Abstract
Nup475 is a nuclear zinc-binding protein of unknown function that is induced in mammalian cells by growth factor mitogens. Nup475 contains two tandemly repeated sequences YKTELCX8CX5CX3H (Cys3His repeats) that are thought to be zinc-binding domains. Similar sequences have been found in a number of proteins from various species of eukaryotes. To determine the metal binding properties and secondary structure of the putative zinc-binding domains of Nup475, we have used synthetic or recombinant peptides that contain one or two domain sequences. The peptide with a single domain bound 1.0 ± 0.1 equivalents of Co2+, and the peptide with two domains bound 1.7 ± 0.4 equivalents of Co2+. Both peptides bound Co2+ and Zn2+ with affinities similar to those of classical zinc finger peptides. In each case, the Co2+ complex exhibited strong d-d transitions characteristic of tetrahedral coordination. For structural studies by nuclear magnetic resonance spectroscopy, we used a more soluble two-domain peptide that had a single amino acid substitution in a nonconserved amino acid residue in the second Cys3His repeat. The mutant peptide unexpectedly showed loss of one of its metal binding sites and displayed ordered structure for only the first Cys3His sequence. On the basis of the nuclear magnetic resonance data, we propose a structure for the Nup475 metal-binding domain in which the zinc ion is coordinated by the conserved cysteines and histidine, and the conserved YKTEL motif forms a parallel sheet-like structure with the C terminus of this domain. This structure is unlike that of any previously described class of metal binding domain.
Keywords: zinc fingers, growth factor-induced proteins
Nup475 [also known as Tris-tetraprolin (1) and TIS11 (2)] contains two tandem repeats of a consensus amino acid sequence YKTELCX8CX5CX3H that is thought to be a novel zinc-binding motif (3). The Nup475 gene, originally identified as one of a set of immediate early genes induced in response to serum or mitogenic growth factors in murine BALB/c 3T3 fibroblasts (3), is expressed in a variety of tissues including regenerating liver, thymus, intestine, and early placenta and in various cultured cells in response to a wide variety of extracellular stimuli, including platelet-derived growth factor, insulin-like growth factor I, interferon-γ, forskolin, and depolarizing agents (3); fibroblast growth factor, epidermal growth factor, and nerve growth factor (2); phorbol esters (4); and insulin (1). In the colonic crypt of rats, a model of proliferation, differentiation, and senescence, the Nup475 messenger RNA has been localized to the postmitotic crypt (5). Initial investigations established that the Nup475 protein is a zinc-binding nuclear protein (3); based on these properties, it was suggested that Nup475 is a transcriptional regulator. However, its actual function remains unknown.
On the basis of sequence similarity to the YKTELCX8CX5CX3H consensus sequence (Cys3His repeat), a number of related proteins in mammals (6, 7, 8), yeast (9, 10), nematodes (C. Mello, personal communication), and Drosophila (11, 12) have been identified. A recent data base search for homologous sequences expanded this family to 14 proteins containing 27 Cys3His domains (Fig. 1) not including proteins that are so highly conserved that they represent the homologous proteins in different species. A majority of the proteins contain two domains with the exception of the Drosophila protein Unkempt (13), that contains five domains and others that contain one domain each.
Figure 1.

Alignment of Nup475 Cys3His domain homologs. Nup475-related Cys3His domains were identified through a Genquest Q search of the Swiss Prot data base using the Cys3His protein sequences of Nup475 and those of known family members to probe the database. Note the conservation of cysteines, histidines, glycines, and aromatic residues. The first seven proteins are previously described members of this family, the last seven are the result of our database search. A striking feature of the latter group is the number of proteins known to be involved in some form of posttranslational RNA metabolism or as part of RNA viruses, although in no case has this motif been shown to directly bind RNA. Not included are the highly conserved human, rat, and bovine homology of Nup475/TTP/TIS11, TIS11b, and TIS11d; only the murine proteins are shown. (GenBank accession nos. for the proteins shown: Nup475, M58691M58691; TIS11b, M58566M58566; TIS11d, M58564M58564; YTIS11p, S76619S76619; DTIS11, U13397U13397; Unkempt, Z11527Z11527; zfs1+, D49913D49913; TIScc1, X81194X81194; human U2AF35, M96982M96982; human U2AFBP-RS, S69507S69507; Drosophila melanogaster Suppressor of sable, M57889M57889; human respiratory synticial virus A2, M11486M11486; turkey rhinotracheitis virus, X63408X63408; and bovine respiratory synticial virus, M82816M82816).
