as a unique mechanism for inactivating CARM1-regulated estrogen-dependent gene expression.
Keywords: estrogen receptor, methylation, transcription, histone
The transcriptional potency of the nuclear receptor (NR) superfamily of transcription factors is highly regulated by their cognate ligands, which trigger conformational changes that foster association with coactivators or corepressors (1, 2). To date, a large subset of the identified coactivators is involved in regulation of chromatin structure, either by covalently modifying histones or remodeling the chromatin template (3). Examples of histone-modifying enzymes include the p160 and p300 families of histone acetyltransferases, whereas chromatin modification enzymes include human SWI/SNF ATP-remodeling complexes (2, 4). Association of these coactivators with hormone-responsive elements increases the accessibility of the chromatin template for the basal transcriptional machinery, leading to the initiation of transcription (5). The p160 family of proteins is among the earliest identified NR coactivators, including the three related proteins SRC-1, SRC-2/glucocorticoid receptor interacting protein 1 (GRIP1)/TIF2, and SRC-3/pCIP/ACTR/AIB1/TRAM1 (4, 5). The NR-associated p160 proteins serve as a binding platform for other coactivators such as p300/CBP (6), coactivator-associated arginine methyltransferase 1 (CARM1) (7), and coiled-coiled coactivator (8).
Methylation of histones on arginines is catalyzed by protein arginine methyltransferases (PRMTs) and correlates with gene activation (9, 10). Several PRMTs, including PRMT1, PRMT2, and CARM1/PRMT4, are involved in estrogen receptor (ER)-mediated transcriptional activation (11–13). CARM1, identified as a p160 family GRIP1-interacting protein, is required for ER transactivation (7). CARM1 methylates histone H3 at R2, R17, and R26 (14), and methylation of these sites has been associated with ER-target gene pS2 activation (12, 15). Although CARM1 cooperates with PRMT1 and p300/CBP in ER-mediated transcriptional activation (13, 16), all three proteins depend on the p160 proteins for their ER coactivator function, suggesting that they act as secondary rather than primary coactivators. In addition to histone H3, CARM1 methylates the cofactor CBP/p300 at multiple sites (17–19). Importantly, loss of CARM1 in the mouse embryos leads to abrogation of the estrogen response and reduction in expression of some ER target genes (20), highlighting the functional importance of this enzyme in ER-regulated gene expression.
Transcriptional regulation of ER-dependent gene expression by protein arginine methylation is a reversible and tightly controlled process, which might be regulated at either substrate (e.g., histone) or enzymatic levels. Kinetic ChIP analyses reveal elevated methyl-H3R17 and recruitment of CARM1 on the pS2 promoter upon activation of the estrogen-responsive pS2 gene, whereas a contemporaneous loss of dimethyl-H3R17 marks ER disengagement and the release of CARM1 from the promoter (21). Recent studies show that monomethylated arginines within histone H3 can be converted by arginine deimination to citrulline, leading to repression of the pS2 gene (22, 23). Therefore, deimination of H3 prevents the transcriptional activation mediated by arginine methylation. At the enzymatic level, the substrate specificity of CARM1 can be altered by associating with other proteins, including subunits of SWI/SNF, forming a NUMAC complex (24). In the context of the complex, but not when present alone, CARM1 acquires the ability to methylate nucleosomal histones. Both CARM1 and subunits of SWI/SNF have been shown to be recruited to the ER target gene pS2 promoter in a ligand-dependent manner (15, 21, 25). CARM1 might coordinate methylation and remodeling events on endogenous ER target genes, or perhaps it is involved in the recruitment or stabilization of SWI/SNF to ER target gene promoters (24). However, it remains unclear as to how CARM1's function is regulated.
In the present study, we report that the methyltransferase activity of CARM1 is modulated through phosphorylation at a specific serine residue, which is conserved among CARM1 from different species, but not conserved among all other PRMTs. Mutation of this serine to alanine in mouse CARM1 converts it to a constitutively active form, whereas mutation to glutamic acid generates a dominant-negative enzyme. Studies of these CARM1 mutants lead to the conclusion that phosphorylation of CARM1 prevents its binding to the methyl donor S-adenosylmethionine (AdoMet), thereby inhibiting its enzymatic activity. Furthermore, the serine to glutamic acid CARM1 mutant exhibits diminished ER transactivation activity, suggesting that phosphorylation of CARM1 would negatively regulate ER-dependent gene expression.
Results
CARM1 Phosphorylation Was Observed During Mitosis.
