Abstract
Curli are functional amyloid fibers assembled by enteric bacteria such as Escherichia coli and Salmonella spp. In E. coli, the polymerization of the major curli fiber subunit protein CsgA into an amyloid fiber depends on the minor curli subunit protein, CsgB. The outer membrane-localized CsgB protein shares ≈30% sequence identity with the amyloid-forming protein CsgA, suggesting that CsgB might also have amyloidogenic properties. Here, we characterized the biochemical properties of CsgB and the molecular basis for CsgB-mediated nucleation of CsgA. Deletion analysis revealed that a CsgB molecule missing 19 amino acids from its C terminus (CsgBtrunc) was not outer membrane-associated, but secreted away from the cell. CsgBtrunc was overexpressed and purified from the extracellular milieu of cells as an SDS-soluble, nonaggregated protein. Soluble CsgBtrunc assembled into fibers that bound to the amyloid-specific dyes Congo red and thioflavin-T. CsgBtrunc fibers were able to seed soluble CsgA polymerization in vitro. CsgBtrunc displayed modest nucleator activity in vivo, as demonstrated by its ability to convert extracellular CsgA into an amyloid fiber. Unlike WT CsgB, CsgBtrunc was only able to act as a nucleator when cells were genetically manipulated to secrete higher concentrations of CsgA. This work represents a unique demonstration of functional amyloid nucleation and it suggests an elegant model for how E. coli guides efficient amyloid fiber formation on the cell surface.
Keywords: amyloid, nucleation, aggregation, seeding Congo red
Enteric bacteria such as Escherichia coli and Salmonella spp. produce amyloid fibers called curli that are the major proteinaceous component of a complex extracellular matrix. The extracellular matrix participates in many aspects of E. coli and Salmonella spp. physiology and pathogenicity. For example, curli and the extracellular matrix convey resistance to desiccation and long-term survival, host cell adhesion and invasion, and immune system activation (1–7). Curli are also critical determinants of biofilm formation, where they most likely mediate the initial step of surface attachment (6, 8, 9). Curliated bacteria colonize chemically diverse surfaces such as plant tissues, stainless steel, and glass (10–12).
Curli are assembled by a unique and highly regulated extracellular nucleation/precipitation pathway. At least six proteins, encoded by the divergently transcribed csgBA and csgDEFG operons (csg, curli-specific genes), are dedicated to curli formation in E. coli (8, 13). The major curli subunit protein, CsgA, is nucleated into a fiber by the minor fiber subunit, CsgB (14, 15). As a result, both subunits are incorporated into the fiber (16). Polymerization is thought to occur after secretion of the subunits to the extracellular space, because CsgA and CsgB do not have to be expressed from the same cell to assemble curli. In a process called interbacterial complementation, CsgA secreted from a ΔcsgB donor cell can be assembled into a fiber on the surface of a CsgB-producing acceptor cell (8, 15, 17). Once polymerized, curli fibers exhibit the biochemical and structural properties of amyloids (17).
Amyloid fiber formation is traditionally associated with human neurodegenerative conditions including Alzheimer's, Parkinson's, and the prion diseases (18). The proteins associated the eukaryotic amyloid diseases' transition from natively soluble conformations into SDS-insoluble, nonbranching, β-sheet fibers. This conformational change is preceded by a lag phase during which cytotoxic intermediates are formed (19, 20).
Curli represent a novel twist to the conventional view of amyloidogenesis because typical disease-associated amyloid formation is considered an off-pathway protein folding event, whereas curli are the product of a highly regulated and directed process. However, the fundamental similarities between curli- and disease-associated amyloid biogenesis suggest that amyloid-fiber polymerization in general occurs via a conserved protein-folding process. Thus, understanding how functional amyloids, such as curli, mediate efficient amyloid nucleation on the cell surface will provide insight into disease-associated amyloidogenesis and cytotoxicity.
Like disease-associated amyloids, curli are nonbranching, β-sheet-rich fibers that are resistant to protease digestion and denaturation by 1% SDS (17, 21–24). Amyloid fibers, including curli, are propagated by the addition of soluble precursors to the growing fiber tip in a process called seeding (25–27). For example, the polymerization of purified CsgA in vitro is characterized by a 1- to 2-h lag phase that precedes rapid fiber elongation (27). The lag phase can be significantly shortened by the addition of preformed CsgA fibers (27). The CsgA fiber seeds are predicted to provide a specific template that guides fiber elongation and allows polymerization to proceed with a shorter lag phase. Amyloid seeding is usually observed between proteins and fibers with identical or nearly identical amino acid sequences (28).
Although CsgA polymerization can be ameliorated by fiber seeds in vitro, assembly of CsgA polymers on the bacterial cell surface depends on the CsgB nucleator protein. The molecular and structural mechanisms of CsgB-mediated nucleation are not known. We hypothesize that CsgB may be an amyloidogenic protein that provides a seed or template to CsgA that promotes curli formation. Consistent with this hypothesis, CsgA and CsgB are ≈30% identical at the amino acid level, and in silico molecular modeling predicts that they adopt a similar cross β-strand structure (16, 24). Both CsgB and CsgA contain a conserved 5-fold sequence homology within their amino acid sequences (8, 15, 16, 24). In CsgA, these regions of homology, called repeating units, have recently been demonstrated to be amyloidogenic (27).