That Nup475 is physiologically important is indicated by a recent preliminary report showing that mice homozygous for a disrupted Nup475 gene develop postnatal cachexia, arthritis, conjunctivitis, dermatitis, and a marked increase in myeloid cells (14). A chromosomal translocation in a human acute T-cell leukemia in a related protein (the human TIS11d) fuses the N terminus of this protein, including the Cys3His repeats, to a fragment of the T-cell receptor β-chain (15), suggesting that the fusion protein may have a role in leukemogenesis. Deletion of a Saccharomyces cerevisiae homolog, YTIS11p, leads to changes in cellular metabolism, causing alkalinization of the medium around the colony when glucose is the carbon source; intact Cys3His repeats were essential to complement the metabolic defect. The N terminus of this protein also contains a potent transcriptional activation domain (9), suggesting that the protein might function as a transcriptional activator. Two proteins, the Schizosaccharomyces pombe zfs1 (10) and the Drosophila melanogaster TIScc1 (12), were isolated by their ability to suppress abnormal mating and cell cycle regulatory phenotypes, respectively, when expressed in mutant S. pombe strains. Both have two Cys3His motifs, and the zfs1+ protein has been reported to bind DNA, although the details of this binding have yet to be described. Sequence-specific DNA or RNA binding has not been described for this class of proteins. Where a function is known at the molecular level, Nup475 family members appear to be involved in some form of posttranslational RNA metabolism or, in the case of some of the proteins with a single Cys3His motif, as part of the nucleocapsid of RNA viruses. However, none of these proteins has been shown to bind RNA directly.
Although it has been assumed that the Cys3His motif is a novel zinc binding domain, only Nup475 has been shown experimentally to bind Zn2+ (3). To characterize the metal binding properties and structure of the Nup475 Cys3His domain, we have used peptides corresponding to the putative metal-binding domains of Nup475 to demonstrate that this domain binds metals with high affinity and to determine by nuclear magnetic resonance spectroscopy the secondary structure of this novel domain.
MATERIALS AND METHODS
Homology Searches.
Putative homologs were identified from the Swiss Prot data base (16) using the Genquest Q server (Oak Ridge National Laboratories, Oak Ridge, TN). This method tolerated smaller amino acid test sequences than other data base search methods. The Nup475 Cys3His amino acid sequences and similar sequences in related proteins were used to probe the data base.
Peptide Synthesis.
The peptide Nup475SD (Fig. 2), containing the first Cys3His motif of Nup475 (amino acids 93–123) (3), was synthesized, purified, and maintained in an anaerobic atmosphere using methods described (17) except that peptide synthesis was performed on PEG-polystyrene matrix using Arginine(Pmc) amino acids (both from Millipore).
Figure 2.
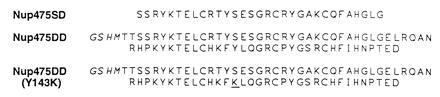
Sequences of the synthetic peptide Nup475SD and the recombinant, thrombin-cleaved wild-type Nup475DD and Nup475DD(Y143K) mutant two-domain peptides. Additional residues conferred by the pET28b vector in the two-domain proteins are italicized; the residue mutated from tyrosine-to-lysine in Nup475DD(Y143K) is underlined.
Cloning of Two-Domain Nup475 Construct into Escherichia coli Expression Plasmids and the Generation of Mutants.
Primers were designed to encode Nup475 amino acids 91–165 with 5′ NdeI and 3′ stop codon/PstI sites. A Nup475 cDNA plasmid provided the template for PCR synthesis (18). The PCR product was cloned into the NdeI and PstI sites of the E. coli expression plasmid pG5 and the sequence confirmed (pMW54). This insert was further subcloned into similarly cut pET28b (Novagen) for expression (pMW55) and metal affinity chromatography as a hexahistidine fusion protein. The thrombin-cleaved protein product of this plasmid was known as Nup475DD Fig. 2). The pMW54 insert was shuttled into the XbaI and PstI sites of Bluescript KS(−) (pMW59) for site-directed mutagenesis. Codons corresponding to amino acid residues Tyr143, Leu144, and Ile157 in the full-length protein were mutated to those for serine or lysine using the method of recombinant-circle PCR to form plasmids pMW60 to pMW65 (19). Confirmed sequences were digested with NdeI/PstI and cloned into similarly cut pET28b for expression (pMW66-70). The protein product of one of the mutants (pMW62) had favorable solubility characteristics and corresponded to Tyr143 of the full-length protein mutated to lysine (Fig. 2). This protein, dubbed Nup475DD(Y143K), was the subject of further NMR and spectroscopic studies.