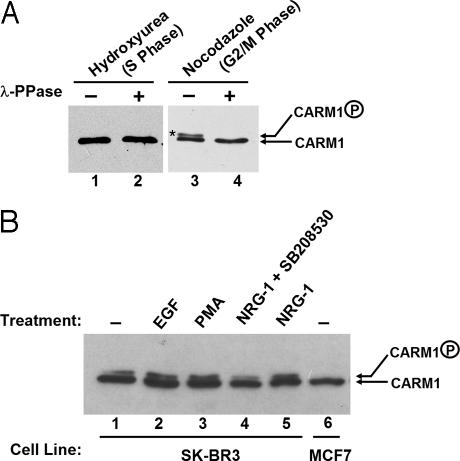
During mitosis, chromatin remodeling coactivators, such as BRG1 and BRM that are the ATPase components of human SWI/SNF complexes, are inactive and excluded from condensed chromatin (26, 27). Because CARM1 directly interacts with BRG1 (24), and BRG1 and BRM were previously shown to be phosphorylated and inactivated during mitosis (26, 27), we wondered whether phosphorylation is also a mechanism to regulate CARM1 activity during mitosis. To test whether CARM1 is phosphorylated during the cell cycle, HeLa cells were treated with hydroxyurea or nocodazole to enrich S or G2/M population cells, followed by immunoprecipitation of CARM1 from the cell extracts. On Western blotting film, a slower migrating form of CARM1 was observed with samples treated with nocodazole (Fig. 1B, lane 3). This slower migrating form disappeared when the samples were pretreated with λ-phosphatase before performing the Western blotting experiment (Fig. 1B, lane 4). Because the slower migrating band is sensitive to λ-phosphatase treatment, we deduced that the slower migrating form represented the phosphorylated CARM1. Phosphorylation of CARM1 during mitosis was also observed in MCF7 cells (data not shown). Furthermore, phosphorylated CARM1 was also detectable in SK-BR3 breast cancer cells that express high levels of Her2/Neu/ErbB2 (Fig. 1B, lanes 1–5). The phosphorylation level appears to be affected upon treatment with some growth factors (Fig. 1B, lanes 2–5). In contrast, no CARM1 phosphorylation was detectable in untreated MCF7, a cell line derived from a pleural effusion of a human breast adenocarcinoma (Fig. 1B, lane 6). It is still not clear what causes the different CARM1 phosphorylation levels among these cancer cells.
Fig. 1.
CARM1 phosphorylation is observed in vivo. (A) CARM1 phosphorylation was observed during mitosis in HeLa cells. CARM1 was immunoprecipitated from HeLa cells treated with 1 mM hydroxyurea (S-phase arrest) (lanes 1 and 2) or 0.5 μg/ml nocodazole (M-phase arrest) (lanes 3 and 4). The CARM1 immune complex was divided into halves; one half was treated with 2.5 μg of λ-phosphatase (Upstate Biotech) (lanes 2 and 4) in supplied buffer, and the other half was treated with buffer alone (lanes 1 and 3). Samples were loaded on 7% SDS/PAGE and transferred to nitrocellulose membrane. CARM1 was detected by using anti-CARM1 antibody (Upstate Biotech). The slower migrating CARM1 representing phosphorylated-CARM1 is denoted by an *. (B) CARM1 phosphorylation can be detected in SK-BR3 cells but not in MCF7 breast cancer cells. SKBR3 cells were treated with DMSO (lane 1), 50 ng/ml EGF (lane 2), 200 ng/ml phorbol 12-myristate 13-acetate (PMA) (lane 3), and 10 ng/ml NRG-1 (lane 5) for 15 min before harvesting. SB208530, a specific inhibitor of p38 kinase, was preincubated with cells for 1 h before addition of NRG-1 (lane 4). Cells were lysed in RIPA lysis buffer (150 mmol/liter NaCl/1% Nonidet P-40/0.5% deoxycholic acid/0.1% SDS/50 mM Tris·HCl, pH 8.0/2 μg/ml aprotinin/10 μg/ml leupeptin/1 mg/ml sodium orthovanadate/1 mmol/liter PMSF). Equal amounts of total proteins were loaded on 7% SDS/PAGE and transferred to nitrocellulose membrane. CARM1 was detected by using anti-CARM1 antibody (Upstate Biotech).
Human CARM1 Is Phosphorylated at a Conserved Serine Residue.
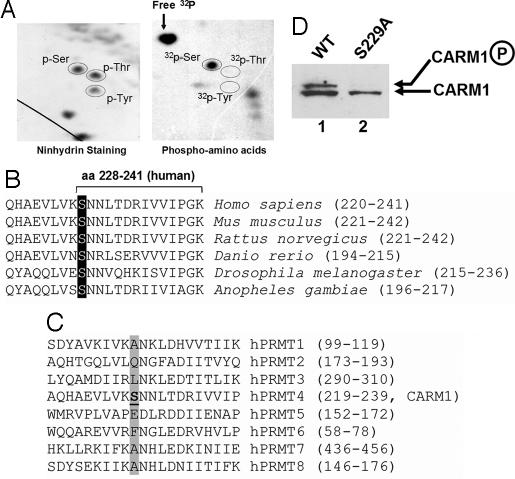
We went on to identify the phosphorylation sites on CARM1. HeLa cells were cultured in phosphate-free media and supplemented with [32P] phosphoric acid in the presence of nocodazole. After 6 h of labeling, CARM1 was immunoprecipitated as described (24). Phospho-amino acid analysis was performed to determine the phosphorylation residues. Fig. 2A shows that CARM1 was phosphorylated on serine residue(s) but not threonine or tyrosine residues. The phosphorylated CARM1 was subsequently subjected to mass spectrometric analysis, in which only one phospho-peptide encompassing amino acids 228–241 of human CARM1 (Fig. 2B) was identified. Because Ser-228 (S228) is the only serine within this peptide, we conclude that Ser-228 is the CARM1 phosphorylation site during mitosis. Sequence alignment demonstrates that this serine is conserved among CARM1 from different species (Fig. 2B), but is not conserved among human PRMT molecules (Fig. 2C). Notice that, the equivalent residue of human CARM1 Ser-228 in mouse and rat is Ser-229. To confirm that the identified serine residue represents the main phosphorylation site on CARM1, we developed cell lines stably expressing wild-type or mutant CARM1 in mouse embryonic fibroblast (MEF) −/− cells. Western blot of the cell extracts with anti-CARM1 antibodies showed that CARM1 phosphorylation was abrogated when the serine residue was mutated to alanine (Fig. 2D), indicating that the identified serine represents the main phosphorylation site in vivo.
Fig. 2.
