Abstract
Specification of sea urchin embryo micromeres occurs early in cleavage, with the establishment of a well defined regulatory state. The architecture of the gene regulatory network controlling the specification process indicates that transcription of the initial tier of control genes depends on a double-negative gate. A gene encoding a transcriptional repressor, pmar1, is activated specifically in micromeres, where it represses transcription of a second repressor that is otherwise active globally. Thus, the micromere-specific control genes, which are the target of the second repressor, are expressed exclusively in this lineage. The double-negative specification gate was logically required from the results of numerous prior experiments, but the identity of the gene encoding the second repressor remained elusive. Here we show that hesC is this gene, and we demonstrate experimentally all of its predicted functions, including global repression of micromere-specific regulatory genes. As logically required, blockade of hesC mRNA translation and global overexpression of pmar1 mRNA have the same effect, which is to cause all of the cells of the embryo to express micromere-specific genes.
Keywords: skeletogenic micromeres, transcriptional repression
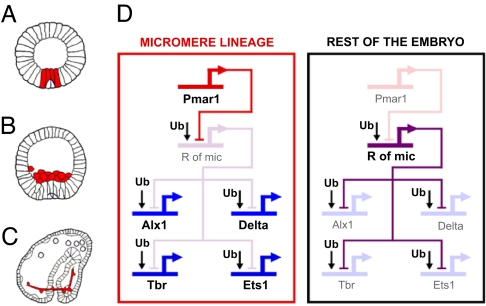
The genomic regulatory code for specification of endomesoderm in the sea urchin embryo is represented as a gene regulatory network (GRN), which explains the mechanism by which distinct regulatory states are deployed in different territories of the developing embryo (for reviews, see refs. 1–4; for current version see http://sugp.caltech.edu/endomes). One portion of this GRN pertains to the specification of the micromeres, which arise at the unequal fourth cleavage at which the four micromeres are segregated off from the vegetal pole of the egg. The large daughter cells of the micromeres arising at the next cleavage are the founder cells of the skeletogenic micromere lineage. This lineage is the sole normal source of the embryonic biomineral skeleton, a distinct synapomorphic feature of echinoid embryos and larvae, and it also produces essential short-range signals required for other aspects of endomesoderm specification (5–7). Three particular developmental events that are relevant for what follows are (i) the expression of the Delta-signaling ligand on the surfaces of the micromere descendants during the early blastula stage (Fig. 1A); (ii) their ingression into the blastocoel at the late blastula stage (Fig. 1B), after which they are known as primary mesenchyme cells; and (iii) their expression of the biomineralization and cytoskeletal genes, which enable them to generate the skeleton (Fig. 1C) (8).
Fig. 1.
Key elements of the GRN model for the early specification of the skeletogenic micromere lineage. The model is based on ref. 9, with subsequent updates (10), as reviewed in refs. 1 and 4. (A–C) Sea urchin embryo drawings (adapted from ref. 2) at the early blastula stage (12–15 h after fertilization; A), at the mesenchyme blastula stage (24 h; B), and at the late gastrula stage (48 h; C). The cells of the skeletogenic micromere lineage at each stage are depicted in red. (D) The GRN model (corresponding to cleavage and blastula stages). Active genes are represented in strong color and bold font. Inactive genes are represented in dim color. Within the micromere lineage, pmar1 is active and represses the predicted gene r of mic. The delta, alx1, ets1, and tbr genes are allowed to be zygotically expressed in this domain. In the rest of the embryo, r of mic keeps delta, alx1, ets1, and tbr silent.