E. coli Expression.
Expression plasmids were introduced into competent BL21(DE3)pLysS cells (Novagen) with kanamycin and chloramphenicol selection as per the manufacturer’s protocol. Unlabeled cells were grown in Luria–Bertani medium to an absorbance at 600 nm of 0.6–0.9 at 37°C, at which time expression was induced for 3 hr with isopropyl β-d-thiogalactoside to 1 mM and ZnCl2 to 100 μM. Solubilities were crudely assessed by sonication and Tris·Tricine SDS/PAGE of comparable amounts of pellet and supernatant (20). Minimal medium expression of the Nup475DD(Y143K) mutant was performed using Mops minimal medium (21), omitting the thymine and uracil, using 4 g/liter glucose and growing the cells at 37°C throughout. The night before induction, 1/500 dilutions of Luria broth starter cultures were grown in the Mops minimal media with 0.06% glycerol as the sole carbon source, and the glucose was added early the next morning. 15N labeling was accomplished by substituting 1 g of 15NH4Cl per liter for the NH4Cl; 13C/15N labeling was performed by additionally substituting U-13C-glucose and U-13C-glycerol in the medium. Induction was performed as above and expression was maximal at about 4 hr. Purification, cleavage, and analysis of the labeled protein was performed as mentioned above; both the 15N and 13C/15N had >95% incorporation of the label assessed by matrix-assisted laser desorption mass spectrometry (22).
Protein Purification.
Five or six frozen E. coli pellets representing 500-ml culture medium each were suspended in 40 ml binding buffer/pellet (6 M guanidinium chloride/40 mM sodium phosphate, pH 8.1/500 mM NaCl), and lysed by passage three times through a French pressure cell. The lysate was spun at 20,000 × g in a Sorvall SS-34 rotor for 60 min and the supernatant poured over 15-ml settled bed volume Chelating Fastflow Sepharose (Pharmacia) previously charged with 15 ml of 0.3 M zinc sulfate, washed as per the manufacturer’s protocol, and equilibrated with binding buffer. The bound protein was washed with an additional 60 ml binding buffer, 60 ml wash buffer (same composition, but pH 6.15) and eluted in buffer of the same composition but at pH 3.5.
Reduction and HPLC Purification of Purified E. coli Protein and Cleaved Peptide.
The eluted protein fraction was adjusted to a pH between 7 and 8 with 10 N NaOH dropwise, an estimated 10-fold excess of DTT was added, and the mixture was incubated at 55°C for 2 hr. The solution was allowed to cool to ambient temperature, the pH was reduced to 1.0–1.5 with concentrated HCl dropwise (to elute bound metal), and the protein was purified on a Rainin C4 reverse-phase HPLC column with a 0–40% gradient of acetonitrile containing 0.1% trifluoroacetic acid. Collected peaks were pooled and dried in a Savant Speedvac concentrator under a 95% nitrogen/5% hydrogen atmosphere to avoid oxidation. All manipulations after this step were performed in this atmosphere whenever possible.
Fusion Protein Cleavage.
The fusion protein pellets were reconstituted in helium-degassed MilliQ water (Millipore) to approximately 1.0–1.7 mg/ml concentration with 1/10 final volume of 10× thrombin cleavage buffer added (20 mM Tris·HCl, pH 8.3/150 mM NaCl/2.5 mM CaCl2; Novagen). The pH was titrated to between 7 and 8 with 1.0 M Hepes base. Test cleavage of a small amount of product with biotinylated thrombin (Novagen) was performed and analyzed by on a Tris·Tricine SDS gel; typically 1 Novagen unit of thrombin was sufficient to cleave 1 mg fusion protein in 1–2 hr at ambient temperature with minimal nonspecific proteolysis. Preparative reactions were stopped with the addition of phenylmethylsulfonyl fluoride to 1 mM; passed over a 1 ml Zn2+-charged Chelating FF Sepharose column, and the flow-through and pH 8.1 and pH 6.15 washes were pooled, reduced, and again HPLC-purified as above. Speedvac-dried pellets were analyzed by Tris·Tricine SDS/PAGE (20), mass spectrometry (22), and 5–5′dithiobis(2-nitrobenzoic acid)/visible spectrometry (23).