Residue Ser-228 is identified as the main phosphorylation site on human CARM1. (A) Phosphorylation of CARM1 during mitosis was observed on serine residues, but not on tyrosine or threonine residues. The 32P-labeled phosphorylated CARM1 was trypsin-digested, acid-treated, and analyzed by phosphoamino acid analysis on a 2D gel. (B) Phosphorylation of human CARM1 is located at Ser-228, which is conserved among human, mouse, rat, zebra fish, fruit fly, and mosquito. The GenBank accession numbers are as follows: Homo sapiens, NP_954592; Mus musculus, NP_067506; Rattus norvegicus, NP_001029259; Danio rerio, XP_701531; Drosophila melanogaster, NP_649963; Anopheles gambiae, XP_318375). (C) The phosphorylation site on CARM1 is not conserved among human PRMT molecules, but next to the conserved asparagine (or aspartate) residue. Notice that hPRMT4 is the alternative name for human CARM1. (D) CARM1 phosphorylation was not detectable with lysates prepared from MEF3−/− cells stably expressing CARM1 S229A mutant (lane 2). Whole-cell lysates from MEF3−/− CARM1 null cells stably expressing either WT or the S229A mutant CARM1 were separated on SDS/PAGE and transferred to nitrocellulose for Western blot. CARM1 was probed with anti-CARM1 antibodies.
The S229E Mutation in Mouse CARM1 Inhibits Its MTase Activity.
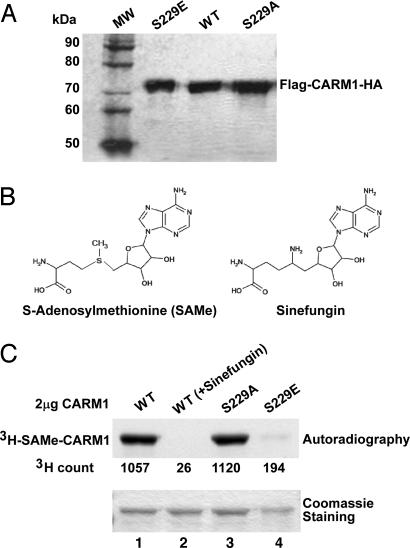
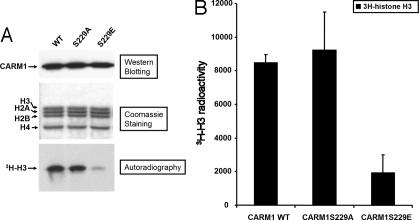
CARM1's arginine MTase activity is absolutely required for CARM1 function in transcription (7). To investigate the effect of CARM1 phosphorylation on its MTase activity, we first examined whether mutation on the phosphorylation site could affect its binding to MTase substrates. We constructed two expression constructs for expressing mutant CARM1 S229A (serine-to-alanine mutation) and S229E (serine-to-glutamic acid), respectively. Whereas the serine-to-alanine mutation was expected to abolish phosphorylation, the serine-to-glutamic acid CARM1 mutant was expected to behave like phosphorylated CARM1, as the side chain of glutamic acid mimics the side chain of phosphorylated serine. These two mutant proteins and wild-type CARM1 were expressed in baculovirus-infected sf9 insect cells and purified as described (17) (Fig. 3A). With the purified proteins, we performed a UV-cross-linking experiment (28) with the substrate analog AdoMet (Fig. 3B Left) in the presence or absence of the competitor compound sinefungin (Fig. 3B Right). As shown in Fig. 3C, wild-type CARM1 cross-linked with 3H-AdoMet and formed adduct 3H-AdoMet-CARM1 (Fig. 3C, lane 1). The binding is inhibited by the nonlabeled competitor sinefungin (Fig. 3C, lane 2). The S229A mutant, but not the S229E mutant protein, could cross-link efficiently with 3H-AdoMet (Fig. 3C, lanes 3 and 4). These results indicated that the S229E mutation affected the binding to the MTase substrates. We further investigated whether such mutation would affect the MTase activity by using an in vitro methylation assay (Fig. 4). Indeed, we observed that the purified S229E mutant has significantly lower MTase activity compared with wild-type or S229A proteins (Fig. 4A). Quantitative results (Fig. 4B) showed that the MTase activity of mutant S229E is about one-fourth of wild-type CARM1. Because the S229E mutation, but not the S229A, had an effect on MTase substrate binding and MTase activity, as the side chain of glutamic acid mimics the side chain of phosphorylated serine, we deduced that CARM1 phosphorylation negatively regulates MTase activity.
Fig. 3.
The mutant mouse CARM1 S229E is defective in binding to substrate S-AdoMet. (A) The recombinant mouse CARM1 (WT and mutants S229E and S229A) were expressed in baculovirus-infected Sf9 cells and purified by affinity chromatography. Samples were run on 7% SDS/PAGE and stained with Coomassie. (B) The chemical structures of AdoMet and Sinefungin. (C) WT and mutant S229A, but not mutant S229E, mouse CARM1 could be cross-linked with AdoMet. Sinefungin, a competitive inhibitor, was preincubated with CARM1 before incubation with 3H-AdoMet in lane 2. Coomassie staining shows the relative amount of protein used in the photo-affinity labeling experiment. 3H count represents the amount of 3H-AdoMet cross-linked with CARM1.
Fig. 4.
The mutant mouse CARM1 S229E shows reduced MTase activity in in vitro methylation assays. (A) In vitro methylation assays. (Top) Western blotting shows the relative amount of WT and mutant (S229A and S229E) mouse CARM1 used in the assay. (Middle) Coomassie staining shows the relative amount of substrate histone used in the assay. (Bottom) Autoradiograph of the methylated 3H-H3. (B) Quantitation of methylated 3H-H3 based on two independent experiments. The 3H-histone H3 bands were excised from the dried gel, immersed in scintillation buffer, and counted for 3H.
