Abstract
Antibodies against the malaria vaccine candidate apical membrane antigen-1 (AMA-1) can inhibit invasion of merozoites into RBC, but antigenic diversity can compromise vaccine efficacy. We hypothesize that polymorphic sites located within inhibitory epitopes function as antigenic escape residues (AER). By using an in vitro model of antigenic escape, the inhibitory contribution of 24 polymorphic sites of the 3D7 AMA-1 vaccine was determined. An AER cluster of 13 polymorphisms, located within domain 1, had the highest inhibitory contribution. Within this AER cluster, antibodies primarily targeted five polymorphic residues situated on an α-helical loop. A second important AER cluster was localized to domain 2. Domain 3 polymorphisms enhanced the inhibitory contribution of the domain 2 AER cluster. Importantly, the AER clusters could be split, such that chimeras containing domain 1 of FVO and domain 2 + 3 of 3D7 generated antisera that showed similarly high level inhibition of the two vaccine strains. Antibodies to this chimeric protein also inhibited unrelated strains of the parasite. Interstrain AER chimeras can be a way to incorporate inhibitory epitopes of two AMA-1 strains into a single protein. The AER clusters map in close proximity to conserved structural elements: the hydrophobic trough and the C-terminal proteolytic processing site. This finding led us to hypothesize that a conserved structural basis of antigenic escape from anti-AMA-1 exists. Genotyping high-impact AER may be useful for classifying AMA-1 strains into inhibition groups and to detect allelic effects of an AMA-1 vaccine in the field.
Keywords: Plasmodium, apical membrane antigen-1, invasion
Antigenic diversity has been implicated in the failure of several licensed and test vaccines (1). The “Combination B” malaria vaccine, containing merozoite surface protein-2 (msp-2) of Plasmodium falciparum 3D7 strain as one of its components, successfully reduced the prevalence of the 3D7 msp-2 genotype but had no impact on the prevalence of parasites with the FC27 msp-2 genotype (2). Understanding the molecular basis of strain specificity and the resulting antigenic escape is therefore important for vaccine development.
Apical membrane antigen-1 (AMA-1) is one of the leading malaria vaccine candidates. Immunization with AMA-1 induces antibodies that inhibit invasion, conferring protection in animals (3). P. falciparum AMA-1 vaccines based on 3D7 and FVO strain are currently in efficacy human trials (4, 5). Despite the strong preclinical evidence favoring its vaccine candidacy, there are >60 polymorphic sites on AMA-1 protein. Among the 50 Thai isolates sequenced, there were 27 haplotypes. Similarly, of the 50 Nigerian sequences there were 45 haplotypes, and of the 68 Papuan New Guinean sequences there were 27 haplotypes (6–8). The strain variability of AMA-1 is a cause of concern to vaccinologists.
Strain-specific differences are reported among field antisera by ELISA (9, 10) or by using a functional assay of parasite growth and invasion inhibition (GIA) (11). Allelic replacement experiments have directly implicated sequence polymorphism in antigenic escape (12), and cross-strain GIAs suggest that the extent of escape correlates sequence distance between the vaccine and the target strain (13). In the rodent malaria challenge model, polymorphism of AMA-1 has been unequivocally linked to vaccine failure (14). Human sera against the WRAIR 3D7 AMA-1 vaccine, which inhibits invasion of the homologous 3D7 strain, showed little or no inhibition of the heterologous FVO strain (5). In an attempt to overcome the polymorphism problem, one group is following a coimmunization strategy, and antibodies to a bi-allelic 3D7+FVO vaccine show high-level inhibition of both the vaccine alleles (4, 13). However, the extent of global haplotype diversity within AMA-1 has hindered the rational selection of haplotypes for the multiallelic mixture approach and is likely to complicate allelic shift analyses in the upcoming efficacy trials unless the most important escape residues are identified.
The nature and distribution of AMA-1 polymorphisms seems to have strong structural basis. Only ≈10% of AMA-1 residues are polymorphic, and these polymorphisms are concentrated in a relatively small hypervariable region on domain 1 (6, 7). Distant polymorphisms cluster in three dimensional space and are located on one side of the AMA-1 crystal structure: “the polymorphic face” (15–17). Additionally, all of the polymorphic sites do not have an equal contribution toward antigenic escape. For example, parasite strain D10 and 3D7 are equally susceptible to inhibition by anti-3D7 AMA-1 antisera, despite the 9-aa differences between 3D7 and D10 AMA-1 (11, 13). We hypothesize that polymorphisms located within important inhibitory epitopes confer most of the escape advantage to the parasite. We term these critical polymorphic sites as “antigenic escape residues” (AER). The objective of this study is to determine the relative inhibitory contribution of various polymorphic clusters to map the structural location of AER of the 3D7 AMA-1 vaccine.
There are 24-aa differences between 3D7 and FVO strain AMA-1: 18 located on domain 1, 4 on domain 2, and 2 on domain 3. To determine the relative inhibitory contribution of these 24 polymorphic sites, we produced chimeric FVO AMA-1 proteins displaying 3D7 specific polymorphic clusters. The chimeras were used to selectively deplete 3D7-recognizing antibodies, in GIA reversal experiments, and the resulting reversal of inhibition was used as a readout to map the AER.
Results
Chimeric AMA-1 Proteins Had Comparable Purity and Contained Elements of Correct Structure.