UV/Visible Spectroscopy of Nup475SD, Nup475DD, and Nup475DD(Y143K) Peptides with Cobalt and Zinc.
Spectroscopic studies of cobalt titrations and zinc back-titrations were performed in teflon-stoppered cuvettes on a Perkin–Elmer Lambda 9 spectrophotometer as described (18), at protein concentrations of 20–50 μM in 1 ml of titration buffer (100 mM NaCl/50 mM Hepes, pH 7.10); 1 cm path length. All manipulations were performed in an anaerobic atmosphere. Zinc back-titrations were performed with a 300-fold excess of cobalt in the cuvette. The data were volume-corrected and curves were fit using nonlinear least squares analysis as described (17) .
NMR Studies of the Nup475DD and Nup475DD(Y143K) Peptides.
Cleaved protein pellets were reconstituted in helium-degassed 90% MilliQ water/10% 2H2O or 100% 2H2O, the protein concentration estimated by tyrosine absorbance at 276 nm (ɛ= 1400 M−1·cm−1), and 2.2 equivalents of ZnCl2 added per equivalent of protein. The pH was adjusted by the stepwise addition of 1 M deuterated Tris (minimum 50 mM final concentration) to a pH of 5.8. The final volume was adjusted to 600 μl with the solvent. NMR studies were performed on a Varian Unity Plus 500-MHz spectrophotometer using a triple resonance gradient probe at 20°C. All data was processed using felix software (Biosym Technologies, San Diego) on Silicon Graphics (Mountain View, CA) workstations.
Two-dimensional double quantum-filtered correlated spectroscopy (24), phase-sensitive nuclear Overhauser effect spectroscopy (NOESY) collected by the hypercomplex method (25, 26), and TOCSY (total correlation spectroscopy) (27, 28) experiments were performed on the unlabeled Nup475DD and unlabeled NUP475DD(Y143K) peptide using solvent presaturation for the H2O samples. Three-dimensional 15N-TOCSY-HSQC (heteronuclear single quantum correlation spectroscopy) (29) and flip-back 1H-15N-NOE spectroscopy-HSQC (30, §) spectra were acquired on a uniformly labeled Nup475DD(Y143K) peptide. Three-dimensional 13C-15N HNCACB (31, 32) and 13C-HCCH-TOCSY (33) spectra were acquired on a uniformly 15N, 13C-labeled NUP475DD(Y143K). All NOE experiments were collected with a mixing time of 200 ms.
RESULTS AND DISCUSSION
One-Dimensional NMR Spectroscopy.
One-dimensional 1H NMR spectra were obtained for peptides Nup475SD and Nup475DD (Fig. 2) with and without 1.1 equivalents of Zn2+ (data not shown). Both peptides showed marked changes upon zinc binding in chemical shifts, suggesting significant conformational changes. Unfortunately, both the single (Nup475SD) and double (Nup475DD) domain proteins at concentrations of 1 mM or higher became significantly less soluble upon the addition of Zn2+. Nup475DD at 0.3 mM concentration appeared soluble and showed good chemical shift dispersion, suggesting a well-folded structure but the metal-bound form showed relatively broad lines, suggesting aggregation (Fig. 3). Two-dimensional data suitable for structural analysis could not be obtained from this sample.
Figure 3.
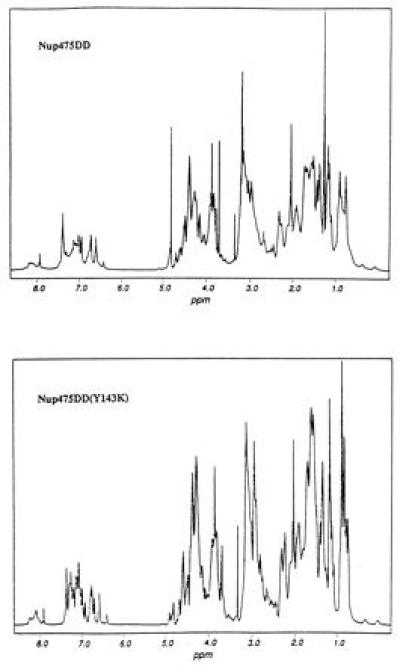
One-dimensional proton NMR in D2O of wild-type two-domain protein Nup475DD and the Nup475DD(Y143K) mutant. Line-broadening in case of the wild-type sample is consistent with aggregation. The comparable overall shape of the two spectra suggests that the protons are in similar chemical environment and are therefore in the same overall protein fold.