Immediately after the fourth-cleavage micromeres are born, they express a gene, pmar1, in response to maternally localized factors (9). This gene encodes a transcriptional repressor of the paired homeodomain family. In the GRN model, pmar1 serves as the linchpin of a proposed double-negative gate controlling the institution of the micromere regulatory state. The second component of this gate is predicted to be a pmar1 target gene that encodes another transcriptional repressor. This gene must also be zygotically expressed, but it would be transcribed everywhere in the embryo except in the micromere lineage, where it is subject to repression by Pmar1. There are eight targets predicted for it in the GRN, of which the most important for present purposes are the genes encoding the Delta ligand and three regulatory genes, tbr, ets1, and alx1. These three genes lie upstream of all the rest of the micromere regulatory apparatus. In this manner, the double-negative gate would ensure expression of this apparatus exclusively in the micromere lineage. Because its identity was unknown, the second repressor has been referred to in the GRN as Repressor of Micromeres (R of mic). Its existence and properties are specifically implied by the two following perturbation experiments (9, 10). First, if expression of pmar1 is forced to occur globally (by injection into the egg of the mRNA), then the delta, tbr, ets1, alx1, and downstream genes are transcribed in all cells of the embryo, and all cells thereby adopt skeletogenic micromere lineage fate. Second, exactly the same outcome follows if an mRNA encoding a dominantly repressive Engrailed fusion of the Pmar1 protein is injected. It follows that the pmar1 gene product naturally acts as a repressor (also indicated by its sequence); that delta, tbr, ets1, and alx1 are controlled by ubiquitous activators; and that localization of expression of these genes to the micromere lineage in normal embryos depends on their repression by R of mic everywhere else in the embryo (Fig. 1D).
To prove the existence of the double-negative gate for micromere lineage specification in the GRN model, it is necessary to find the gene playing the role of the predicted R of mic and establish that its expression and functions are also as predicted. The r of mic gene should encode a transcriptional repressor, and it should have three distinct characteristics: (i) Its zygotic expression should be transcriptionally repressed by Pmar1; (ii) it should be zygotically expressed everywhere except in the micromere lineage by the time zygotic expression of delta, alx1, ets1, and tbr starts; and (iii) the outcome of knocking down its expression should be similar to forcing global Pmar1 expression (i.e., all cells of the embryo should adopt micromere lineage specification and express delta, alx1, ets1, and tbr).
Results
Genomic Screen for Candidate r of mic Genes.
The Strongylocentrotus purpuratus genome sequence enabled consideration of all sea urchin genes encoding transcription factors in our search for r of mic. The total number of annotated transcription factors in this genome, excluding C2H2 zinc finger genes, is 283 (11), and the total number of predicted C2H2 zinc fingers (some of which encode transcription factors) is 377 (12). The levels of mRNA expression at several developmental time points were measured for all of these 660 genes (12–16). We selected as r of mic candidates all putative regulatory genes for which at least 200 transcripts were detected per embryo at 12 h after fertilization, when delta, alx1, ets1, and tbr are all zygotically transcribed. This threshold is conservative (low) given that r of mic must be expressed in most of the embryo, or in 100 to 150 cells, at this time. This selection resulted in a list of ≈100 candidate genes. We excluded those that are maternally and not zygotically expressed up to 12 h after fertilization and all previously studied transcription factors for which enough information was available to confirm that they could not be r of mic. The surviving list now contained 46 candidates [see supporting information (SI) Table 1].
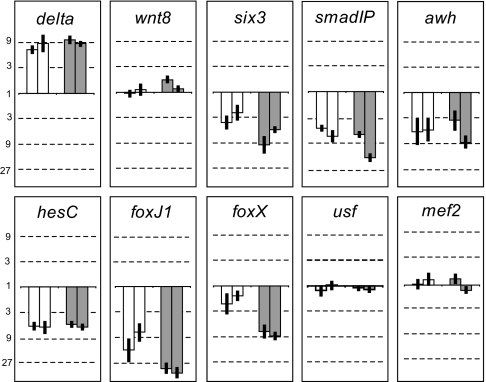
Because r of mic should be transcriptionally repressed by Pmar1, we screened the 46 candidate genes for down-regulation on forced expression of Pmar1 in the whole embryo [Fig. 2 and SI Fig. 6; mRNA overexpression (MOE)]. The effect of pmar1 MOE on the level of transcripts of each r of mic candidate gene was measured at 9 and 12 h after fertilization by using quantitative PCR. The delta gene was included in the screen as a control. As expected, in the two experiments performed, delta was significantly up-regulated (≥3-fold changes in transcript levels were considered significant) at both 9 and 12 h after fertilization (Fig. 2). This result indicated that r of mic must have been down-regulated at both time points in these two experiments. Five of the 46 regulatory genes tested were found to be significantly down-regulated at both time points in the two experiments performed. These genes were six3, smadIP, awh, hesC, and foxJ1 (Fig. 2).