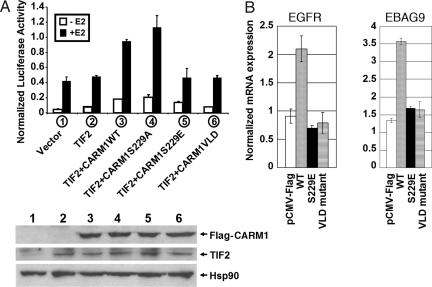
Mouse CARM1 S229E Mutant Does Not Stimulate ER-Dependent Gene Expression in Vivo.
The activation of some ER-controlled genes depends on synergistic cooperation among different classes of coactivators, including histone acetyltransferases, protein arginine MTases, and chromatin remodeling factors (21). In transient transfection assays, CARM1 and GRIP1/TIF2 have been shown to act synergistically to activate ER-dependent transcription (16). Because MTase activity is essential for transactivation of ER-dependent transcription (7) and we observed that CARM1 phosphorylation negatively regulates the MTase activity, we investigated whether CARM1 phosphorylation affects CARM1 transactivation of ER-dependent transcription in vivo. We performed cotransfection experiments with different CARM1 expression constructs and a TK-(ERE)3-luciferase reporter in MEF20−/−, a MEF cell line derived from CARM1 knockout mice (20). Fig. 5A shows a comparison of the effect of different CARM1 mutants on ER transactivation. The CARM1 mutant VLD189–191-AAA (CARM1VLD) had been previously characterized as a MTase-deficient mutant for transactivation of ER-dependent transcription (7), thus serving as a control in this experiment. Both wild-type CARM1 and the S229A mutant were found to transactivate 17β-estradiol (E2)-induced reporter activity, whereas the S229E mutant and CARM1VLD mutant exhibited little transactivation activity (Fig. 5A Upper). Western blotting confirmed the relatively equal level of CARM1 expression in transfected MEF20−/− cells (Fig. 5A Lower). These results suggested that the S229E mutation in CARM1 has impaired the ability to stimulate E2-induced ER transcription, implying that phosphorylation of CARM1 would compromise the ability of CARM1 to regulate ER-dependent gene expression. Transient transfection method was used to examine the effect of CARM1 mutants on endogenous ER target gene expression. Empty vector, wild-type CARM1, S229E, or VLD mutant were cotransfected with ERα to MEF20−/− cells and treated with E2 for 4 h. The expression level of two ER target genes, EGFR and EBAG9 (29), were determined by quantitative RT-PCR. Fig. 5B shows that, unlike wild-type CARM1, the S229E mutant and the dominant negative mutant CARM1VLD189–191-AAA (7) fail to stimulate Egfr and Ebag9 transcription, suggesting that phosphorylation of CARM1 could be a mechanism to regulate ER target gene expression in vivo.
Fig. 5.
Mouse CARM1 mutant S229E is defective in the activation of ER-dependent transcription. (A) Plasmids for TIF2 (200 ng), Flag-CARM1 (200 ng), and ER (2.5 ng) were transfected into MEF cells from CARM1 knockout mice with TK-(ERE)x3-LUC reporter plasmids (200 ng) and plasmids encoding β-gal as an internal control. (Upper) Relative luciferase activity was normalized to the level of β-gal. (Lower) The cell lysates used for luciferase assays were analyzed for CARM1, TIF2, and Hsp90 expression by Western blotting. The expressed TIF2 and Flag-tagged CARM1 proteins in transfected cells were detected with anti-GRIP1/TIF2 (Santa Cruz Biotechnology, Santa Cruz, CA) and anti-Flag (Sigma, St. Louis, MO) antibodies, respectively. Hsp90 serves as a control showing the relative amount of loaded samples. (B) Empty vector and CARM1 expression constructs were cotransfected with ERα into CARM1−/− MEF cells. After 4 h of 17β-estradiol treatment, the expression levels of two endogenous ER-target genes, EGFR and EBAG9, were measured with quantitative RT-PCR.
Mouse CARM1 S229E Mutant Failed to Form Dimers in Vivo.
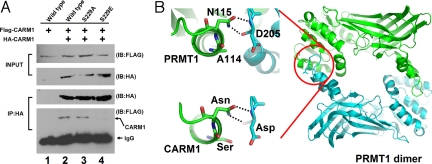
A common feature among the known PRMT structures is that PRMTs form dimers or multimers that are essential for AdoMet binding and thus essential for enzymatic activity (30–33). CARM1 has also been shown to form dimers and multimers in solution (34). In the crystal structure of PRMT1 (30), the polar interaction between the side chain of Asn-115 and Asp-205 main-chain atoms is critical for dimer formation (Fig. 6B), and the formation of dimers is required for the binding of the AdoMet (30). The corresponding residues in mouse CARM1 are Asn-230 and Asp-323. Our identified phosphorylation site on mouse CARM1 (Ser-229) corresponds to an alanine residue (Ala-114) in PRMT1 (Fig. 2C). As the dimerization regions are conserved among PRMT homologs (33), we modeled the CARM1 dimer structure based on the crystal structure of PRMT1 (Fig. 6B). We found that Asn-230 and Asp-323 would interact with each other, similar to the polar interaction between Asn-115 and Asp-205 in PRMT1. When Ser-229 is phosphorylated, the relatively larger side chain of phospho-Ser residue would interfere with the interaction between Asn-230 and Asp-323. Similar interference would remain if Ser-229 is mutated to Glu, which also has a relatively larger side chain. Therefore, phosphorylation at Ser-229 in mouse CARM1 could interfere with its dimerization, thus possibly resulting in diminished AdoMet binding and the abrogation of enzymatic activity.