Domains 1, 2, 3, and 2 + 3 were selectively switched from FVO to 3D7 type in chimeric proteins D1, D2, D3, and D2+3, respectively. Proteins C1, C2, C3, C1+C2, and C1-L were chimeras in which clusters of 3D7-specific polymorphisms within domain 1 were substituted for the FVO type. Polymorphisms included within a cluster were based on the spatial proximity in three-dimensional space [Fig. 1 and supporting information (SI) Table 1]. The recombinant chimeric proteins were expressed in Escherichia coli, refolded, and purified to homogeneity. All nine chimeras, the 3D7 and the FVO AMA-1 proteins, migrated as monomeric bands at ≈54 kDa (Fig. 2A). Proteins were >95% pure by densitometry of Coomassie blue-stained bands. Positive reactivity with a P. falciparum AMA-1, conformation dependent, monoclonal antibody 4G2dc1, suggested correct folding of the chimeras (28) (Fig. 2B). Furthermore, the presence of disulphide bonded tertiary structures were confirmed by mobility difference of the proteins observed by SDS/PAGE, under reducing and nonreducing conditions (SI Fig. 8).
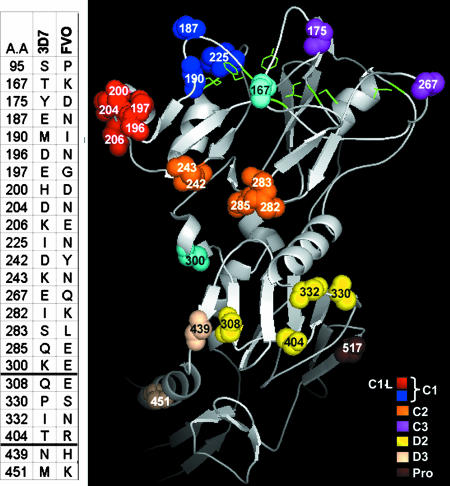
Fig. 1.
Clusters of AMA-1 polymorphic sites. (Left) The table shows the 24 polymorphic differences between 3D7 and FVO AMA-1. Dark lines separate domain boundaries. (Right) Polymorphic face of P. falciparum AMA-1 (15, 16, 28). Residues selectively switched from FVO to 3D7 type are shown as solid amino acids. Polymorphisms were switched domain-wise: D1 (red plus blue plus orange plus purple plus sky blue), D2 (yellow), or D3 (wheat). Polymorphic clusters within domain 1 were also switched: C1 (red plus blue), C2 (orange), C1-L (red), C3 (purple), and C1+C2 (red plus blue plus orange). The C-terminal proteolytic cleavage site of AMA-1 (Pro; brown) (23) and the side-chains of the hydrophobic trough residues (green) (15) are also shown. Domain 1 residues not included in the cluster chimeras are colored sky blue. Figures were made by using PyMOL software (www.pymol.org).
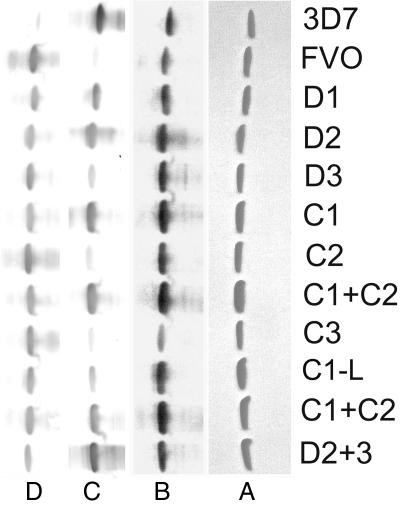
Fig. 2.
Purity and reactivity of AMA-1 chimeras. (A) Chimeric AMA-1 proteins analyzed for purity by nonreduced SDS/PAGE stained with Coomassie blue. (B) Western blot with mAb 4G2dc1. Anti-3D7 and anti-FVO AMA-1 rabbit sera pools (1:10,000 dilution) were mixed with an equal volume of 0.5 mg/ml of FVO or 3D7 AMA-1 proteins, respectively. The cross-reactive antibodies were blocked, and the free strain-specific anti-3D7 (C) and anti-FVO (D) antibodies were used in a Western blot. Densitometric analysis of Fig. 2C blot is shown in SI Fig. 9. Two batches of C1+C2 were loaded on these gels.
Domain 1 and Domain 2 Are the Prime Targets of Strain-Specific Antibodies.
The chimeras were screened for gain in reactivity to a 3D7 specific polyclonal antibody reagent by using Western blot (Fig. 2C). A densitometric scan of the blot was also done (SI Fig. 9). The 3D7 specific antibodies showed minimal cross-reactivity with the FVO AMA-1 scaffold. Among the domain chimeras D1 and D2 showed high reactivity as compared with D3. The D2+3 chimera showed higher reactivity than D2 or D3 (SI Fig. 9). Among the cluster chimeras, C1+C2 and C1 showed high reactivity as compared with C1-L, C2, and C3. The FVO AMA-1-based chimeras were also tested for loss of reactivity to a strain-specific anti-FVO reagent (Fig. 2D). Although cross-reactivity of anti-FVO with the FVO AMA-1 scaffold (common in all chimeras) could be a complicating factor, chimeric proteins reacted similarly with anti-FVO, suggesting that strain-specific anti-FVO are more cross-reactive than strain-specific anti-3D7.