Fig. 2.
pmar1 MOE screen. Graphs showing fold change in mRNA expression for r of mic candidate genes on overexpression of pmar1 mRNA. delta and wnt8 were used as positive and negative controls, respectively. A fold change of 1 (solid line) indicates no change. The numbers situated above 1 indicate a fold increase, whereas the numbers situated below 1 indicate a fold decrease (in logarithmic scale). A 3-fold or greater change was considered to be significant. Open and filled bars represent data from samples at 9 and 12 h of development, respectively. For each type of bar, the two bars correspond to two independent batches of embryos. Error bars represent the standard deviation from three independent measurements on the same sample. Results for 8 of the 46 r of mic candidate genes are shown here (see SI Fig. 6 for data on the remaining 38 candidates).
Among these five transcriptional regulatory genes, hesC particularly caught our attention. Its level of mRNA expression at 9 and 12 h is highest of all five (data not shown). In addition, HesC is a basic helix–loop–helix transcription factor belonging to the Hairy/E(spl) family, and almost all transcription factors of this family are known to function as repressors (17). That HesC belongs to this family is supported by a phylogenetic analysis (16) and by the fact that it contains the two characteristic domains of the family: the C-terminus WRPW motif (used to recruit TLE/Grg/Groucho and mediate transcriptional repression) (18, 19) and the Orange domain. We therefore focused on hesC.
Temporal and Spatial Expression of HesC.
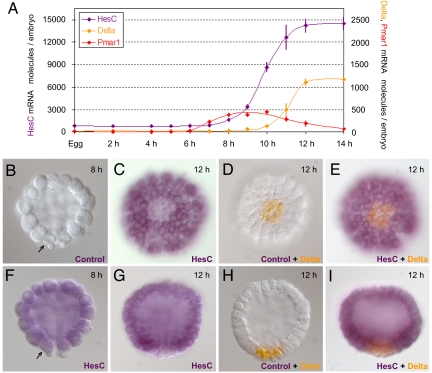
The spatial and temporal patterns of expression predicted for the r of mic gene are unique. The time course of hesC expression was determined at 1- to 2-h intervals by means of quantitative PCR (Fig. 3A). This experiment showed that hesC is maternally expressed, but only at low levels. The level of hesC transcript then increases steeply between 8 and 12 h after fertilization, indicating zygotic transcription. To compare the temporal expression of hesC to that of up- and downstream genes in the double-negative gate, we measured the levels of pmar1 and delta mRNA in the same embryo samples (Fig. 3A). As would be expected for r of mic, the zygotic expression of hesC starts before delta is detected, and it occurs while pmar1 mRNA is present.
Fig. 3.
HesC temporal and spatial expression pattern. (A) Measurements of hesC mRNA molecules per embryo (purple) are compared with those of pmar1 (red) and delta (orange) at the indicated developmental time points. Error bars represent the standard deviation from three individual measurements on the same sample. (B–I) Images of embryos on which WMISH (B, C, F, and G) or double-WMISH (D, E, H, and I) was performed. The developmental stage of each embryo is indicated at the upper right corner. (B and F–I) Side views, with a vegetal side at the bottom. (C–E) Vegetal views. The arrows in B and F point at one of the two visible skeletogenic micromere cells. Probes used are indicated at the lower right corner. Control, probe used to control for nonspecific staining.
Whole-mount in situ hybridization (WMISH) provided strong evidence. At 8 h, the steep zygotic expression of hesC had just started and at this time, hesC mRNA was found essentially everywhere in the embryo, including the micromere lineage (Fig. 3F; compare to control in Fig. 3B). The two small cells at the vegetal pole of the embryo, which showed weaker staining than the rest of the embryo, are the small micromeres, that do not belong to the skeletogenic micromere lineage, which is the subject of this article. At 12 h, the steep zygotic increase in hesC expression attained its plateau value (Fig. 3A), and delta mRNA was already present. The expression of hesC had changed dramatically, in that this transcript disappeared from a set of 12 cells at the vegetal pole (Fig. 3 C and G), whereas it continued to be expressed everywhere else. Exactly 12 cells expressed delta mRNA at this time (Fig. 3 D and H), and the same number of cells are now in the micromere lineage. To confirm that the 12 cells lacking hesC mRNA expression corresponded to the 12 micromere lineage cells, we performed double-WMISH. As shown in Fig. 3 E and I, every cell of the embryo expressed either hesC (purple) or δ (orange), but no cell expressed both genes. Therefore, zygotic expression of hesC had occurred everywhere in the embryo except the micromere lineage, precisely as predicted for r of mic.