Site-directed mutagenesis of codons of nonconserved hydrophobic residues (by inspection of the homologous sequences in Fig. 1) was undertaken to generate a two-domain sample soluble enough for NMR structure determination. Residues in the second domain corresponding to Tyr143, Leu144, and Ile157 of the full-length protein were mutated to Ser and Lys, and the crude solubilities of these six proteins were assessed on a Tris·Tricine SDS protein gel loaded with comparable amounts of supernatant and pellet from isopropyl-β-thiogalactoside-induced E. coli cultures (data not shown). One of these mutants, the Tyr to Lys mutant [Nup475DD(Y143K), see Fig. 2], had favorable solubility and expression profiles and was chosen for further study. The one-dimensional 1H NMR spectrum of the Nup475DD(Y143K) mutant peptide exhibited a pattern of chemical shift dispersion nearly superimposable on the wild-type two-domain protein spectrum, but did not suffer from the line broadening of the parent molecule (Fig. 3). It was soluble at 1.7 mM at pH 5.8 and yielded many NOE crosspeaks.
Metal Binding.
The interaction of Co2+ with the single domain Nup475SD, the double domain Nup475DD, and the mutant double domain peptide Nup475DD(Y143K) were examined by a series of cobalt titrations and zinc back-titrations. All three peptides bound Co2+ and produced the characteristic strong d-d transitions seen in tetrahedrally coordinated complexes of this metal ion (Fig. 4). The position of these transitions is most consistent with S3N coordination. Thus, these spectra support the proposed three Cys and one His metal binding ligands suggested by the sequences. The Nup 475SD (Fig. 4a) and the Nup 475DD(Y143K) peptides (Fig. 4c) bound a single Co2+ with upper limit dissociation constants for cobalt of Kd Co2+ ≤ 10−7 M and for zinc of Kd Zn2+ ≤ 10−11 M. The Nup475DD (Fig. 4b) saturably bound 1.7 ± 0.4 equivalents of cobalt with an average dissocation constant with an upper boundary of Kd Co2+ ≤ 10−7 M. A well-defined dissocation constant (rather than an upper bound) for any of these peptides was not determined because of tight metal binding at pH 7. All of these dissociation constants indicate affinities for metal similiar to those of other well-characterized Zn2+-binding proteins, including a TFIIIA-type zinc finger peptide, CP-1 (Kd Co2+ = 3.8 × 10−6 M, Kd Zn2+ = 3 × 10−9) (34) or the Rauscher murine leukemia virus CX2CX4HX4C gag zinc finger peptide (Kd Co2+ = 2 × 10−8 M, Kd Zn2+ = 1 × 10−12 M) (35). This supports the notion that the Nup475 domains are capable of binding metal with high affinity and with physiologically meaningful parameters. Like other zinc-binding proteins with tetrahedral binding sites, zinc was preferred over cobalt by several orders of magnitude.
Figure 4.
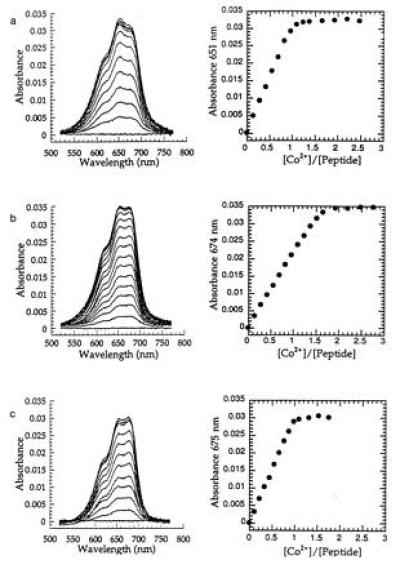
Absorption spectra of Co2+ complexes of proteins and peak absorbance vs. equivalents of metal bound of Nup475SD (a); Nup475DD (b); and Nup475DD(Y143K) (c). The Nup475SD spectra shown are at 43.9 μmol peptide, the Nup475DD spectra are at 26.2 μmol protein, and the Nup475(Y143K) spectra are at 30.1 μmol protein. The overall shapes of the d-d absorbances are similar to previously reported TFIIIA-type zinc fingers mutated to have Cys3His tetrahedral coordination (17). One of the effects of the Tyr to Lys mutation in Nup475(Y143K) (c) is that the mutant binds only one equivalent of metal, unlike the wild-type protein from which it was derived (b). Zinc back-titrations were performed in the same system using a 300-fold excess of cobalt.