Domain 1, Domain 2, and Domain 2 + 3 Polymorphic Sites Are Located Within Inhibitory Epitopes.
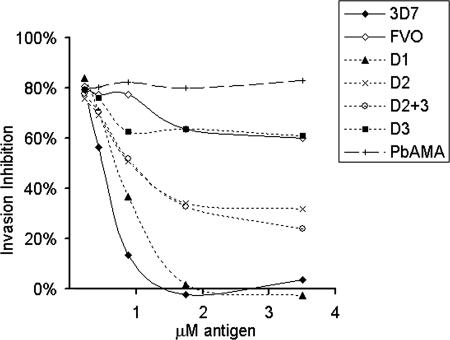
By displaying 3D7 polymorphisms onto the FVO AMA-1 scaffold, we determined the inhibitory contribution of various polymorphic clusters, as indicated by their ability to deplete strain-specific antibodies and reverse parasite growth inhibition. Fig. 3 shows results from a typical reversal assay, showing domain-wise inhibitory contribution of 3D7 specific polymorphisms. At 7% concentration, an anti-3D7 AMA-1 pool from 3 rabbits showed ≈80% inhibition of 3D7 parasites in a GIA (Fig. 3). Addition of the rodent Plasmodium berghei AMA-1 had no effect on inhibition. Homologous 3D7 AMA-1 protein depleted both cross-reactive and strain-specific antibodies against AMA-1 and resulted in a dose dependent and complete reversal of inhibition. In comparison, the FVO AMA-1, which differs from 3D7 AMA-1 at 24 polymorphic sites, depleted only the cross-reactive antibodies, causing significantly less reversal from 80% to 60%. Of note, the relationship between inhibition and antibody concentration is nonlinear (SI Fig. 10). The 80–60% reduction in inhibition results in ≈60% drop in antibody concentration. Thus, ≈40% of antibodies in the polyclonal serum pool were 3D7 strain-specific.
Fig. 3.
Five serial dilutions (x axis) of the 3D7, the FVO, or the domain chimeras were compared for their ability to reverse invasion inhibition (y axis), mediated by 7% anti-3D7 AMA-1 serum pool against the 3D7 parasite target. P. berghei AMA-1 (PbAMA) was used as the negative control antigen.
The D3 chimera caused baseline reversal that was comparable with FVO protein (Fig. 3; D3 compared with FVO) therefore, the 60% strain-specific inhibition was due to antibodies against domain 1 and domain 2 or domain 2 + 3 polymorphisms. D1 chimera caused complete reversal of inhibition, similar to 3D7 AMA-1 (Fig. 3; D1, 3D7). This observation suggested that all of the polymorphic inhibitory epitopes were located on domain 1. However, this was not the case as the depletion of domain 2 and domain 2 + 3 antibodies by D2 and D2+3 also showed reversal of inhibition (Fig. 3; D2+3, D2) from 80% down to 25–30%. This observation suggested that anti-AMA-1 antibody-mediated parasite inhibition was complex and resulted from different antibody components: 25–30% inhibition was mediated by domain 1 specific antibodies, whereas 30–35% inhibition resulted from a combination of domain 1 and domain 2-specific antibodies. Also, the slightly higher reversal by D2+3 as compared with D2, and the higher reactivity of D2+3 chimera as compared with the D2 and D3 (Fig. 2C and SI Fig. 9), suggested a role of domain 3 polymorphisms when displayed along with domain 2 polymorphisms. Overall the domain 1 polymorphisms played a dominant role in binding to inhibitory antibodies.
Strain-Specific Inhibitory Antibodies to Domain 1 Target a Cluster of 13 Polymorphic Residues.
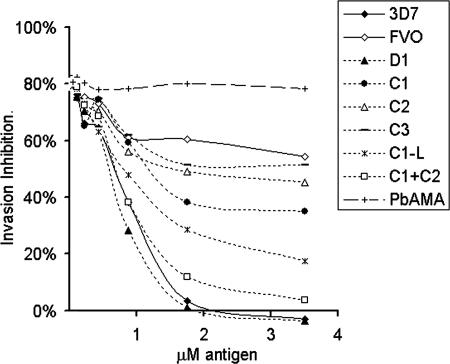
The 18 domain 1 polymorphisms were further divided into 3 clusters C1, C2, and C3 (Fig. 1) and tested in a GIA reversal assay. Fig. 4 shows results from one such assay. Chimera C1+C2 mediated reversal that was similar to D1 and 3D7 AMA-1 protein, indicating that the majority of the domain 1 inhibitory epitopes were in the C1 and C2 clusters (Fig. 4, compare C1+C2, D1, and 3D7). C3, because of its low strain-specific antigenicity (Fig. 2C), showed reversal that was slightly greater than baseline reversal by FVO protein (Fig. 4, compare FVO and C3). Further fragmentation of the C1+C2 cluster into C1 (eight polymorphisms) and C2 (five polymorphisms) showed significantly less reversal as compared with C1+C2. Mixing experiments showed that equimolar C1 and C2 proteins caused significantly less reversal than C1+C2 (data not shown). Therefore, inhibitory strain-specific antibodies to domain 1 recognize a surface footprint that incorporates both C1 and C2. This observation may also explain why a previous study (12) concluded that polymorphisms on the C1 cluster alone (residues 178–260) did not significantly contribute to the absence of cross-strain inhibition in the 3D7-W2mef model.