Functional Analysis of hesC.
The predicted function of r of mic is to repress micromere lineage specification. Thus, if hesC is r of mic, blocking its translation should result in all cells of the embryo becoming specified similarly to skeletogenic micromeres, the same as when global pmar1 expression is forced to occur (9). We used a morpholino antisense oligonucleotide (MASO) targeting hesC mRNA for this experiment. The striking effect of this perturbation on the morphology of the developing embryos is shown in Fig. 4. Up to the blastula stage, hesC MASO embryos were indistinguishable from unperturbed embryos (Fig. 4 A and C). In both, ingression of primary mesenchyme cells into the blastocoel started ≈20 h after fertilization (data not shown). However, in unperturbed embryos, primary mesenchyme cell ingression had been completed by 24 h after fertilization (Fig. 4B), whereas in hesC MASO embryos, ingression of cells continued until the blastocoel was essentially full (Fig. 4D). All, or almost all, cells of hesC MASO embryos thus behave in a way normally unique to the micromere lineage. Importantly, at all three stages, hesC MASO embryos look strikingly similar to pmar1 MOE embryos (Fig. 4 C–F; data not shown for 20-h stage).
Fig. 4.
Morphology of HesC MASO embryos. (A–D) Images of embryos that were either unperturbed (A and B) or perturbed (C and D) by HesC MASO. (E and F) Pmar1 MOE embryos from a different batch are also shown for comparison. (A, C, and E) Blastula stage embryos 16 h after fertilization. (B, D, and F) Late mesenchyme blastula stage embryos 24 to 26 h after fertilization.
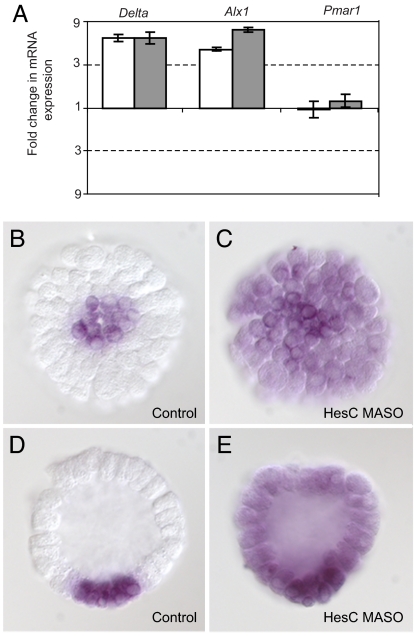
We next assessed the effect of HesC MASO perturbation on the levels of mRNA of delta, alx1, ets1, and tbr. If hesC is r of mic, the prediction (Fig. 1D) is that these genes will now be allowed to be expressed in all cells, and their level of transcript should therefore increase, as occurs in pmar1 MOE embryos (9, 10). Fig. 5 and SI Table 2 show this result. By 12 h after fertilization, the amount of transcript of delta and alx1 had increased 4- to 7-fold above normal in the two experiments performed (Fig. 5A), and by 24 h, that of ets1 and tbr had similarly increased (SI Table 2). The level of expression of pmar1 was not affected, indicating that the up-regulation of these genes was not caused by any change in pmar1 (Fig. 5A).
Fig. 5.
Effect of HesC MASO on micromere lineage genes. (A) Fold change in delta, alx1, and pmar1 mRNA expression in HesC MASO embryos relative to unperturbed embryos (12 h after fertilization). Fold change representation is as in Fig. 2. Open and filled bars indicate two independent batches of embryos. (B–E) Images of embryos (12 h after fertilization) on which WMISH was performed by using delta probe. Vegetal view (B) and side view (D) of an unperturbed embryo and vegetal view (C) and side view (E) of an HesC MASO embryo are shown.
The derepression of micromere lineage specification occurs in all cells of hesC MASO embryos. This finding is illustrated for the delta marker, as shown in Fig. 5 B–E. In unperturbed embryos, delta mRNA is localized to the micromere lineage (Fig. 5 B and D), whereas in hesC MASO embryos, it is detected throughout the whole embryo (Fig. 5 C and E). Thus, HesC functions to repress micromere lineage specification in all cells other than the micromere lineage, the defining characteristic of the predicted r of mic.