To determine the molecular mechanism of curli fiber nucleation, we have biochemically and genetically characterized CsgB. We demonstrate CsgB-mediated nucleation in vitro and show that CsgB is itself an amyloidogenic protein, providing insight into how these functional amyloid fibers are assembled.
Results
CsgBtrunc Forms Amyloid-Like Fibers in Vitro.
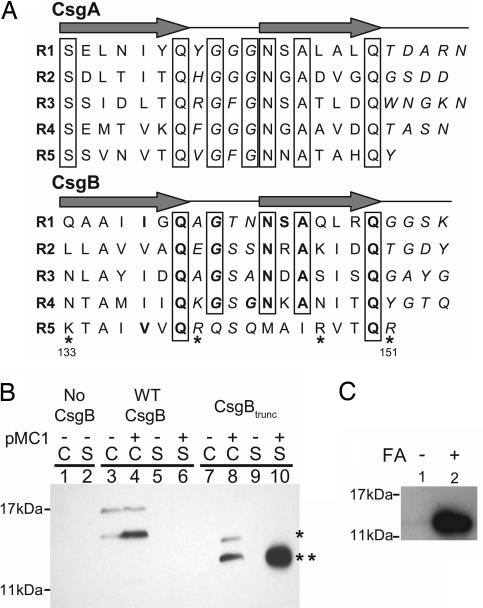
The similarity in amino acid content and the in silico structural predictions of CsgA and CsgB suggest that these proteins may share biochemical properties (Fig. 1A). Numbering from the indicated serine residues, the first four repeating units from CsgA and CsgB contain at least five identical amino acids: a glutamine at position 7, a glycine at position 9, an asparagine at position 12, an alanine at position 14, and another glutamine at position 18 (Fig. 1A). Thus, the consensus sequence of the β-strand-loop-β-strand structure for each CsgA repeating unit would be Ser X5 Gln X1 Gly X1 Gly Asn X1 Ala X3 Gln, whereas the consensus sequence of the first four repeating units of CsgB are X6 Gln X1 Gly X2 Asn X1 Ala X3 Gln (Fig. 1A). Although sequence alignments comparing CsgA with CsgB reveal a high degree of amino acid conservation between the first four repeating units, the fifth repeating unit (amino acids 133–151) of CsgB is less conserved (Fig. 1A). The fifth repeating unit of CsgB does not complete the medial glycine, asparagine, or alanine stacks conserved in all five CsgA repeating units or in the first four repeating units of CsgB. Furthermore, 4 of the 19 amino acids in the C-terminal domain of CsgB are positively charged amino acids, a higher percent of charged amino acids than any of the other repeating units found within either protein (Fig. 1A). Because the imperfect repeating units of CsgA are amyloidogenic and the first four repeating units of CsgB share a high degree of conservation to CsgA, we hypothesized that the first four repeating units of CsgB may also be amyloidogenic.
Fig. 1.
Biochemical characterization of CsgB and CsgBtrunc. (A) The imperfect repeating units in CsgA and CsgB (R1–R5) are predicted to form β-strand-loop-β-strand structures. Amino acids comprising the β-strand are located below the arrows, and amino acids predicted to comprise the loops are italicized. Bolded letters represent amino acids conserved in CsgB and CsgA at each position relative to the start of each repeating unit. Boxed letters represent amino acids conserved throughout the repeating units in both proteins. Positively charged amino acids in the fifth repeating unit of CsgB are marked with asterisks. (B) Western blot analysis of WT CsgB (*) and CsgBtrunc (**) expressed in the Δcsg strain LSR12. The csgB constructs were coexpressed with the isopropyl β-d-thiogalactoside-inducible csgG containing plasmid pMC1 or the corresponding empty vector, pTrc99A. Cultures were grown in LB to an OD600 of 0.9 and induced with 250 μM isopropyl β-d-thiogalactoside. Postinduction cell suspensions were normalized by OD600 and separated by centrifugation into cell (C) and supernatant fractions (S). The blot was probed by using αCsgB peptide antisera. Lane 1 contains LSR12/pTrc99A/pNH3 cells, and lane 2 contains corresponding supernatant; lanes 3 (C) and 5 (S) contain LSR12/pTrc99A/pNH4; lanes 4 (C) and 6 (S) contain LSR12/pMC1/pNH4; lanes 7 (C) and 9 (S) contain LSR12/pTrc99A/pNH2; and lanes 8 (C) and 10 (S) contain LSR12/pMC1/pNH2. (C) Western blot of purified CsgBtrunc probed with αCsgB peptide antibody. Two samples containing equal concentrations of purified soluble CsgBtrunc were incubated at room temperature overnight before centrifugation for 1 min in an Eppendorf microcentrifuge (Eppendorf-5 Prime, Boulder, CO). The supernatant was decanted and the pellet was resuspended in 2× SDS loading buffer (lane 1) or pretreated with 90% formic acid (FA) and dried before resuspension in 2× SDS loading buffer (lane 2).
To test the hypothesis that the first four repeating units of CsgB contained amyloid-like properties, a csgB allele missing the C-terminal repeating unit (called csgBtrunc; containing a C-terminal deletion of amino acids from 133–151) was cloned into the expression vector, pNH2. WT csgB and csgBtrunc were expressed in the Δcsg strain LSR12 that contained a plasmid-encoding CsgG, the proposed curli outer membrane secretion protein (29, 30). Western blot analysis revealed that WT CsgB was cell-associated, but CsgBtrunc was secreted away from the cells in a CsgG-dependent manner (Fig. 1B). Both WT CsgB and CsgBtrunc were SDS-soluble when expressed in the absence of CsgA (Fig. 1B). However, WT csgB expression was considerably reduced compared with csgBtrunc, and repeated attempts to purify WT CsgB were unsuccessful. Because outer membrane-associated WT CsgB was not amendable to purification, we focused on characterizing the biochemical properties of CsgBtrunc.