Fig. 6.
S229E mutant CARM1 is impaired in dimerization. (A) Coimmunoprecipitation of HA- and FLAG-tagged CARM1 from MEF20−/− cell extracts transiently transfected with constructs expressing HA- and FLAG-tagged wild-type or mutant CARM1. Cell extracts were directly used in the Western blot experiment (top two blots) to confirm the expression of HA- and FLAG-tagged CARM1. In experiments shown in the lower two blots, HA-CARM1 was first immunoprecipitated from cell extracts with anti-HA antibodies. Then the coimmunoprecipitated FLAG-tagged CARM1 was probed with anti-FLAG antibodies. FLAG-CARM1 was not detected when constructs expressing S229E mutant CARM1 were used (comparing lane 4 with lanes 2 and 3 in the lowest blot). (B) Mouse CARM1 residue S229 lies close to the dimerization interface in the remodeled CARM1 structure. PRMT1 forms homodimers in crystal structure (Right), and the Asn-Asp (N115-D205) interaction was assumed to be critical for stabilizing the dimer status. Residue A114 in PRMT1 (Upper Left) is equivalent to S229A mutant in mouse CARM1 (Lower Left).
To investigate the possibility that CARM1 phosphorylation affects its dimerization and because of the lack of purified phosphorylated CARM1, we tested whether mouse CARM1 S229E is defect in dimerization. In Fig. 6A, we performed coimmunoprecipitation experiments with MEF−/− cells cotransfected with constructs expressing FLAG- and HA-tagged CARM1. After CARM1 was immunoprecipitated with anti-HA antibodies, we probed the immunoprecipitates with anti-FLAG antibodies. When constructs expressing wild-type or S229A mutant CARM1 were used, FLAG-tagged CARM1 was detected from the HA immunoprecipitates (Fig. 6A, lanes 2 and 3). However, with constructs expressing S229E mutant CARM1, no FLAG-tagged CARM1 was detected (Fig. 6A, lane 4 in the bottom blot) in HA immunoprecipitates, although it was detectable in the cell extracts (Fig. 6A, lane 4 in the top blot). Because serine-to-glutamic acid mutation mimics serine phosphorylation, our results support the hypothesis that CARM1 phosphorylation at Ser-229 in mouse interferes with the formation of the CARM1 dimers. The concomitant loss of CARM1 dimerization and MTase activity in the S229E mutant suggests that MTase activity may depend on dimerization. Because we still do not have purified phosphorylated CARM1 and the kinase involved in CARM1 phosphorylation is still unknown, a direct relationship between dimerization and MTase activity has yet to be tested.
Discussion
Understanding how histone arginine methylation is dynamically regulated is important because this “mark” is strongly associated with the activation of gene expression (9). Here, we report on the regulation of protein arginine methylation and ER-dependent transcription by phosphorylation.
Phosphorylated CARM1 was observed from enriched mitotic cells (Fig. 1A), and the phosphorylation site was identified at Ser-229 (mouse/rat, or Ser-228 in human) through phospho-amino acid analysis (Fig. 2). The slower migrating form of CARM1 is subtle but clear and is sensitive to phosphatase treatment (Fig. 1B). The phosphorylated serine we identified is conserved among species, suggesting that the CARM1 phosphorylation might also be evolutionarily conserved. We confirmed that phosphorylation at the identified serine residue represents the main phosphorylation site in CARM1 in vivo (Fig. 2D). The cellular kinases that phosphorylate CARM1 remain to be identified. Phosphorylated CARM1 were detected in COS7 cells and SK-BR3 breast cancer cells, similar to mitotic cells. Phosphorylated CARM1 was not detected in MCF7 cells initially used (Fig. 1B); however, low levels of p-CARM1 can be detected with a different clone of MCF7 cells.
We examined the effect of phosphorylation on CARM1 MTase activity both in vitro and in vivo. We demonstrated that mutation of the phosphorylation site affects the binding to MTase substrate (Fig. 3). Because the S229E mutation is meant to mimic phosphorylation and this mutation inhibits MTase substrate binding whereas wild-type and the S229A mutant did not, we deduced that phosphorylation might negatively regulate MTase activity. Because of the lack of phospho-specific CARM1 antibodies, we have not been able to enrich enough phosphorylated CARM1 to directly analyze its MTase activity. Without knowing the phosphorylation kinase, we also failed to generate phosphorylated CARM1 in vitro. Nevertheless, we compared the MTase activities of wild-type CARM1, S229A, and S229E mutant CARM1 (Fig. 4) and observed reduced MTase activity with S229E mutant CARM1.
To further examine the effect of phosphorylation on CARM1's transactivation of endogenous ER target gene expression, we initially attempted to isolate CARM1 null MEF cells stably expressing all CARM1 mutants. Unfortunately, neither CARM1 S229E- nor CARM1VLD-expressing clones could be selected, although stable clones expressing wild-type CARM1 and CARM1 S229A were successfully obtained. This result indicated that MEF cells could survive with complete loss of CARM1 but not with the constitutive expression of MTase-deficient CARM1 mutants (CARM1 S229E and CARM1VLD). Thus, we performed cotransfection transcription assays (Fig. 5). Our results showed that S229E mutant CARM1 failed to stimulate ER-dependent gene expression in vivo. These results are consistent with our hypothesis that phosphorylation of CARM1 negatively regulates its MTase activity and thereby the transcriptional activity. However, there are other explanations for our discovery that S229E mutant CARM1 failed to stimulate ER-dependent gene expression. For example, phosphorylation of S229 might prevent CARM1 interaction with the coactivator TIF2 or SWI/SNF. Further experiments are required to exclude or confirm such hypotheses.