Discussion
The GRN Model Prediction and the Evidence.
As regions of a GRN approach completion, the levers of logic can be used to generate precise predictions of missing components. As described elsewhere, the portion of the sea urchin endomesoderm GRN that pertains to specification and initial differentiation of the skeletogenic micromere domain now includes every regulatory gene predicted in the genomic sequence that is expressed specifically in the micromere lineage up to the onset of gastrulation (see http://sugp.caltech.edu/endomes), although some linkages among these genes may remain to be determined. Further, the GRN does not identify genes expressed ubiquitously in the embryo, which provide inputs to micromere genes, and it includes only a sample of the terminal skeletogenic differentiation genes. Nonetheless, it should include all, or almost all, genes encoding transcription factors that are causally responsible for specification of the skeletogenic regulatory state. From the GRN analysis came the prediction of the double-negative gate shown in Fig. 1D (9), and we have now identified the predicted missing component of this gate: The r of mic gene of the GRN model is hesC. In retrospect, the exact match between the predicted behavior of r of mic and the observed behavior of hesC is remarkable. Both the unique pattern of expression of hesC, which is not reproduced by any other known gene in this embryo, and the unique effects of preventing its expression are those required by the double-negative gate model in Fig. 1D. No additional players are likely to be inserted in the specification gate of Fig. 1D because manipulation of either component, pmar1 overexpression or hesC underexpression, suffices to transform the whole embryo into cells specified as skeletogenic mesenchyme. In either perturbation, all cells (i) express the regulatory state of the skeletogenic micromere lineage (i.e., they transcribe the delta, alx1, tbr, and ets1 genes, normally at this stage specific to the skeletogenic micromere lineage; in the pmar1 overexpression, they even express terminal differentiation genes, such as sm50, not examined here); (ii) ingress into the blastocoel; and (iii) assume a mesenchymal form (Figs. 4 and 5 and SI Table 2) (9, 10).
Although this remains to be finally authenticated by identification of the cis-regulatory target sites, HesC interactions with the target genes of Fig. 1D are likely to be direct, as is likely to be the interaction of Pmar1 with the hesC regulatory apparatus. First, both genes encode proteins that contain transcriptional repression domains (see Results for HesC and ref. 9 for Pmar1). Second, the kinetics with which the gate operates almost precludes any intervening steps. In sea urchin embryos at 15°C, it requires ≈2–3 h for a regulatory gene to be activated, its product to be translated and transported to the nucleus, and a target gene to respond (20). We show here (Fig. 3A) that zygotic expression of hesC starts only ≈2 h after that of pmar1, and the zygotic expression of delta starts only ≈2 h after that of hesC. Third, we have carried out a cis-regulatory analysis of the delta gene, the key relevant results of which are summarized in SI Fig. 7. For the present discussion, the most important findings are: (i) the cis-regulatory module controlling delta expression in the micromere lineage responds to pmar1 overexpression, just as does the endogenous delta gene (i.e., by ubiquitous ectopic expression), and also responds in just the same way to hesC MASO, again like the endogenous gene; however (ii), a region can be isolated from it that causes ubiquitous expression in unperturbed embryos and is not subject to repression, whereas other regions repress the ectopic expression and are subject to pmar1 control. These results exclude the possibility that there is an indirect effect such that HesC represses another gene that is in turn responsible for delta activation because then the cis-regulatory DNA regions responsible for activation and repression would not be physically separable. Because HesC has a WRPW sequence, like all of its orthologues, it must act as a repressor, and we have shown that it does not interfere with pmar1 expression. These observations strongly indicate repressive interaction at the delta cis-regulatory module.
The Double-Negative Gate.
The main feature of this mechanism is the use of two repressors in regulatory tandem, and nonlocalized activators to produce a highly confined spatial pattern of gene expression. This mechanism is not uncommon; for example, the dorsal–ventral GRN for the early Drosophila embryo (21) affords several examples that are in essence similar. An alternative first step would be the highly localized expression of activators. This mechanism is common in later development, but in the early embryo, the boundaries of expression domains are often controlled negatively by the activation of repressors (1). In the sea urchin embryo, the known maternal regulatory transcripts are all globally distributed (12, 15, 16). Early on, before territorial regulatory states have been established, regional activation of repressors in response to initial anisometric cues is as parsimonious a strategy as regional activation of activators (1). An additional advantage of the double-negative gate is that it provides, de facto, the active repression of regulatory states outside the correct domain of their expression. Thus, it acts as an exclusion effect (22), actively ensuring silence of target genes in ectopic locations while ensuring their expression in correct locations.