The surprising result of our amino acid substitution corresponding to Tyr143 to Lys was that the Nup475DD(Y143K) peptide bound only one equivalent of metal even at 300-fold excess of cobalt, implying that the effect of the tyrosine-to lysine mutation is the significant reduction of the affinity of the second domain for metal ions. That this second domain saturably binds metal in the wild-type Nup475DD cobalt titrations supports the notion that this is a functional metal-binding site in the wild-type protein. The lack of binding in the Nup475DD(Y1443K) mutant peptide was unexpected because the mutation occurred at a nonconserved amino acid position.
Multidimensional NMR Spectroscopy Studies of Nup475DD(Y143K).
Sequential assignments for the Nup475DD(Y143K) peptide were accomplished from a combination of 15N-TOCSY-HSQC, flip-back 1H-15N-NOESY-HSQC, 13C-15N HNCACB, and 13C-HCCH-TOCSY experiments. Although the peptide was 77 amino acids long, assignments could be made only for residues 9–43, implying that the remaining part of the peptide (which includes the second Cys3His repeat) was loosely structured or unstructured. This is consistent with the cobalt binding data shown in Fig. 4. Fig. 5 shows some of the data used in assigning the NMR spectra for residues 9–43 and reveals that many NOE crosspeaks were observed. These crosspeaks along with additional long range NOEs suggest a structure consisting of a series of strands and turns. A schematic structure is shown in Fig. 6. The expected Cys and His metal binding ligands have crosspeaks including those between the Cys15 and Cys30 HN′s and the Hδ ring proton of His34 that indicate they are in close proximity and in a central location. Turns are indicated from cross strand NOEs seen between protons on Thr17, Tyr18, Gly22, and Arg23; on Tyr26, Gly27, and Cys30; and on Gln31, Phe32, and Ala33. The N-terminal residues (which correspond to the Y10KTEL14 motif) show crosspeaks to residues Ala33 and His34 and as well as crosspeaks to the structured sequence between residues 39–41. One of the functions of the YKTEL sequence appears to be to stabilize the overall protein fold through these interactions. This novel structure is not at all like the β-sheet/α-helical structure of the TFIIIA-like zinc fingers, or indeed that of any other class of structurally characterized zinc-binding motif. A full structure determination of the folded portion of Nup 475DD(Y143K) is in progress.
Figure 5.
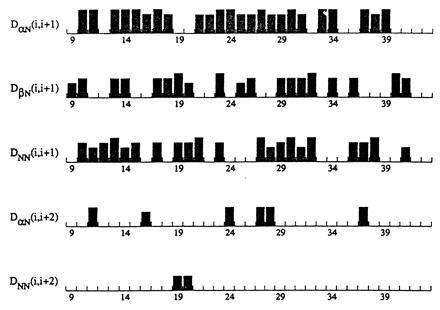
Sequential NOEs for amino acids 9–41 of the Nup475(Y143K) peptide. Numbering refers to the cleaved recombinant peptide, with the first amino acid after the cleavage site numbered 1. The height of the column represents the relative intensity of the cross peak. Not shown are HNCACB connectivities that run the span of the entire segment shown.
Figure 6.
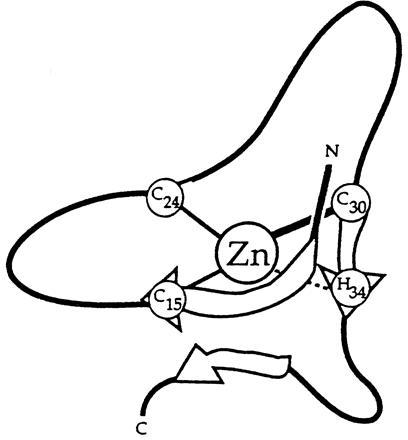
Schematic of the proposed secondary structure of the first Nup475 domain. The central zinc atom is coordinated by the three spatially conserved cysteines and histidine. Note the two turns and the suggestion of a parallel sheet formed by the conserved Y10KTEL14 motif and the C30QFAH34 residues surrounding the coordinating histidine.