Fig. 4.
Five serial dilutions (x axis) of the 3D7, FVO AMA-1, or the domain 1 cluster chimeras were compared for their ability to reverse invasion inhibition (y axis), mediated by 7% anti-3D7 AMA-1 serum pool against the 3D7 parasite target. P. berghei AMA-1 (PbAMA) was used as the negative control antigen.
An additional chimera, C1-L, displaying a short 3D7 loop containing five highly polymorphic sites, was also tested. Surprisingly, C1-L (Fig. 4; C1-L) showed reversal that was at least as high or in some experiments higher than C1. This observation indicated that polymorphisms on the C1-L loop had the highest inhibitory contribution within C1+C2. Residues 167 and 300 (in sky blue in Fig. 1) that were a part of D1 chimera were not included in domain 1 cluster chimeras because of their relative isolation on primary and tertiary structure. Because C1+C2 polymorphisms accounted for the majority of the strain-specific inhibitory activity of D1, the inhibitory role of residues 167 and 300 was presumed to be minor.
AER Map of 3D7 AMA-1.
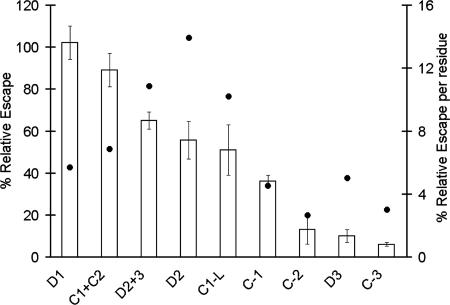
Depletion of strain-specific antibodies by recombinant proteins prevent their binding to the parasite resulting in reversal of inhibition. In the field, polymorphic differences between vaccine and challenge strain are thought to reduce binding of strain-specific antibodies, resulting in antigenic escape. Therefore the in vitro reversal of inhibition by chimeric proteins is an indirect strategy to map the AER. Because 3D7 AMA-1 protein depletes antibodies to all of the 24 3D7 polymorphic sites, its corresponding reversal represents 100% “relative escape” (RE). Likewise, FVO AMA-1 does not deplete any 3D7-specific antibodies (it depletes only cross-reactive antibodies); its reversal represents 0% RE. Fig. 5 shows the mean RE ± SD of the chimeras from three independent GIA reversal experiments, plotted on the 100–0% RE scale. RE per polymorphic residue was also calculated by dividing the mean RE by the number of 3D7-specific residues being displayed by that chimera.
Fig. 5.
Anti-3D7 AMA-1 serum pool (7% concentration)-mediated inhibition was reversed by a 1.75 μM concentration of 3D7, FVO, or the chimeric AMA-1 proteins. The percent invasion was calculated as (% parasitemia in test well/% parasitemia in adjuvant control well). RE (y axis) was determined by subtracting % invasion in the presence of FVO AMA-1 antigen from % invasion in the presence of the test antigen and then adjusting RE for 3D7 AMA-1 to 100% and FVO AMA-1 to 0% (not plotted). This calculation allowed the reversal of inhibition due to cross-reactive antibodies to be canceled out. Mean RE ± SD of three independent experiments is shown. RE/residue (dots) = mean RE/number of residues displayed by a particular chimera.
There was no significant difference between the RE of 3D7, D1 and C1+C2 (3D7 vs. D1, P = 0.99; 3D7 vs. C1+C2, P = 0.5). This observation suggested that C1+C2 contained the most important AER of the 3D7 AMA-1 vaccine, because depletion of antibodies to its 13 polymorphic sites accounted for ≈100% escape in the 3D7-FVO model. Within C1+C2, the loop C1-L had the highest impact, 51% RE, which was higher than 36% RE for C1; however, this difference failed to reach statistical significance (P = 0.2). The five polymorphic sites within C1-L were, therefore, most likely the most important component of the C1+C2 AER cluster. Outside of domain 1, the 65% RE for D2+3 was slightly higher than the 52% RE for D2, but not significantly different (P = 0.8). This observation confirmed that the four polymorphic sites within D2 form the second most important AER cluster, and that domain 3 polymorphic sites enhanced the overall inhibitory contribution of domain 2 polymorphisms. RE for C2, C3, and D3 were not significantly different from heterologous FVO AMA-1 (FVO vs. D3, C2, C3; P = 0.6, 0.3, and 0.9, respectively).
When RE per residue was considered (Fig. 5; dots), D2, D2+3, and C1-L clusters contained AER with the highest impact (≥10% RE per residue). D1, C1+C2, C1, and D3 AER had mid-level impact (5–7%), and C2 and C3 clusters contained polymorphisms with the lowest impact (≈3% RE per residue). Because the D3 polymorphisms by themselves are not contributing to inhibition, the most significant AER can be assigned to D2 and C1-L.
GIA with Antibodies Raised Against Chimeric Proteins Confirm the AER Map.