The potential mechanism that the MTase activity of CARM1 is regulated by phosphorylation is still not clear. We observed that a serine-to-glutamic acid mutation, which mimics the serine phosphorylation, results in the defect of dimer formation (Fig. 6A). This observation is consistent with our modeled CARM1 structure (Fig. 6B) in which the relatively larger side chain of phospho-Ser residue would interfere with a potential residue interaction. The corresponding side-chain interaction in PRMT1 has been proposed to be critical for its dimer formation. Nevertheless, more direct evidence is needed to confirm whether CARM1 phosphorylation regulates its MTase activity through interfering with its dimerization.
Our finding that protein arginine methylation by CARM1 is modulated by phosphorylation provides a link between two major protein epigenetic modification pathways. Cha et al. (35) recently reported that Akt-mediated phosphorylation of EZH2 suppresses methylation of Lys-27 in histone H3 by impeding EZH2 binding to histone H3. These results are analogous to our findings that phosphorylation of CARM1 impedes substrate binding and suppresses its MTase activity. Taken together, it seems that protein methylation, either on lysine or arginine residue, can be regulated by phosphorylation through a similar mechanism.
Materials and Methods
Plasmids, Baculovirus, Retrovirus, and Protein Purification.
CARM1 was subcloned from pSG5-CARM1 plasmid (gift from Michael R. Stallcup, University of Southern California, Los Angeles) to pFastbac-HTb (Invitrogen, Carlsbad, CA). Recombinant CARM1 proteins were expressed in SF9 cells via the baculovirus system and purified through Ni-NTA resin. CARM1 S229A and S229E mutant constructs were generated by using a site-directed mutagenesis kit (Stratagene, La Jolla, CA) and the mutant proteins were similarly purified. Histones were purified from HeLa cells as described (34). For generating retrovirus constructs, CARM1 was subcloned into pLHCX (Clontech, Mountain View, CA) between HindIII and ClaI sites. Phenix cells were transfected with pLHCX-CARM1 to produce retrovirus, which was then used to infect MEF−/− cells. Expression of CARM1 was confirmed by Western blotting.
Phosphoamino Acid Analysis.
HeLa cells were pregrown in phosphate-free DMEM and labeled for 6 h with [32P]orthophosphate (0.8 mCi/ml; 2 ml; ICN Pharmaceutical, Costa Mesa, CA) in the presence of 15 μg/ml nocodazole. Cells were lysed and immunoprecipitated with anti-CARM1 peptide antibody (Upstate Biotech, Lake Placid, NY) or CARM1 antibody preneutralized with synthetic peptide corresponding to amino acids 595–608 of mouse CARM1 (SPMSIPTNTMHYGS). Immune complexes were resolved by 7% SDS/PAGE. The gel was dried on a 3-mm paper filter (Whatman, Middlesex, U.K.) and exposed for autoradiography. The phosphoamino acid analysis procedure was performed as described (36). By comparison of CARM1 immunoprecipitation and control immunoprecipitation, the predicted phosphorylated CARM1 band was excised from the paper and extracted from the gel by using ammonium bicarbonate buffer followed by trichloroacetic acid precipitation. The precipitated CARM1 was oxidized in performic acid and digested with trypsin. The bicarbonate buffer was evaporated by several rounds of lyophilization, and the tryptic peptide was mixed with 1 μg of phosphoamino acid standards containing phosphoserine, threonine, and tyrosine and spotted on a TLC plate (EM Science, Lawrence, KS). Peptides were resolved by electrophoresis and chromatography in two dimensions. The plate was dried and developed with a solution of ethanol containing 0.2% ninhydrin and subsequently heated in an oven at 100°C for 30 min and exposed to autoradiography.
Mass Spectrometric Analysis of Phosphorylated CARM1 Peptide.
CARM1 was immunoprecipitated from 15 mg of HeLa cell extracts made from nocodozole- or hydroxyurea-treated cells. Nonphosphorylated and phosphorylated CARM1 was separated on 7% SDS/PAGE and Coomassie-stained. CARM1 was excised from the gel after destaining, and the digested mixture was analyzed on a MALDI-TOF instrument that allowed the determination of the masses of the peptides in the mixture with high accuracy (10–50 ppm). A Mascot search algorithm (Matrix Science, Boston) was performed to search peptide mass fingerprinting.
Photoaffinity Labeling of CARM1 and Competition by Sinefungin.
UV cross-linking of S-adenosyl-l-[methyl-3H] methionine to CARM1 was performed as described (28). Ten micrograms of CARM1, 3.0 μM [3H]AdoMet (15 Ci/mmol, 66 μM; Amersham Bioscience, Piscataway, NJ), and 5 mM DTT were mixed in 1× PBS and exposed to UV light (254 nm) at a distance of 1 cm for 30 min at 4°C. When sinefungin was used, 200 μM sinefungin (EMD Biosciences, San Diego) was included in the reaction before the addition of [3H]AdoMet. A UVGL-58 UV cross-linker was used (UVP, Upland, CA). After cross-linking, samples were separated on 7% SDS/PAGE, Commassie-stained, destained, and immersed into Amplify solution (Amersham Bioscience) for 15 min before drying. The dried gel was exposed to XAR film (Kodak, Rochester, NY) overnight.