CsgBtrunc was affinity-purified from cell-free supernatants. Immediately after purification, CsgBtrunc was SDS-soluble and migrated to its predicted molecular weight in an SDS/PAGE gel (supporting information (SI) Fig. 6A). However, after incubation at room temperature for 24 h, a precipitate appeared in CsgBtrunc-containing samples, and CsgBtrunc no longer migrated into an SDS/PAGE gel, indicating that CsgBtrunc had assembled into SDS-insoluble aggregates. CsgBtrunc migrated into an SDS/PAGE gel when the aggregates were pretreated with 90% formic acid (Fig. 1C).
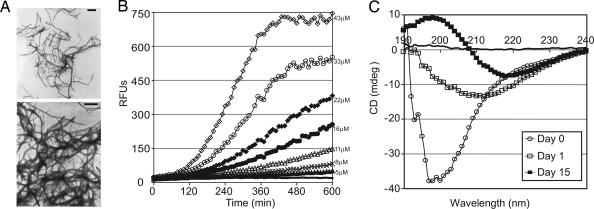
The ultrastructure of CsgBtrunc aggregates was investigated by transmission electron microscopy (TEM) and with the amyloid-specific dyes thioflavin T and Congo red (31–34). TEM analysis of purified CsgBtrunc revealed the presence of highly ordered, amyloid-like fibers (Fig. 2A). When freshly purified CsgBtrunc was mixed with the amyloid-specific dye, thioflavin T fluorescence increased in a concentration-dependent manner after an ≈100-min lag phase (Fig. 2B). Amyloid formation was also assessed by using the diazo dye Congo red, which, in the presence of an amyloid fiber, produces a unique spectral pattern called a “red shift” (35). CsgBtrunc aggregates mixed with Congo red had a maximum absorbance at 501 nm. The maximum absorbance of freshly purified CsgBtrunc was 485 nm, whereas the maximum absorbance of the nonamyloid protein BSA mixed with Congo red was 491 nm (SI Fig. 6B).
Fig. 2.
Amyloid-like properties of CsgBtrunc aggregates. (A) TEM analysis of soluble CsgBtrunc incubated at room temperature overnight reveals fibrous aggregates. (Scale bars: 500 nm.) (B) Freshly purified CsgBtrunc was mixed with the amyloid-specific dye thioflavin T and incubated at room temperature for 600 min at the following concentrations: 43 μM (open diamonds), 33 μM (open circles), 22 μM (filled diamonds), 16 μM (filled circles), 11 μM (open triangles), 8 μM (“x”s), 5 μM (filled triangles), 0 μM (solid line). Relative florescence emitted at 495 nm was measured every 10 min after excitation at 438 nm. (C) Far-UV spectral analysis of 22 μM CsgBtrunc immediately after purification (open circles), after incubation for 24 h (open squares) at room temperature, or after incubation at room temperature for 15 days (filled squares). The potassium-phosphate-buffer (KPi)-only control is represented by a solid black line.
The structural changes that occur during CsgBtrunc polymerization were measured by using circular dichroism (CD). The CD spectra of freshly purified CsgBtrunc displayed a trough at 198 nm, which is characteristic of proteins that adopt a random coil structure (Fig. 2C, open circles). After incubation of CsgBtrunc at room temperature for 24 h, the CD spectrum revealed a trough at 212 nm (Fig. 2C, open squares). Upon prolonged incubation, the CD analysis of CsgBtrunc revealed a trough at 218 nm, indicative of a β-sheet-rich conformation (Fig. 2C, filled squares). Collectively, the CsgBtrunc aggregates are SDS insoluble, β-sheet-rich fibers that interact with the amyloid specific dyes thioflavin T and Congo red. These biochemical features are consistent with the hypothesis that CsgBtrunc forms amyloid-like aggregates.
CsgBtrunc Can Nucleate CsgA in Vitro.