Evolutionary Implications.
Sea urchins are the only echinoderm class that produces an embryo/larva skeleton from a precociously specified micromere lineage. Thus, the regulatory apparatus for skeletogenic micromere specification, including the double-negative gate, arose in this lineage. An idea proposed earlier is that generation of the larval skeleton evolved as a cooption of the gene regulatory program for the production of the adult calcite skeleton (9, 23). The hesC–pmar1 double-negative gate provides in principle a particularly economical means for highjacking the downstream skeletogenic regulatory machinery. Part of the circuitry is likely to have been already available. The Hairy/E(spl) family factors are used to repress the delta gene across the Bilateria (e.g., in both insect and vertebrate nervous system development) (24, 25). Sea urchin HesC repression of delta may indicate the inclusion in the cooption process of an ancient widespread plug-in (i.e., a conserved GRN linkage that is used in multiple, entirely unrelated, developmental contexts) (26). Now that the regulatory players are all in hand and most of their roles known, it should be possible to experimentally explore the evolution of the sea urchin skeletogenic specification by synthetically recreating the regulatory steps that led to its existence.
Materials and Methods
Animals, pmar1 MOE, and hesC MASO.
pmar1 was overexpressed (i.e., its expression was forced in all cells of the embryos) by microinjecting pmar1 mRNA into fertilized eggs. Microinjection solutions were prepared containing 25 ng/μl of pmar1 mRNA and 0.12 M KCl.
Translation of HesC transcripts was blocked by microinjection of hesC MASO into fertilized eggs. MASO was synthesized (Gene Tools, Philomath, OR) complementary to the sequence of the first 25 bp of the coding region of hesC. The sequence of the oligonucleotide was: 5′-GTTGGTATCCAGATGAAGTAAGCAT-3′. Microinjection solutions were prepared containing 0.12 M KCl and 100, 250, or 500 μM hesC MASO.
Gametes from S. purpuratus were microinjected as described (27). We aimed at microinjecting a volume of ≈10 pl. Unperturbed embryos from the same batch were used as a control. Living embryos were visualized at chosen developmental time points on an Axioscope 2 Plus microscope (Carl Zeiss, Hallbergmoos, Germany) equipped with the recording device AxioCam MRm (Carl Zeiss).
Quantification of mRNA.
An RNeasy Micro Kit (74004; Qiagen, Valencia, CA) was used to isolate RNA from samples of ≈100 embryos as described in the manufacturer's manual. cDNA was prepared from these samples by RT-PCR. The iScript cDNA Synthesis Kit (170–8891; Bio-Rad, Hercules, CA) was used for this purpose.
Quantitative PCR was conducted as described (27) by using primer sets designed to produce amplicons of 125 to 150 bp (for primer sequences, see http://sugp.caltech.edu/resources/methods/q-pcr.psp). Amplification reactions were analyzed on an ABI 7900HT Fast Real-Time PCR System by using SYBR Green chemistry (iScript One-Step RT-PCR Kit; Bio-Rad). Levels of ubiquitin mRNA are known to remain relatively constant (≈220,000 molecules per embryo) during the relevant developmental stages (28, 29) and were used as an internal standard to determine the levels of mRNA per embryo of all other genes.
WMISH.
Digoxigenin (DIG)-labeled RNA probes were prepared as described (30). A DIG-labeled HesC probe was transcribed from the HesC cDNA clone yde51c06 (CX199264; from a S. purpuratus EST library) kindly provided by James Coffman (Mount Desert Island Biological Laboratory, Salisbury Cove, ME). A sense DIG-labeled control probe was transcribed from the same clone, which does not recognize any known or predicted transcript. Dinitrophenol (DNP)-labeled RNA delta probe was prepared as described (31) by using the same plasmid as used for the DIG-labeled delta probe of ref. 9.