Transient Transfection of CARM1 MEF−/− Cells, Coimmunoprecipitation, and Luciferase Assay.
CARM1 knockout MEFs (20) were cultured in DMEM supplemented with 10% FBS and plated into 12-well dishes in phenol red-free DMEM supplemented with 10% charcoal-stripped FBS 1 day before transfection. Transfections were performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. For coimmunoprecipitation assays, cells were transfected with 200 ng of pCMX-FLAG-CARM1 and/or pSG5-HA-CARM1 constructs expressing either wild-type or mutants CARM1 with either FLAG or HA tag. CARM1 were detected with anti-HA or anti-FLAG antibodies. For luciferase assays, cells were transfected with 5 ng of pCMX-hERα, 200 ng of CMX-TIF2, and/or 200 ng of CMX-CARM1 wild type or mutants and 100 ng of β-gal constructs. The medium was replaced with fresh medium containing 100 nM estradiol or ethanol 1 day before luciferase levels were measured and normalized with β-gal activities.
ER Endogenous Gene Expression in Transiently Transfected MEF20−/− by Real-Time PCR.
Approximately 24 h before transfection, 400,000 MEF20−/− cells were seeded into 6-cm culture dishes. The cells in each dish were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. For each dish, 8 μg of pCMV-Flag, 2 μg of pCMV-ERα and 6 μg of pCMV-Flag-Carm1 WT, 2 μg of pCMV-ERα and 6 μg of pCMV-Flag-CARM1 S229E, 2 μg of pCMV-ERα, and 6 μg of pCMV-Flag-CARM1 VLD mutant were transfected. Five hours posttransfection, the cells were washed once with PBS and grown in phenol red-free DMEM supplemented with 10% charcoal-striped FBS. Forty-eight hours posttransfection, the cells were treated with 100 nM estradiol and harvested 4 h later. Total RNA was extracted with TRIzol (Invitrogen) according to the manufacturer's protocol. The total RNA was treated with RQ1 RNase-Free DNase (Promega, Madison, WI), and the first-strand cDNA was synthesized with random primers and SuperScript III reverse transcriptase (Invitrogen). The quantitative RT-PCR was performed with a SYBR GREEN PCR Master Mix (Applied Biosystems, Foster City, CA) and analyzed with DNA Engine OPTICON 2 (BioRad, Hercules, CA). The values were corrected by 36B4, which is an invariant control (37). The primer pairs used were as follows: Egfr QF1 (5′-CTGTACCTATGGATGTGCTG-3′) and Egfr QR1 (5′-ACCACCACTATGAAGAGGAG-3′) for Egfr; Ebag9 QF1 (5′-CTAAGCAGACAGATGTGGAG-3′) and Ebag9 QR1 (5′-GTCATGTCCTTGAAGTAGTCAG-3′) for Ebag9; 36B4-forward (5′-AGATGCAGCAGATCCGCAT-3′) and 36B4-reverse (5′-GTTCTTGCCCATCAGCACC-3′) for 36B4. All PCR conditions were as follows: initial denaturation at 95°C for 15 min, 35 cycles of 95°C for 15 s, 55°C for 30 s, 72°C for 30 s followed by final extension at 72°C for 8 min.
Acknowledgments
We thank C. Park and F. Wolfgang for peptide analysis, J. Meisenhelder for help with phosphoamino acid analysis, M. Bedford (University of Texas M. D. Anderson Cancer Center, Houston) for CARM1 knockout MEFs, M. R. Stallcup for pSG5-CARM1 plasmid, R. Evans for support and advice in the early stage of the project, and M. Hoffmann, E. Powell, H. Chen, J. Mertz, and L. Chen for critical reading of the manuscript. This study was supported by National Institute of Health Grant P30CAD1452 and a grant from the Pardee Foundation (to W.X.) and National Institutes of Health Grant GM068680 (to X.C.). P.K. was supported by National Institutes of Health Grant T32 CA009135.