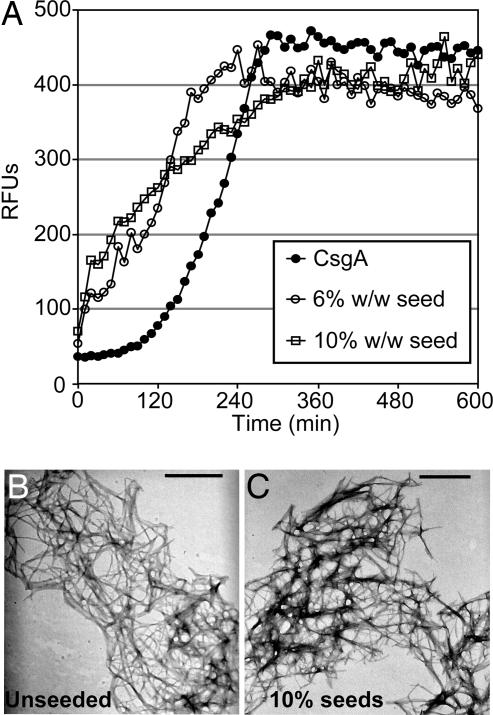
The biochemical analysis of CsgBtrunc demonstrated that the first four repeating units of CsgB contained an amyloidogenic domain. We next asked if CsgBtrunc fibers provided a nucleation surface for CsgA polymerization. CsgA's transition into an amyloid fiber in vitro is characterized by distinguishable lag, growth, and stationary phases. The lag phase can be shortened by the addition of preformed CsgA fibers (27). When mixed with thioflavin T, the relative florescence units of a 30-μM solution of freshly purified CsgA increased after a 100-min lag phase (Fig. 3A, filled circles). The lag phase of CsgA polymerization was dramatically shortened when 10% wt/wt or 6% wt/wt CsgBtrunc aggregates were added to the reaction (Fig. 3A open squares and circles, respectively). As a control for nonspecific seeding of CsgA, we used amyloid fibers derived from the islet amyloid polypeptide protein (36). The lag phase of CsgA was not reduced when 17% wt/wt of islet amyloid polypeptide seeds were added (SI Fig. 7), suggesting that CsgA seeding is a phenomena specific to the proteins that compose the curli fiber, CsgB and CsgA. TEM analysis of the aggregates produced when CsgA was seeded by CsgBtrunc revealed fibers with a similar structure to those produced by CsgA self-polymerization (compare Fig. 3B with Fig. 3C). These results suggest that CsgBtrunc promotes the conversion of soluble CsgA into an insoluble fiber in a specific manner, a process defined as nucleation.
Fig. 3.
CsgBtrunc can nucleate CsgA in vitro. (A) Thioflavin T kinetic plot monitoring the polymerization of 30 μM CsgA (filled circles) and 30 μM CsgA in the presence of 10% wt/wt (open squares) or 6% wt/wt (open circles) CsgBtrunc aggregates. (B) TEM of 30 μM CsgA. (C) TEM of the fibers produced by polymerization of 30 μM CsgA seeded by 10% CsgBtrunc. (Scale bars: 500 nm.)
CsgBtrunc Is Secreted from the Cell and Is Sufficient for Nucleation in Vivo.
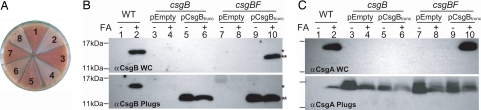
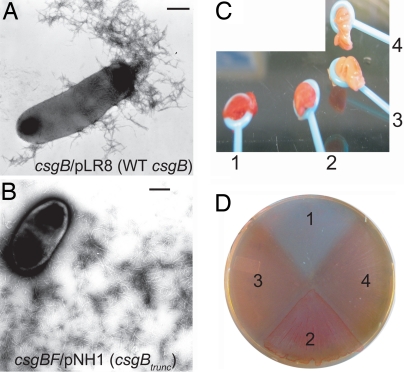
To determine whether CsgBtrunc had retained nucleator activity in vivo, csgBtrunc was expressed under the control of the csgBA promoter in plasmid pNH1. Colonies of E. coli producing curli fibers stain red when grown on Congo red indicator plates. A csgB mutant strain harboring pNH1 (csgBtrunc), pLR8 (WT csgB), or pLR2 (empty vector) was streaked on Congo red indicator plates and incubated at 26°C. After 48 h of growth on Congo red indicator plates, WT and csgB/pLR8 cells stained red, whereas csgB/pNH1 (csgBtrunc) cells were light red (Fig. 4A compare areas 1, 3, and 4). Western blotting was used to localize CsgBtrunc expressed by cells grown under curli-inducing conditions. WT CsgB was detected in whole cell (WC) samples by Western blot analysis, whereas CsgBtrunc was not (Fig. 4B Upper compare WC lanes 2, 5, and 6). However, CsgBtrunc was detected in samples in which the underlying agar was collected along with the WCs (Fig. 4B Lower agar plug lanes 2, 5, and 6). Therefore, CsgBtrunc is not cell-associated, confirming our previous finding in Fig. 1B that, unlike WT CsgB, CsgBtrunc is secreted away from the cell as an SDS-soluble protein.
Fig. 4.
In vivo nucleation properties of CsgBtrunc. (A) Congo red-containing yeast extract casamino acids (YESCA) plates comparing the Congo red binding phenotypes of csgB and csgBF harboring an empty vector control (pLR2), WT csgB on a plasmid (pLR8), or csgBtrunc (pNH1). Cells: area 1, WT/pLR2; area 2, csgB/pLR2; area 3, csgB/pLR8; area 4, csgB/pNH1; area 5, csgBF/pNH1; area 6, csgBF/pNH1; area 7, csgBF/pLR8; area 8, csgBF/pLR2. All strains contain pTrc99A except area 6, which contains pCsgF. (B Upper) WC lysate samples probed with αCsgB peptide antiserum reveal WT CsgB (*) and CsgBtrunc (**). Samples were treated with (+) or without (−) formic acid (FA). (B Lower) WCs and the underlying agar (plugs) were collected from YESCA plates and probed for CsgB. (C Upper) Western blots of WCs probed with αCsgA antiserum. (C Lower) Western blots of WCs and underlying agar (plug) samples probing for CsgA. The WT cells in lanes 1 and 2 contain empty vectors pLR2 and pTrc99A. All other strains in A and B also contain the empty vector pTrc99A in addition to either pLR2 (pEmpty) or pNH1 (pCsgBtrunc).