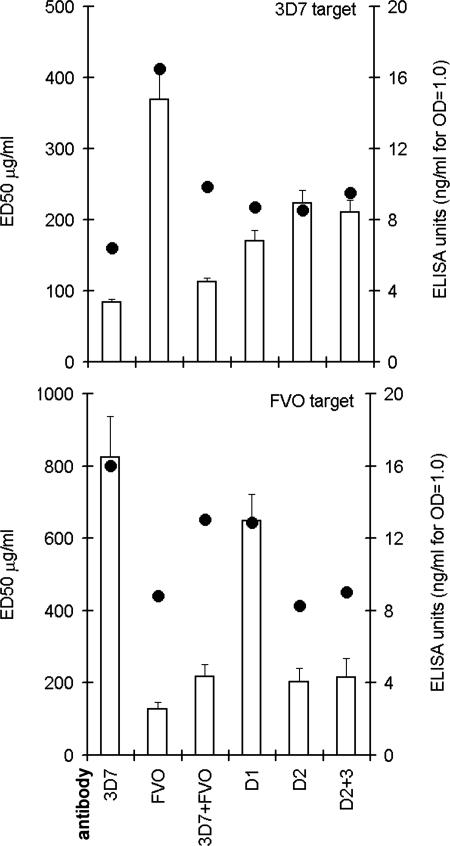
Groups of three rabbits were vaccinated with 100 μg each 3D7 AMA-1, FVO AMA-1, chimeras D1, D2, D2+3, or the biallelic mixture of 3D7+ FVO AMA-1 (200 μg of total protein). Sera from three rabbits within the group were pooled, and AMA-1-specific IgG were affinity purified and concentrated to an equivalent 4 mg/ml. A SDS/PAGE confirmed similar purity of the IgG fractions (SI Fig. 11). Three independent GIA experiments were conducted against both the 3D7 and FVO target (SI Fig. 12), and the average ED50 (effective dose for 50% invasion inhibition) was plotted (Fig. 6, left y axis). ELISA was also performed, by using 3D7 or FVO AMA-1-coated plates. ELISA unit was defined as ng/ml IgG needed for OD415 = 1.0 (Fig. 6, right y axis).
Fig. 6.
Left y axis (bars), concentration of IgG required for 50% invasion inhibition (ED50) was determined against 3D7 and FVO target parasites. The mean ED50 (± SD) was plotted from three separate GIA experiments (SI Fig. 12). Right y axis (circles), ELISA units (ng/ml IgG that resulted in OD415 = 1.0) determined by using the homologous 3D7 or FVO AMA-1 coat antigens.
Inhibition of 3D7 target.
The anti-3D7 AMA-1 showed the highest reactivity against the homologous 3D7 antigen (lowest ELISA units). Comparatively, antibodies against the domain chimeras showed lower ELISA reactivity. Anti-3D7 also had the lowest ED50 against the 3D7 strain (84 μg/ml) by GIA. Anti-D1 (170 μg/ml), anti-D2 (223 μg/ml), and anti-D2+3 (211 μg/ml) showed significantly higher ED50 (P value compared with anti-3D7, <0.001). There was no significant difference in the ED50 for anti-D2 and anti-D2+3. As predicted by the AER map, the high-level strain-specific inhibition of 3D7 parasite is primarily due to inhibitory targets on domain 1 and domain 2. The biallelic vaccine showed the best inhibitory activity, despite the lower ELISA reactivity, its ED50 was comparable with the 3D7 vaccine.
Inhibition of FVO target.
Anti-FVO showed the highest reactivity against the homologous FVO antigen by ELISA. Interestingly, IgG against domain chimeras containing FVO-specific domain 1 (i.e., anti-D2 and anti-D2+3) showed ELISA reactivity comparable with anti-FVO. Not surprisingly therefore, the ED50 for anti-D2 (214 μg/ml) and anti-D2+3 (215 μg/ml) were statistically not different from the 127 μg/ml ED50 for anti-FVO (P ≥ 0.5). In comparison, antisera against vaccines that did not contain FVO domain 1, i.e., anti-3D7 (824 μg/ml) and anti-D1 (649 μg/ml), showed significantly higher ED50 (P < 0.0001). Therefore, unlike the 3D7 target, the inhibition of FVO parasite depends only on the domain 1 genotype of the vaccine. A GIA reversal experiment further corroborated this finding, where only the D1 chimera (but not D2, D2+3, or D3) showed reduced ability to reverse anti-FVO AMA-1-mediated inhibition of the FVO parasite (SI Fig. 13). As with the 3D7 target, the biallelic vaccine showed superior inhibitory activity; despite the lower ELISA reactivity, its ED50 was comparable with the FVO vaccine.
Chimeric Proteins as Vaccine Candidates.
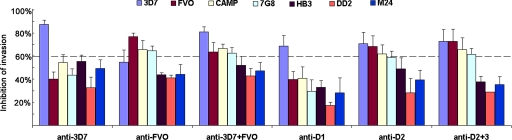
Efficacy of an AMA-1 vaccine primarily depends on the quantity and quality of the induced antibodies. Whole sera from the rabbits were tested in a GIA against unrelated P. falciparum strains CAMP, 7G8, HB3, DD2, and M24 (Fig. 7 and SI Fig. 14). Observations in Aotus trials suggest that ≥60% inhibition in a GIA correlates protection (18). We have made similar observations in a 3D7 AMA-1 vaccine trial, in Aotus monkeys, against a closely homologous FCH4 strain challenge (our unpublished data). By using ≥60% inhibition at 20% serum concentration, as a cutoff inhibition which correlates protection (dotted line, Fig. 7), the GIA data were analyzed. Anti-3D7 inhibited only the 3D7 strain. Anti-FVO inhibited the FVO strain and also CAMP and 7G8 strains. Anti-3D7+FVO (biallelic vaccine), inhibited 3D7, FVO, CAMP, and 7G8 strains. Among the chimeric vaccines, anti-D2+3 induced antibodies that almost matched the inhibitory spectrum of the biallelic vaccine, inhibiting the 3D7, FVO, CAMP, and 7G8 strains. A confirmatory GIA with affinity purified anti-AMA-1 confirmed the high-level cross-strain inhibitory activity of anti-D2+3 (SI Fig. 15). These results demonstrate that it is possible to generate cross-reacting antiserum by mixed vaccination, or by using AMA-1 chimeras.