WMISH was performed by using a standard method, as described in refs. 32 and 33, with minor modifications (Sagar Damle and E.H.D., unpublished data). Hybridization reaction and washes were carried out at 65°C. Concentration of probe in hybridization reaction was 1 ng/μl. Antibody incubation was carried out containing a 1,000-fold dilution of anti-DIG antibody (Fab fragments; Roche Diagnostics, Indianapolis, IN) for DIG-labeled probes or anti-DNP antibody-alkaline phosphatase (Mirus, Madison, WI) for DNP-labeled probes.
Double-WMISH protocol (from Sagar Damle and E.H.D., unpublished data) was based on the above protocol for WMISH and the double-WMISH protocol described earlier (34). Steps before the hybridization reaction were as described above for WMISH. The hybridization reaction was carried out containing two probes (1 ng/μl each): a DNP-labeled probe and a DIG-labeled probe. Anti-DIG antibody was used for the first antibody incubation. The first staining reaction (purple) was then carried out as described above for the WMISH protocol, with NBT (N-6876; Sigma–Aldrich, St. Louis, MO)/BCIP (B-8503; Sigma–Aldrich). The staining reaction and antibody activity were stopped as in ref. 34. Anti-DNP antibody-alkaline phosphatase was used in the second antibody incubation. The second staining reaction (orange) was similar to the first one, except that INT (1–8377; Sigma–Aldrich)/BCIP was used instead of NBT/BCIP.
Supplementary Material
Acknowledgments
We thank Jina Yun for her enormous help in the pmar1 screen; Mary Wahl for carrying out the experiment in SI Table 2; Dr. Qiang Tu for help in alignments and phylogenetic analyses; Dr. Joel Smith for helpful comments on the manuscript; Dr. Jim Coffman for providing an HesC cDNA clone; Pat Leahy for taking care of the sea urchins used for this work; and Sagar Damle for his extremely useful instruction in performing WMISHs and double-WMISHs. This work was supported by National Institutes of Health Grant HD-37105 (to E.H.D.).
Abbreviations
- DIG
digoxigenin
- DNP
dinitrophenol
- GRN
gene regulatory network
- MASO
morpholino antisense oligonucleotide
- MOE
mRNA overexpression
- WMISH
whole-mount in situ hybridization.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705324104/DC1.
References
- 1.Davidson EH. The Regulatory Genome: Gene Regulatory Networks In Development and Evolution. New York: Academic; 2006. [Google Scholar]
- 2.Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, et al. Science. 2002;295:1669–1678. doi: 10.1126/science.1069883. [DOI] [PubMed] [Google Scholar]
- 3.Oliveri P, Davidson EH. Curr Opin Genet Dev. 2004;14:351–360. doi: 10.1016/j.gde.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Levine M, Davidson EH. Proc Natl Acad Sci USA. 2005;102:4936–4942. doi: 10.1073/pnas.0408031102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McClay D, Peterson R, Range R, Winter-Vann A, Ferkowicz M. Development (Cambridge, UK) 2000;127:5113–5122. doi: 10.1242/dev.127.23.5113. [DOI] [PubMed] [Google Scholar]
- 6.Sweet H, Hodor P, Ettensohn C. Development (Cambridge, UK) 1999;126:5255–5265. doi: 10.1242/dev.126.23.5255. [DOI] [PubMed] [Google Scholar]
- 7.Sweet HC, Gehring M, Ettensohn CA. Development (Cambridge, UK) 2002;129:1945–1955. doi: 10.1242/dev.129.8.1945. [DOI] [PubMed] [Google Scholar]
- 8.Livingston BT, Killian C, Wilt F, Cameron A, Landrum M, Ermolaeva O, Sapojnikov V, Maglott D, Buchanan A, Ettensohn C. Dev Biol. 2006;300:335–348. doi: 10.1016/j.ydbio.2006.07.047. [DOI] [PubMed] [Google Scholar]
- 9.Oliveri P, Carrick DM, Davidson EH. Dev Biol. 2002;246:209–228. doi: 10.1006/dbio.2002.0627. [DOI] [PubMed] [Google Scholar]