The localization defect of CsgBtrunc might account for its inability to nucleate CsgA into a fiber in vivo. Because the local concentration of CsgA would be predicted to be highest close to the cell surface, and CsgBtrunc is secreted away from cells into the extracellular space, we reasoned that CsgBtrunc would have a decreased chance of interacting with CsgA. To increase the probability that CsgBtrunc would interact with CsgA, we used two genetically modified csgB strains that secrete more CsgA. The first strain overexpressed CsgG, proposed to be a central component of the curlin secretion apparatus (29). Overexpression of CsgG has been demonstrated to increase the secretion of CsgA (30). When grown under curli-expressing conditions on Congo red indicator plates, csgB cells containing pNH1 (csgBtrunc) and pMC1 (csgG) stained darker red than csgB cells containing pNH1 (csgBtrunc) or pMC1 (csgG) alone (SI Fig. 8). The second strain we used was a csgF mutant. A csgF mutant secretes more CsgA, although the precise role of this protein is still unresolved (17). A csgBF mutant was transformed with pNH1 (csgBtrunc). CsgBtrunc was able to partially restore Congo red-binding to a csgBF strain (Fig. 4A area 5). Congo red binding depended on the expression of CsgBtrunc as the csgBF/pLR2 (empty vector) remained white on Congo red indicator plates (Fig. 4A area 8). Congo red binding was also dependent on CsgA; csgFBA/pNH1 did not bind Congo red (data not shown).
Another measure of fiber formation is the SDS solubility of both CsgA and CsgB. CsgA and CsgB that have assembled into an amyloid fiber are resistant to depolymerization by SDS and need to be treated with formic acid to liberate the monomers (22). At 48 h, CsgA became SDS-insoluble in csgBF/pNH1 (csgBtrunc) cells (Fig. 4C Upper; compare WC lanes 9 and 10). CsgBtrunc was also SDS-insoluble at 48 h when expressed in csgBF cells (Fig. 4B Upper; compare WC lanes 9 and 10). However, a substantial amount of both CsgBtrunc and CsgA were SDS-soluble when the underlying agar was collected along with the cells (Fig. 4 B Lower and C Lower agar plugs lanes 9 and 10). In contrast, CsgB and CsgA were completely SDS-insoluble in WT cells (Fig. 4 B Lower and C Lower; compare agar plug lanes 1 and 2). TEM was used to visualize the fibers produced in csgB/pLR8 (WT csgB) and csgBF/pNH1 (csgBtrunc). The csgBF/pNH1 (csgBtrunc) cells consistently produced fibers that were more diffuse and extended further away from the cell than csgB/pLR8 (WT csgB) cells (compare Fig. 5 A and B). To summarize, the csgBF/pNH1 (csgBtrunc) cells stained light red on Congo red indicator plates, produced SDS insoluble CsgBtrunc and CsgA, and assembled curli-like fibers, indicating that the first four repeating units of CsgB are sufficient for nucleator activity. Although CsgBtrunc clearly mediates CsgA nucleation in vivo, it does so less efficiently than WT CsgB.
Fig. 5.
Characterization of fibers nucleated by CsgBtrunc in vivo. (A) TEM of csgB/pLR8. (B) TEM of csgBF/pNH1. (Scale bars: 500 nm.) (C) Cells were mechanically scraped off of a Congo red-containing YESCA plate by using disposable inoculating loops to compare the Congo red-binding phenotypes of WT/pLR2 (empty vector; labeled “1”), csgBF/pNH1 (csgBtrunc; labeled “2”), csgFBA/pNH1 (csgBtrunc; labeled “3”), or csgFBA/pLR2 (empty vector; labeled “4”). (D) The Congo red-containing YESCA plate used to cultivate cells in Fig. 4C after cells have been removed. In D, the numbers indicate the region of the Congo red plate where the following strains had grown: 1, WT/pLR2; 2, csgBF/pNH1; 3, csgFBA/pNH1; and 4, csgFBA/pLR2.
We further investigated the Congo red binding phenotype of the csgBF/pNH1 (csgBtrunc) strain as it compared with WT by mechanically scraping the cells off the Congo red YESCA plate after 48 h of growth. When scraped off the plate, WT cells were stained a deep red color (Fig. 5C area 1). In contrast, the csgBF/pNH1 (csgBtrunc) appeared pink (Fig. 5C area 2). The nonfiber-producing csgFBA/pNH1 (csgBtrunc) appeared light brown (Fig. 5C area 3). The portion of the plate where the WT cells had grown was white/very light red (Fig. 5D area 1). The cell-associated fibers produced by the WT cells bound the Congo red dye leaving little dye in the agar plate. Surprisingly, the portion of the Congo red agar plate where the csgBF/pNH1 (csgBtrunc) grew appeared bright red, outlining the areas of cellular growth (Fig. 5D area 2). These data suggest a portion of the fibers produced by the csgBF/pNH1cells were localized to the agar plate.
Discussion
Nucleus formation is proposed to be the rate-limiting step during amyloid fiber formation (37). Despite years of research on the eukaryotic amyloids, curli are the only known amyloid pathway to have a dedicated nucleator protein in vivo. Mounting evidence suggests that amyloid precursors are cytotoxic, whereas the fibers are an innocuous final product of the misfolding event (38–46). Functional amyloidosis may have evolved specialized mechanisms to eliminate the build up of the cytotoxic intermediates. For example, PMEL17, a mammalian functional amyloid, self aggregates so quickly that no toxic intermediates are hypothesized to form (47). Using a dedicated nucleator may be an alternative strategy to decrease exposure to cytotoxic intermediates. In the curli system, CsgA polymerization in vitro contains a lag phase that can be shortened by CsgBtrunc, consistent with the idea that CsgBtrunc facilitates the direct conversion of CsgA to a mature amyloid fiber.