Fig. 7.
Individual rabbit sera from each vaccine group were tested in a GIA by using multiple serum dilutions against 3D7, FVO, CAMP, 7G8, HB3, DD2, and M24 strains (SI Fig. 14). The mean invasion (+SD) of three rabbits at 20% serum dilution was plotted.
Within the sequence limits of the AMA-1 constructs, the CAMP, HB3, 7G8, M24, and DD2 strains differ from 3D7 AMA-1 at 19, 24, 26, 24, and 22 residues, respectively (SI Table 2). These strains are closer to FVO AMA-1, differing at 16, 21, 16, 22, and 19 residues, respectively. This is the likely reason for higher cross-strain inhibitory activity of anti-D2 and D2+3 (and not the anti-D1 chimera).
Discussion
Determining the inhibitory contribution of 24 polymorphic residues of the 3D7 AMA-1 vaccine has direct relevance to vaccine development efforts. The highest impact AER on 3D7 domain 1 mapped to an α-helix-containing loop, C1-L. This loop includes residue 197 that is critical for the binding of an inhibitory mAb 1F9 (19). Parasite strains that differ from a vaccine within the C1-L polymorphisms are likely to show maximal escape. Mapping of a second important AER cluster on domain 2 was surprising, because population analyses suggest that only domain 1 and domain 3 are under strong diversifying selection (6–8). Domain 2 and C1-L genotyping may provide an important readout of allelic effects in upcoming vaccine trials. Domain 3 polymorphisms, by themselves, do not seem to be a major target of inhibitory antibodies, particularly when the whole ectodomain is used as an immunogen, and similar findings have been previously reported (20, 21). However, we consistently observed that domain 3 polymorphisms enhanced the overall inhibitory activity of domain 2 polymorphisms. Critically, domain 1 and domain 2 + 3 AER could be combined from different strains to generate chimeric proteins that induced inhibitory response against both the vaccine strains.
To determine whether the high-impact AER genotype can predict antigenic escape, cross-strain GIA data in Fig. 7 was correlated with AMA-1 sequences (SI Table 2). Anti-FVO showed biologically significant inhibition against FVO, 7G8 and CAMP, these strains belong to the “FVO inhibition group” (Fig. 7). Within the “high-impact” AER of C1-L and domain 2 clusters, the 7G8 and CAMP share a high level of similarity compared with the escape strains. Although no other members of the “3D7 inhibition group” were identified, it is reported that D10 represents one such strain (11, 13). As predicted by the AER results, none of the 9-aa differences between 3D7-D10 strains map to C1-L, and only one difference is within the domain 2 AER cluster (SI Table 2). These data suggest that examination of AER genotypes will be useful for strain selection of multiallelic vaccines, or to predict escape frequency during efficacy trials.
C1+C2 AER are present on loops with their hydrophilic side-chains extending away from the conserved PAN domain core (Fig. 1). Domain 2 AER also localized to loops, although much shorter loops (Fig. 1). Structural plasticity allows loops to accommodate radical charge changes without impacting the core folding. Presumably, radical changes can most effectively diminish antibody binding, and this requirement coupled with the AT codon bias of P. falciparum leaves only a few highly restricted choices of mutations that are positively selected for by the immune system. This reasoning could explain the limited repertoire of amino acid substitutions (most sites are dimorphic) and the resulting balancing selection reported among field isolates (6, 22).
AER within the C1+C2 cluster map adjacent to the hydrophobic trough of AMA-1 (Fig. 1). The domain 2 AER map in close spatial proximity to a proteolytic processing site at residue 517 (Fig. 1) (23). We have previously shown that bivalent polyclonal antibodies to AMA-1 inhibit invasion by steric blocking of functional sites and by inhibiting processing and trafficking of AMA-1 due to their cross-linking effect (24, 25). It is possible that strain-specific antibodies to domain 1 block access to the hydrophobic trough and the domain 2 + 3 antibodies cross-link AMA-1 and inhibit the C-terminal cleavage. If steric blocking of functional sites is more efficient at inhibiting invasion (compared with cross-linking), the dominant role of domain 1 AER can be explained.
Although AMA-1 has >60 polymorphic sites, mapping the AER in the 3D7-FVO model provides a model to rationalize the basis of antigenic escape from antibodies against a vaccine strain of AMA-1. In future efficacy trials, genotyping the escape variants would determine whether the high-impact AER clusters identified here universally mediate antigenic escape. We also demonstrate the feasibility of combining epitope mapping with protein engineering approaches to improve the cross-strain inhibitory activity of AMA-1 vaccines.
Methods
Immunization and Affinity Purification of IgG.