An additional surprise was the finding that, in spite of the lack of conservation in the aligned sequences of the homologs, the structured part of the C terminus extends well beyond His-34, the last coordinating ligand, to include the sequence 35GLGELR41Q, and possibly beyond. The single domain peptide studied does not include this complete sequence, and the two domain peptides do not include the corresponding sequence for the second domain. These truncations may be responsible for the difficulties with solubility as the C terminal sequences may mask other regions of the structured domains that lead to aggregation when exposed (36). Preliminary studies of a non-mutant two domain construct with a longer C-terminal extension after the second domain revealed marked improvement in solubility over the Nup475DD construct, suggesting that residues that are C terminal to the coordinating histidine of the second Cys3His repeat may serve a role similar to that postulated for the 35GLGELR41Q segment. Further study of this new construct and additional analysis of the structure of the Nup475DD(Y143K) peptide should clarify these findings.
Conclusions.
We propose the Nup475 Cys3His repeat as the prototype of a new class of nuclear zinc-binding domain. This domain coordinates zinc with three conserved cysteines and a single histidine, and has conserved glycine and aromatic residues. The domain appears to be larger than first suggested, including more residues toward the C terminus. The secondary structure of this domain is one of turns between the coordinating residues, followed by a sheet-like structure joining the region N-terminal to the first cysteine to residues adjoining the coordinating histidine. Most likely there is a family of homologs of this domain that share the same or similar cysteine, histidine, glycine and aromatic spacings in the primary sequence and can be expected to bind metal with a similar overall protein fold. The detailed function of the Nup475 protein and the significance of the Cys3His domain remain to be determined. We are in the process of determining the high resolution structure of the Nup475DD(Y143K) peptide and exploring the function of the metal binding region that should shed additional light on the function of Nup475 and related proteins.
Acknowledgments
We gratefully acknowledge Dr. Chitanandra Abeygunawardana and Dr. Susumu Mori for their help with implementing the three-dimensional NMR experiments, and Dr. Gregory Kato and Dr. Timothy Schaefer for helpful discussions. This work was supported in part by the Lucille P. Markey Charitable Trust.
Footnotes
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. §1734 solely to indicate this fact.
Abbreviations: NOESY, nuclear Overhauser effect spectroscopy; TOCSY, total correlation spectroscopy; HSQC, heteronuclear single quantum correlation spectroscopy.
Abeygunawardana, C., Mori, S., van Zijl, P. & Mildvan, A. S., 37th Experimental Nuclear Magnetic Resonance Conference, Monterey, CA, March 27, 1996, p. WT4.
References
- 1.Lai W S, Stumpo D J, Blackshear P J. J Biol Chem. 1990;265:16556–16563. [PubMed] [Google Scholar]
- 2.Varnum B C, Ma Q F, Chi T H, Fletcher B, Herschman H R. Mol Cell Biol. 1991;11:1754–1758. doi: 10.1128/mcb.11.3.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubois R N, McLane M W, Ryder K, Lau L F, Nathans D. J Biol Chem. 1990;265:19185–19191. [PubMed] [Google Scholar]
- 4.Varnum B C, Lim R W, Kujubu D A, Luner S J, Kaufman S E, Greenberger J S, Gasson J C, Herschman H R. Mol Cell Biol. 1989;9:3580–3583. doi: 10.1128/mcb.9.8.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dubois R N, Bishop P R, Graves-Deal R, Coffey R J. Cell Growth Differ. 1995;6:523–529. [PubMed] [Google Scholar]