Curli assembly on the cell surface of E. coli is a highly regulated and elegant process. In a simple model of curli formation, the major curli subunit CsgA is secreted from the cell as a soluble, unpolymerized protein. CsgA would remain unpolymerized unless it contacts a CsgB-expressing cell. In a process called nucleation, CsgB mediates the conversion of CsgA from a soluble protein to an insoluble fiber. This conversion marks the first step of curli biogenesis. Once fiber formation is initiated, CsgA can then use the growing fiber tip as a template for additional polymerization in a process called seeding. We propose CsgB-mediated “nucleation” and fiber mediated “seeding” of CsgA polymerization to be mechanistically similar processes.
A refined model of extracellular nucleation/precipitation is suggested based on our work. The mature CsgB protein contains five imperfect repeating units that share similarities with the five repeating units in CsgA, and which have been shown to be amyloidogenic (24, 27). We found that a C-terminal CsgB truncation mutant lacking the fifth repeating unit (CsgBtrunc) no longer associates with the outer membrane and is secreted into the cellular milieu (Figs. 1B, 4B, and 5D). CsgBtrunc contains an amyloidogenic domain (Fig. 2), which is sufficient to nucleate soluble CsgA in vitro (Fig. 3A). However, CsgBtrunc does not complement a csgB mutant in vivo, possibly because CsgBtrunc is not optimally positioned on the cell surface to act on newly secreted CsgA molecules. This defect is overcome when CsgBtrunc is expressed in csgB genetic backgrounds manipulated to secrete increased levels of CsgA (Fig. 4, SI Fig. 8). Thus, CsgBtrunc is sufficient to perform nucleation in a csgB background when presented with increased levels of CsgA.
Our model predicts that the fifth repeating unit of CsgB may not be essential for nucleation, but might be required for outer-membrane association such that it is optimally localized to nucleate CsgA monomers as they are secreted. However, in vitro polymerization studies with full-length CsgB protein may reveal that the C-terminal repeating unit contributes, either negatively or positively, to the kinetics of fiber formation and is not simply an outer membrane anchor. Therefore, it remains to be seen whether the C terminus of CsgB similarly modulates fiber formation directly, or is merely a localization factor that indirectly aids the nucleation process (i.e., retaining CsgB near the highest concentrations of CsgA). However, these two functions of the CsgB C terminus are not necessarily mutually exclusive, and this would not be the first example of a functional amyloid having a repeating unit(s) that serves two functions. Studies investigating the effect of deleting repeating units from the C terminus of the yeast prion protein Sup35 showed that some repeating units contribute directly to fiber growth, some contribute to the inheritance phenotype, and some contribute to both (48, 49). In our curli model, the C-terminal repeating unit of CsgB anchors the protein to the outer membrane, the optimum locale for interaction with CsgA monomers.
An inherent problem in functional amyloid systems is maintaining control of a self-propagating reaction such that it does not occur at the wrong time or in the wrong place. We propose that the curli system has achieved control by separating the nucleation and elongation properties into two distinct proteins (CsgB and CsgA, respectively) and then dictating when and where those proteins interact. The separation of nucleation and polymerization may be a general mechanism to harness functional amyloid formation. This mechanism makes curli an ideal system to study the initial stages of amyloid fiber formation from both an in vivo and in vitro perspective.
Materials and Methods
Bacterial Growth.
To induce curli production, bacteria were grown at 26°C on YESCA plates (17). Curli production was monitored by using Congo red-YESCA plates (17). Antibiotics were added to the growth media in the following concentrations: kanamycin, 50 μg/ml; chloramphenicol, 25 μg/ml; or ampicillin, 100 μg/ml.
Bacterial Strains and Plasmids.
Strains and plasmids used in this study are found in SI Table 1. Primer sequences used in this study can be found in SI Table 2. Plasmid pNH1, containing csgBtrunc under the control of the csgBA promoter, was constructed by amplifying csgBtrunc using the CsgBtrunc CF and CsgBtrunc CR primers, digesting the PCR product with NcoI and BamHI, and ligating into pLR2. Plasmid pNH2 contains a C-terminally his-tagged version of csgBtrunc. The csgBtrunc insert was amplified by using the CsgBFor and CsgBtruncRhis primers and was ligated into the NdeI and EcoRI sites of the IPTG-inducible plasmid pMC3. WT csgB was his-tagged and cloned into pMC3 by using the WTCsgBFor and WTCsgBRhis primers resulting in plasmid pNH4. The pNH3 empty vector plasmid was constructed by gel-purifying the NcoI–BamHI double digestion product of pNH2. The digestion product was blunt ended, ligated, and transformed into LSR12. The csgFBA mutant was constructed in the MHR 592 background, and the csgBA operon was deleted by using the Lambda Red system and the following primers: CsgBAREDP1 and CsgAREDP2 (50). Strain LSR12 is the flp-mediated deletion of the curli operon in LSR6.
CsgA Purification.
CsgA was purified as previously described (27).
Thioflavin T Assay.
Thioflavin T assays were performed as previously described (27). To create CsgBtrunc seeds, freshly purified CsgBtrunc was incubated at room temperature for at least 24 h with shaking. Before use, seed samples were sonicated by using a sonic dismembrator (Fisher Model 100; Fisher, Pittsburgh, PA) for three 15-second bursts on ice.