- 10.Ettensohn CA, Illies MR, Oliveri P, De Jong DL. Development (Cambridge, UK) 2003;130:2917–2928. doi: 10.1242/dev.00511. [DOI] [PubMed] [Google Scholar]
- 11.Howard-Ashby M, Materna SC, Brown CT, Tu Q, Oliveri P, Cameron RA, Davidson EH. Dev Biol. 2006;300:27–34. doi: 10.1016/j.ydbio.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Materna SC, Howard-Ashby M, Gray RF, Davidson EH. Dev Biol. 2006;300:108–120. doi: 10.1016/j.ydbio.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 13.Rizzo F, Fernandez-Serra M, Squarzoni P, Archimandritis A, Arnone MI. Dev Biol. 2006;300:35–48. doi: 10.1016/j.ydbio.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Tu Q, Brown CT, Davidson EH, Oliveri P. Dev Biol. 2006;300:49–62. doi: 10.1016/j.ydbio.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 15.Howard-Ashby M, Materna SC, Brown CT, Chen L, Cameron RA, Davidson EH. Dev Biol. 2006;300:74–89. doi: 10.1016/j.ydbio.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 16.Howard-Ashby M, Materna SC, Brown CT, Chen L, Cameron RA, Davidson EH. Dev Biol. 2006;300:90–107. doi: 10.1016/j.ydbio.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 17.Kageyama R, Ohtsuka T, Hatakeyama J, Ohsawa R. Exp Cell Res. 2005;306:343–348. doi: 10.1016/j.yexcr.2005.03.015. [DOI] [PubMed] [Google Scholar]
- 18.Paroush Z, Finley RL, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 19.Grbavec D, Stifani S. Biochem Biophys Res Commun. 1996;223:701–705. doi: 10.1006/bbrc.1996.0959. [DOI] [PubMed] [Google Scholar]
- 20.Bolouri H, Davidson EH. Proc Natl Acad Sci USA. 2003;100:9371–9376. doi: 10.1073/pnas.1533293100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stathopoulos A, Levine M. Dev Biol. 2005;280:482–493. doi: 10.1016/j.ydbio.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Oliveri P, Davidson EH. Science. 2007;315:1510–1511. doi: 10.1126/science.1140979. [DOI] [PubMed] [Google Scholar]
- 23.Hinman V, Nguyen A, Cameron RA, Davidson EH. Proc Natl Acad Sci USA. 2003;100:13356–13361. doi: 10.1073/pnas.2235868100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beatus P, Lendahl U. J Neurosci Res. 1998;54:125–136. doi: 10.1002/(SICI)1097-4547(19981015)54:2<125::AID-JNR1>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 25.Gibert J, Simpson P. Int J Dev Biol. 2003;47:643–651. [PubMed] [Google Scholar]
- 26.Davidson EH, Erwin DH. Science. 2006;311:796–800. doi: 10.1126/science.1113832. [DOI] [PubMed] [Google Scholar]
- 27.Rast JP, Amore G, Calestani C, Livi CB, Ransick A, Davidson EH. Dev Biol. 2000;228:270–286. doi: 10.1006/dbio.2000.9941. [DOI] [PubMed] [Google Scholar]
- 28.Nemer M, Rondinelli E, Infante D, Infante A. Dev Biol. 1991;145:255–265. doi: 10.1016/0012-1606(91)90124-l. [DOI] [PubMed] [Google Scholar]
- 29.Ransick A, Rast JP, Minokawa T, Calestani C, Davidson EH. Dev Biol. 2002;246:132–147. doi: 10.1006/dbio.2002.0607. [DOI] [PubMed] [Google Scholar]
- 30.Minokawa T, Rast JP, Arenas-Mena C, Franco CB, Davidson EH. Gene Expression Patterns. 2004;4:449–456. doi: 10.1016/j.modgep.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 31.Long S, Rebagliati M. BioTechniques. 2002;32:494–498. doi: 10.2144/02323bm04. [DOI] [PubMed] [Google Scholar]
- 32.Croce J, Lhomond G, Gache C. Mech Dev. 2003;120:561–572. doi: 10.1016/s0925-4773(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 33.Bradham CA, McClay DR. Development (Cambridge, UK) 2006;133:21–32. doi: 10.1242/dev.02160. [DOI] [PubMed] [Google Scholar]
- 34.Minokawa T, Wikramanayake A, Davidson EH. Dev Biol. 2005;288:545–558. doi: 10.1016/j.ydbio.2005.09.047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.