- 6.Taylor G A, Lai W S, Oakey R J, Seldin M F, Shows T B, Eddy R J, Blackshear P J. Nucleic Acids Res. 1991;19:3454. doi: 10.1093/nar/19.12.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor G A, Thompson M J, Lai W S, Blackshear P J. J Biol Chem. 1995;270:13341–13347. doi: 10.1074/jbc.270.22.13341. [DOI] [PubMed] [Google Scholar]
- 8.Kaneda N, Oshima M, Chung S Y, Guroff G. Gene. 1992;118:289–291. doi: 10.1016/0378-1119(92)90202-z. [DOI] [PubMed] [Google Scholar]
- 9.Ma Q, Herschman H R. Oncogene. 1995;10:487–494. [PubMed] [Google Scholar]
- 10.Kanoh J, Sugimoto A, Yamamoto M. Mol Biol Cell. 1995;6:1185–1195. doi: 10.1091/mbc.6.9.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma Q, Wadleigh D, Chi T, Herschman H. Oncogene. 1994;9:3329–3334. [PubMed] [Google Scholar]
- 12.Warbrick E, Glover D. Gene. 1994;151:243–246. doi: 10.1016/0378-1119(94)90664-5. [DOI] [PubMed] [Google Scholar]
- 13.Mohler J, Weiss N, Murli S, Mohammadi S, Vani K, Vasilakis G, Song C H, Epstein A, Kuang T, English J, Cherdak D. Genetics. 1992;131:377–388. doi: 10.1093/genetics/131.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor G A, Thompson M J, Lai W S, Blackshear P J. Mol. Endocrinol. 1996;10:140–146. doi: 10.1210/mend.10.2.8825554. [DOI] [PubMed] [Google Scholar]
- 15.Ino T, Yasui H, Hirano M, Kurosawa Y. Oncogene. 1995;11:2705–2710. [PubMed] [Google Scholar]
- 16.Bairoch, A. & Boeckmann, B. (1991) Nucleic Acids Res. 19, Suppl., 2247–2249. [DOI] [PMC free article] [PubMed]
- 17.Krizek B A, Amann B T, Kilfoil V J, Merkle D L, Berg J M. J Am Chem Soc. 1991;113:4518–4523. [Google Scholar]
- 18.Saiki R K, Gelfand D H, Stoffer S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 19.Jones D H, Winistorfer S C. BioTechniques. 1992;12:528–530. [PubMed] [Google Scholar]
- 20.Schagger H, von J G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 21.Serpersu E H, Shortle D, Mildvan A S. Biochemistry. 1986;25:68–77. doi: 10.1021/bi00349a011. [DOI] [PubMed] [Google Scholar]
- 22.Cornish T J, Cotter R J. Anal Chem. 1993;65:1043–1047. doi: 10.1021/ac00056a017. [DOI] [PubMed] [Google Scholar]
- 23.Riddles P W, Blakeley R L, Zerner B. Methods Enzymol. 1983;91:49–60. doi: 10.1016/s0076-6879(83)91010-8. [DOI] [PubMed] [Google Scholar]
- 24.Rance M, Sorensen O W, Bodenhausen G, Wagner G, Ernst R R, Wuthrich K. Biochem Biophys Res Commun. 1983;117:479–485. doi: 10.1016/0006-291x(83)91225-1. [DOI] [PubMed] [Google Scholar]
- 25.Jeener J, Meier B H, Bachmann P, Ernst R R. J Chem Phys. 1979;71:4546–4553. [Google Scholar]
- 26.Macura S, Ernst R R. Mol Phys. 1980;41:95–117. [Google Scholar]
- 27.Levitt M, Freeman R, Frenkiel T. J Magn Reson. 1982;47:328–330. [Google Scholar]
- 28.Bax A, Davis D G. J Magn Reson. 1985;65:355–360. [Google Scholar]
- 29.Zhang O, Kay L E, Olivier J P, Forman K J. J Biomol NMR. 1994;4:845–858. doi: 10.1007/BF00398413. [DOI] [PubMed] [Google Scholar]
- 30.Marion D, Driscoll P C, Kay L E, Wingfield P T, Bax A, Gronenborn A M, Clore G M. Biochemistry. 1989;28:6150–6156. doi: 10.1021/bi00441a004. [DOI] [PubMed] [Google Scholar]
- 31.Wittekind M, Mueller L. J Magn Reson Ser B. 1993;101:201–205. [Google Scholar]
- 32.Muhandiram D R, Kay L E. J Magn Reson Ser B. 1994;103:203–216. [Google Scholar]
- 33.Kay L E, Xu G-Y, Singer A U, Muhandiram D R, Forman-Kay J D. J Magn Reson Ser B. 1993;101:333–337. [Google Scholar]
- 34.Berg J M, Merkle D L. J Am Chem Soc. 1989;111:3759–3761. [Google Scholar]
- 35.Green L M, Berg J M. Proc Natl Acad Sci USA. 1990;87:6403–6407. doi: 10.1073/pnas.87.16.6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dyda F, Hickman A B, Jenkins T M, Engelman A, Craigie R, Davies D R. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]