CD Spectroscopy.
CsgBtrunc samples were assayed in a Jasco (Tokyo, Japan) J-810 spectropolarimeter from 190 to 250 nm in a quartz cell with a 1-mm path length at 25°C.
Electron Microscopy.
A Philips (Amsterdam, The Netherlands) CM12 Scanning Transmission Electron Microscope was used to visualize the fiber aggregates. Samples were absorbed onto formvar-coated copper grids (Ernest F. Fullam, Latham, NY) for 2 min, washed with deionized water, and negatively stained with 2% uranyl acetate for 90 seconds.
SDS/PAGE and Western Blotting.
Bacteria WC lysates were prepared and probed for both CsgB and CsgA by using previously described methods (29). Blots were probed for CsgB first, developed, and then stripped by using 100 mM 2-mercaptoethanol, 2% SDS, and 62.5 mM Tris·HCl, pH 6.7 before reprobing for CsgA.
Supplementary Material
Acknowledgments
We thank Dr. S. Hultgren's (Washington University, St. Louis, MO) laboratory for supplying us with strains and plasmids; members of M.R.C.'s and Dr. Hultgren's laboratories for helpful discussions and review of this manuscript; the Ursula Jakob laboratory for technical assistance while completing the CD experiments; and the German Academic Exchange Service for supplying funding for J.C.S. This work was supported by National Institutes of Health Grant AI073847-01 and Michigan Alzheimer's Disease Research Center award P50-A608671.
Abbreviations
- TEM
transmission electron microscopy
- WC
whole cell
- CD
circular dichroism
- YESCA
yeast extract casamino acids.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703310104/DC1.
References
- 1.Kikuchi T, Mizunoe Y, Takade A, Naito S, Yoshida S. Microbiol Immunol. 2005;49:875–884. doi: 10.1111/j.1348-0421.2005.tb03678.x. [DOI] [PubMed] [Google Scholar]
- 2.Wang X, Rochon M, Lamprokostopoulou A, Lunsdorf H, Nimtz M, Romling U. Cell Mol Life Sci. 2006;63:2352–2363. doi: 10.1007/s00018-006-6222-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gophna U, Barlev M, Seijffers R, Oelschlager TA, Hacker J, Ron EZ. Infect Immun. 2001;69:2659–2665. doi: 10.1128/IAI.69.4.2659-2665.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bian Z, Brauner A, Li Y, Normark S. J Infect Dis. 2000;181:602–612. doi: 10.1086/315233. [DOI] [PubMed] [Google Scholar]
- 5.Bian Z, Yan ZQ, Hansson GK, Thoren P, Normark S. J Infect Dis. 2001;183:612–619. doi: 10.1086/318528. [DOI] [PubMed] [Google Scholar]
- 6.Uhlich GA, Cooke PH, Solomon EB. Appl Environ Microbiol. 2006;72:2564–2572. doi: 10.1128/AEM.72.4.2564-2572.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vidal O, Longin R, Prigent-Combaret C, Dorel C, Hooreman M, Lejeune P. J Bacteriol. 1998;180:2442–2449. doi: 10.1128/jb.180.9.2442-2449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barnhart MM, Chapman MR. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerstel U, Romling U. Res Microbiol. 2003;154:659–667. doi: 10.1016/j.resmic.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Barak JD, Gorski L, Naraghi-Arani P, Charkowski AO. Appl Environ Microbiol. 2005;71:5685–5691. doi: 10.1128/AEM.71.10.5685-5691.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jeter C, Matthysse AG. Mol Plant Microbe Interact. 2005;18:1235–1242. doi: 10.1094/MPMI-18-1235. [DOI] [PubMed] [Google Scholar]
- 12.Ryu JH, Kim H, Frank JF, Beuchat LR. Lett Appl Microbiol. 2004;39:359–362. doi: 10.1111/j.1472-765X.2004.01591.x. [DOI] [PubMed] [Google Scholar]
- 13.Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. Mol Microbiol. 1995;18:661–670. doi: 10.1111/j.1365-2958.1995.mmi_18040661.x.. [DOI] [PubMed] [Google Scholar]
- 14.Bian Z, Normark S. EMBO J. 1997;16:5827–5836. doi: 10.1093/emboj/16.19.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammar M, Bian Z, Normark S. Proc Natl Acad Sci USA. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White AP, Collinson SK, Banser PA, Gibson DL, Paetzel M, Strynadka NC, Kay WW. J Mol Biol. 2001;311:735–749. doi: 10.1006/jmbi.2001.4876. [DOI] [PubMed] [Google Scholar]
- 17.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen FE, Kelly JW. Nature. 2003;426:905–909. doi: 10.1038/nature02265. [DOI] [PubMed] [Google Scholar]