Abbreviations
- AdoMet
S-adenosylmethionine
- CARM1
coactivator-associated arginine methyltransferase 1
- ER
estrogen receptor
- GRIP1
glucocorticoid receptor interacting protein 1
- MEF
mouse embryonic fibroblast
- NR
nuclear receptor
- PRMT
protein arginine methyltransferase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Hermanson O, Glass CK, Rosenfeld MG. Trends Endocrinol Metab. 2002;13:55–60. doi: 10.1016/s1043-2760(01)00527-6. [DOI] [PubMed] [Google Scholar]
- 2.Xu W. Biochem Cell Biol. 2005;83:418–428. doi: 10.1139/o05-057. [DOI] [PubMed] [Google Scholar]
- 3.Rosenfeld MG, Glass CK. J Biol Chem. 2001;276:36865–36868. doi: 10.1074/jbc.R100041200. [DOI] [PubMed] [Google Scholar]
- 4.McKenna NJ, O'Malley BW. Endocrinology. 2002;143:2461–2465. doi: 10.1210/endo.143.7.8892. [DOI] [PubMed] [Google Scholar]
- 5.Narlikar GJ, Fan HY, Kingston RE. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 6.Jaber BM, Mukopadhyay R, Smith CL. J Mol Endocrinol. 2004;32:307–323. doi: 10.1677/jme.0.0320307. [DOI] [PubMed] [Google Scholar]
- 7.Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 8.Kim JH, Li H, Stallcup MR. Mol Cell. 2003;12:1537–1549. doi: 10.1016/s1097-2765(03)00450-7. [DOI] [PubMed] [Google Scholar]
- 9.Stallcup MR. Oncogene. 2001;20:3014–3020. doi: 10.1038/sj.onc.1204325. [DOI] [PubMed] [Google Scholar]
- 10.Wysocka J, Allis CD, Coonrod S. Front Biosci. 2006;11:344–355. doi: 10.2741/1802. [DOI] [PubMed] [Google Scholar]
- 11.Qi C, Chang J, Zhu Y, Yeldandi AV, Rao SM, Zhu YJ. J Biol Chem. 2002;277:28624–28630. doi: 10.1074/jbc.M201053200. [DOI] [PubMed] [Google Scholar]
- 12.Ma H, Baumann CT, Li H, Strahl BD, Rice R, Jelinek MA, Aswad DW, Allis CD, Hager GL, Stallcup MR. Curr Biol. 2001;11:1981–1985. doi: 10.1016/s0960-9822(01)00600-5. [DOI] [PubMed] [Google Scholar]
- 13.Koh SS, Chen D, Lee YH, Stallcup MR. J Biol Chem. 2001;276:1089–1098. doi: 10.1074/jbc.M004228200. [DOI] [PubMed] [Google Scholar]
- 14.Schurter BT, Koh SS, Chen D, Bunick GJ, Harp JM, Hanson BL, Henschen-Edman A, Mackay DR, Stallcup MR, Aswad DW. Biochemistry. 2001;40:5747–5756. doi: 10.1021/bi002631b. [DOI] [PubMed] [Google Scholar]
- 15.Bauer UM, Daujat S, Nielsen SJ, Nightingale K, Kouzarides T. EMBO Rep. 2002;3:39–44. doi: 10.1093/embo-reports/kvf013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee YH, Koh SS, Zhang X, Cheng X, Stallcup MR. Mol Cell Biol. 2002;22:3621–3632. doi: 10.1128/MCB.22.11.3621-3632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu W, Chen H, Du K, Asahara H, Tini M, Emerson BM, Montminy M, Evans RM. Science. 2001;294:2507–2511. doi: 10.1126/science.1065961. [DOI] [PubMed] [Google Scholar]
- 18.Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. Proc Natl Acad Sci USA. 2005;102:3611–3616. doi: 10.1073/pnas.0407159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chevillard-Briet M, Trouche D, Vandel L. EMBO J. 2002;21:5457–5466. doi: 10.1093/emboj/cdf548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yadav N, Lee J, Kim J, Shen J, Hu MC, Aldaz CM, Bedford MT. Proc Natl Acad Sci USA. 2003;100:6464–6468. doi: 10.1073/pnas.1232272100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- 22.Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, et al. Science. 2004;306:279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 23.Cuthbert GL, Daujat S, Snowden AW, Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD, Tempst P, Bannister AJ, Kouzarides T. Cell. 2004;118:545–553. doi: 10.1016/j.cell.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 24.Xu W, Cho H, Kadam S, Banayo EM, Anderson S, Yates JR, 3rd, Emerson BM, Evans RM. Genes Dev. 2004;18:144–156. doi: 10.1101/gad.1141704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DiRenzo J, Shang Y, Phelan M, Sif S, Myers M, Kingston R, Brown M. Mol Cell Biol. 2000;20:7541–7549. doi: 10.1128/mcb.20.20.7541-7549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sif S, Stukenberg PT, Kirschner MW, Kingston RE. Genes Dev. 1998;12:2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muchardt C, Reyes JC, Bourachot B, Leguoy E, Yaniv M. EMBO J. 1996;15:3394–3402. [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng D, Yadav N, King RW, Swanson MS, Weinstein EJ, Bedford MT. J Biol Chem. 2004;279:23892–23899. doi: 10.1074/jbc.M401853200. [DOI] [PubMed] [Google Scholar]
- 29.O'Lone R, Frith MC, Karlsson EK, Hansen U. Mol Endocrinol. 2004;18:1859–1875. doi: 10.1210/me.2003-0044. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X, Cheng X. Structure (London) 2003;11:509–520. doi: 10.1016/s0969-2126(03)00071-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Zhou L, Cheng X. EMBO J. 2000;19:3509–3519. doi: 10.1093/emboj/19.14.3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weiss VH, McBride AE, Soriano MA, Filman DJ, Silver PA, Hogle JM. Nat Struct Biol. 2000;7:1165–1171. doi: 10.1038/82028. [DOI] [PubMed] [Google Scholar]
- 33.Cheng X, Collins RE, Zhang X. Annu Rev Biophys Biomol Struct. 2005;34:267–294. doi: 10.1146/annurev.biophys.34.040204.144452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu W, Cho H, Evans RM. Methods Enzymol. 2003;364:205–223. doi: 10.1016/s0076-6879(03)64012-7. [DOI] [PubMed] [Google Scholar]
- 35.Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT, Ping B, Otte AP, Hung MC. Science. 2005;310:306–310. doi: 10.1126/science.1118947. [DOI] [PubMed] [Google Scholar]
- 36.Meisenhelder J, Hunter T, van der Geer P. In: Current Protocols in Protein Science. Coligan JE, Dunn BM, Ploegh HL, Speicher DW, Wingfield PT, editors. Vol 2. New York: Wiley; 1999. pp. 13.9.1–13.9.27. [Google Scholar]
- 37.Takaishi K, Duplomb L, Wang MY, Li J, Unger RH. Proc Natl Acad Sci USA. 2004;101:7106–7111. doi: 10.1073/pnas.0401715101. [DOI] [PMC free article] [PubMed] [Google Scholar]