Groups of three rabbits were administered three doses of 100 μg each of antigen along with the adjuvant Montanide ISA720 adjuvant (Seppic, Inc., Paris, France) (26). Sera from all three rabbits in a group were pooled (0.7 ml each). The 2.1-ml serum pool was passed over a protein G column (Amersham, Piscataway, NJ). IgG binding and elution buffers (Pierce, Rockford, IL) were used according to the manufacturer's instructions. After neutralization with 1 M Tris (pH 8.0), the IgG were dialyzed against PBS. An AMA-1 affinity column was prepared by binding 5 mg/ml 3D7 and FVO AMA-1 to a cyanogen bromide Sepharose column (Amersham). IgG were passed over this column, and the flow-through was confirmed to be negative for AMA-1 antibodies by ELISA. AMA-1-specific IgG were eluted and dialyzed against PBS. IgG were concentrated by using 10-kDa cutoff membrane to 4 mg/ml. ELISA were done essentially as described (26).
Cloning and Expression of Chimeras.
AMA-1 sequences (residues 83Gly-531Glu) were derived from accession nos. CAC05390 (FVO) and AAB36701 (3D7). E. coli codon optimized 3D7 and FVO AMA-1 genes were used as templates for mutant gene construction. Fig. 1 and SI Table 1 show the color-coded amino acids switched and their respective location on the crystal structure. Mutagenesis was performed either by designing overlapping PCR templates or by using the Genetailor site directed mutagenesis system (Invitrogen, Carlsbad, CA). A rodent malaria parasite P. berghei AMA-1 amino acids 23–476 was also PCR amplified. All gene products were cloned into TOPO 2.1 vector (Invitrogen), sequenced, and transferred to the pET32 based plasmid. Tuner(DE3) E. coli cells (Novagen, San Diego, CA) were used for expression. Proteins were produced by using 1L shake-flask culture, extracted from inclusion bodies, refolded, and purified by using a two-step chromatographic procedure (our unpublished data). Purification method for 3D7 AMA-1 has been described (26). The purified proteins were dialyzed against PBS and tested for purity by SDS/PAGE.
GIA and GIA Reversal.
GIA were performed essentially as described (27). All GIA were done in duplicate wells, except the SI Fig. 15, which was in triplicates. Synchronized cultures at late-ring stage were diluted to 0.25–0.3% parasitemia and 2% hematocrit by using uninfected cells. Final culture volume was 60 μl in a 96-well plate format. Parasites developed for 40 h at 37°C and ring stages formed after the invasion cycle were stained with 1× SYBR green dye (BMA, Rockland, ME) and counted by using a Flow-cytometer BD FACSCalibur. Percent invasion = % parasitemia in the test well/% parasitemia in the adjuvant control serum or the adjuvant control IgG well. Percent inhibition of invasion = 1 − (% parasitemia in test well/% parasitemia in adjuvant control IgG or adjuvant control serum well). Relative escape (RE) = (% invasion in presence of test antigen) − (% invasion in presence of FVO AMA-1). RE was expressed on a 100–0% sliding scale; RE in the presence of 3D7 AMA-1 = 100%; FVO AMA-1 = 0%.
Statistical Analysis.
ANOVA procedure was performed by using the General Linear Model with MINITAB Release 14.12.0 software. P < 0.05 for Tukey simultaneous test was treated as statistically significant. A four-parameter curve-fitting was performed by using Softmax Pro software (Molecular Devices) for ED50 calculation and for the ELISA units calculation.
Modeling of the PfAMA-1 Ectodomain.
Coordinates for PfAMA-1 and PvAMA-1 were aligned as described (28).
Supplementary Material
Acknowledgments
We thank J. David Haynes [Walter Reed Army Institute of Research (WRAIR)] for critical advice, Kathy Moch (WRAIR) for malaria cultures, Craig Morrissette (WRAIR) for statistical support, Alan W. Thomas (Biomedical Primate Research Center, Rijswijk, The Netherlands) for mAb 4G2dc1, Evelina Angov (WRAIR) for the modified pET32 plasmid, and Carole Long and Tom Wellems (National Institutes of Health, Bethesda, MD) for M24 sequence and strain. Funding was provided by the U.S. Agency for International Development Malaria Vaccine Development Program.
Abbreviations
- AMA-1
apical membrane antigen-1
- GIA
growth and invasion inhibition assay
- AER
antigenic escape residues
- RE
relative escape.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0701464104/DC1.