- 19.Lashuel HA, Hartley D, Petre BM, Walz T, Lansbury PT., Jr Nature. 2002;418:291. doi: 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- 20.Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallagher M, Ashe KH. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 21.Olsen A, Jonsson A, Normark S. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- 22.Collinson SK, Emody L, Muller KH, Trust TJ, Kay WW. J Bacteriol. 1991;173:4773–4781. doi: 10.1128/jb.173.15.4773-4781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collinson SK, Doig PC, Doran JL, Clouthier S, Trust TJ, Kay WW. J Bacteriol. 1993;175:12–18. doi: 10.1128/jb.175.1.12-18.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collinson SK, Parker JM, Hodges RS, Kay WW. J Mol Biol. 1999;290:741–756. doi: 10.1006/jmbi.1999.2882. [DOI] [PubMed] [Google Scholar]
- 25.Naiki H, Hasegawa K, Yamaguchi I, Nakamura H, Gejyo F, Nakakuki K. Biochemistry. 1998;37:17882–17889. doi: 10.1021/bi980550y. [DOI] [PubMed] [Google Scholar]
- 26.Serio TR, Cashikar AG, Kowal AS, Sawicki GJ, Moslehi JJ, Serpell L, Arnsdorf MF, Lindquist SL. Science. 2000;289:1317–1321. doi: 10.1126/science.289.5483.1317. [DOI] [PubMed] [Google Scholar]
- 27.Wang X, Smith DR, Jones JW, Chapman MR. J Biol Chem. 2007;282:3713–3719. doi: 10.1074/jbc.M609228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Nuallain B, Williams AD, Westermark P, Wetzel R. J Biol Chem. 2004;279:17490–17499. doi: 10.1074/jbc.M311300200. [DOI] [PubMed] [Google Scholar]
- 29.Robinson LS, Ashman EM, Hultgren SJ, Chapman MR. Mol Microbiol. 2006;59:870–881. doi: 10.1111/j.1365-2958.2005.04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loferer H, Hammar M, Normark S. Mol Microbiol. 1997;26:11–23. doi: 10.1046/j.1365-2958.1997.5231883.x. [DOI] [PubMed] [Google Scholar]
- 31.Vassar PS, Culling CF. Arch Pathol. 1959;68:487–498. [PubMed] [Google Scholar]
- 32.Klunk WE, Pettegrew JW, Abraham DJ. J Histochem Cytochem. 1989;37:1273–1281. doi: 10.1177/37.8.2666510. [DOI] [PubMed] [Google Scholar]
- 33.LeVine H., III Protein Sci. 1993;2:404–410. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.LeVine H., III Methods Enzymol. 1999;309:274–284. doi: 10.1016/s0076-6879(99)09020-5. [DOI] [PubMed] [Google Scholar]
- 35.Klunk WE, Jacob RF, Mason RP. Methods Enzymol. 1999;309:285–305. doi: 10.1016/s0076-6879(99)09021-7. [DOI] [PubMed] [Google Scholar]
- 36.Kayed R, Bernhagen J, Greenfield N, Sweimeh K, Brunner H, Voelter W, Kapurniotu A. J Mol Biol. 1999;287:781–796. doi: 10.1006/jmbi.1999.2646. [DOI] [PubMed] [Google Scholar]
- 37.Rochet JC, Lansbury PT., Jr. Curr Opin Struct Biol. 2000;10:60–68. doi: 10.1016/s0959-440x(99)00049-4. [DOI] [PubMed] [Google Scholar]
- 38.Sousa MM, Cardoso I, Fernandes R, Guimaraes A, Saraiva MJ. Am J Pathol. 2001;159:1993–2000. doi: 10.1016/s0002-9440(10)63050-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lue LF, Kuo YM, Roher AE, Brachova L, Shen Y, Sue L, Beach T, Kurth JH, Rydel RE, Rogers J. Am J Pathol. 1999;155:853–862. doi: 10.1016/s0002-9440(10)65184-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McLean CA, Cherny RA, Fraser FW, Fuller SJ, Smith MJ, Beyreuther K, Bush AI, Masters CL. Ann Neurol. 1999;46:860–866. doi: 10.1002/1531-8249(199912)46:6<860::aid-ana8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 41.Wang J, Dickson DW, Trojanowski JQ, Lee VM. Exp Neurol. 1999;158:328–337. doi: 10.1006/exnr.1999.7085. [DOI] [PubMed] [Google Scholar]
- 42.Moechars D, Dewachter I, Lorent K, Reverse D, Baekelandt V, Naidu A, Tesseur I, Spittaels K, Haute CV, Checler F, et al. J Biol Chem. 1999;274:6483–6492. doi: 10.1074/jbc.274.10.6483. [DOI] [PubMed] [Google Scholar]
- 43.Larson J, Lynch G, Games D, Seubert P. Brain Res. 1999;840:23–35. doi: 10.1016/s0006-8993(99)01698-4. [DOI] [PubMed] [Google Scholar]
- 44.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM, Stefani M. Nature. 2002;416:507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 45.Sirangelo I, Malmo C, Iannuzzi C, Mezzogiorno A, Bianco MR, Papa M, Irace G. J Biol Chem. 2004;279:13183–13189. doi: 10.1074/jbc.M308207200. [DOI] [PubMed] [Google Scholar]
- 46.Malisauskas M, Ostman J, Darinskas A, Zamotin V, Liutkevicius E, Lundgren E, Morozova-Roche LA. J Biol Chem. 2005;280:6269–6275. doi: 10.1074/jbc.M407273200. [DOI] [PubMed] [Google Scholar]
- 47.Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, Kelly JW. PLoS Biol. 2006;4:e6. doi: 10.1371/journal.pbio.0040006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osherovich LZ, Cox BS, Tuite MF, Weissman JS. PLoS Biol. 2004;2:e86. doi: 10.1371/journal.pbio.0020086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parham SN, Resende CG, Tuite MF. EMBO J. 2001;20:2111–2119. doi: 10.1093/emboj/20.9.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Datsenko KA, Wanner BL. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.