References
- 1.Klenerman P, Zinkernagel RM. Nature. 1998;394:482–485. doi: 10.1038/28860. [DOI] [PubMed] [Google Scholar]
- 2.Genton B, Al-Yaman F, Betuela I, Anders RF, Saul A, Baea K, Mellombo M, Taraika J, Brown GV, Pye D, et al. Vaccine. 2003;22:30–41. doi: 10.1016/s0264-410x(03)00536-x. [DOI] [PubMed] [Google Scholar]
- 3.Stowers AW, Kennedy MC, Keegan BP, Saul A, Long CA, Miller LH. Infect Immun. 2002;70:6961–6967. doi: 10.1128/IAI.70.12.6961-6967.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malkin EM, Diemert DJ, McArthur JH, Perreault JR, Miles AP, Giersing BK, Mullen GE, Orcutt A, Muratova O, Awkal M, et al. Infect Immun. 2005;73:3677–3685. doi: 10.1128/IAI.73.6.3677-3685.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polhemus M, Magill AJ, Cummings JF, Kester KE, Ockenhouse CF, Lanar DE, Dutta S, Barbosa A, Soisson L, Diggs C, et al. Vaccine. 2007 doi: 10.1016/j.vaccine.2007.03.012. in press. [DOI] [PubMed] [Google Scholar]
- 6.Polley SD, Chokejindachai W, Conway DJ. Genetics. 2003;165:555–561. doi: 10.1093/genetics/165.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polley SD, Conway DJ. Genetics. 2001;158:1505–1512. doi: 10.1093/genetics/158.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortes A, Mellombo M, Mueller I, Benet A, Reeder JC, Anders RF. Infect Immun. 2003;71:1416–1426. doi: 10.1128/IAI.71.3.1416-1426.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortes A, Mellombo M, Masciantonio R, Murphy VJ, Reeder JC, Anders RF. Infect Immun. 2005;73:422–430. doi: 10.1128/IAI.73.1.422-430.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polley SD, Mwangi T, Kocken CH, Thomas AW, Dutta S, Lanar DE, Remarque E, Ross A, Williams TN, Mwambingu G, et al. Vaccine. 2004;23:718–728. doi: 10.1016/j.vaccine.2004.05.031. [DOI] [PubMed] [Google Scholar]
- 11.Hodder AN, Crewther PE, Anders RF. Infect Immun. 2001;69:3286–3294. doi: 10.1128/IAI.69.5.3286-3294.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Healer J, Murphy V, Hodder AN, Masciantonio R, Gemmill AW, Anders RF, Cowman AF, Batchelor A. Mol Microbiol. 2004;52:159–168. doi: 10.1111/j.1365-2958.2003.03974.x. [DOI] [PubMed] [Google Scholar]
- 13.Kennedy MC, Wang J, Zhang Y, Miles AP, Chitsaz F, Saul A, Long CA, Miller LH, Stowers AW. Infect Immun. 2002;70:6948–6960. doi: 10.1128/IAI.70.12.6948-6960.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crewther PE, Matthew ML, Flegg RH, Anders RF. Infect Immun. 1996;64:3310–3317. doi: 10.1128/iai.64.8.3310-3317.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bai T, Becker M, Gupta A, Strike P, Murphy VJ, Anders RF, Batchelor AH. Proc Natl Acad Sci USA. 2005;102:12736–12741. doi: 10.1073/pnas.0501808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pizarro JC, Vulliez-Le Normand B, Chesne-Seck ML, Collins CR, Withers-Martinez C, Hackett F, Blackman MJ, Faber BW, Remarque EJ, et al. Science. 2005;308:408–411. doi: 10.1126/science.1107449. [DOI] [PubMed] [Google Scholar]
- 17.Chesne-Seck ML, Pizarro JC, Vulliez-Le NB, Collins CR, Blackman MJ, Faber BW, Remarque EJ, Kocken CH, Thomas AW, Bentley GA. Mol Biochem Parasitol. 2005;144:55–67. doi: 10.1016/j.molbiopara.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 18.Singh S, Miura K, Zhou H, Muratova O, Keegan B, Miles A, Martin LB, Saul AJ, Miller LH, Long CA. Infect Immun. 2006;74:4573–4580. doi: 10.1128/IAI.01679-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coley AM, Parisi K, Masciantonio R, Hoeck J, Casey JL, Murphy VJ, Harris KS, Batchelor AH, Anders RF, Foley M. Infect Immun. 2006;74:2628–2636. doi: 10.1128/IAI.74.5.2628-2636.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Healer J, Triglia T, Hodder AN, Gemmill AW, Cowman AF. Infect Immun. 2005;73:2444–24451. doi: 10.1128/IAI.73.4.2444-2451.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lalitha PV, Ware LA, Barbosa A, Dutta S, Moch JK, Haynes JD, Fileta BB, White CE, Lanar DE. Infect Immun. 2004;72:4464–4470. doi: 10.1128/IAI.72.8.4464-4470.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verra F, Hughes AL. Parassitologia. 1999;41:93–95. [PubMed] [Google Scholar]
- 23.Howell SA, Well I, Fleck SL, Kettleborough C, Collins CR, Blackman MJ. J Biol Chem. 2003;278:23890–23898. doi: 10.1074/jbc.M302160200. [DOI] [PubMed] [Google Scholar]
- 24.Dutta S, Haynes JD, Barbosa A, Ware LA, Snavely JD, Moch JK, Thomas AW, Lanar DE. Infect Immun. 2005;73:2116–3122. doi: 10.1128/IAI.73.4.2116-2122.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dutta S, Haynes JD, Moch JK, Barbosa A, Lanar DE. Proc Natl Acad Sci USA. 2003;100:12295–12300. doi: 10.1073/pnas.2032858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dutta S, Lalitha PV, Ware LA, Barbosa A, Moch JK, Vassell MA, Fileta BB, Kitov S, Kolodny N, Heppner DG, et al. Infect Immun. 2002;70:3101–3110. doi: 10.1128/IAI.70.6.3101-3110.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haynes JD, Moch JK, Smoot DS. Methods Mol Med. 2002;72:535–554. doi: 10.1385/1-59259-271-6:535. [DOI] [PubMed] [Google Scholar]
- 28.Collins CR, Withers-Martinez C, Bentley GA, Batchelor AH, Thomas AW, Blackman MJ. J Biol Chem. 2007;282:7431–7441. doi: 10.1074/jbc.M610562200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.