Abstract
Enlargement of normal terminal duct lobular units (TDLUs) by hyperplastic columnar epithelial cells is one of the most common abnormalities of growth in the adult female human breast. These hyperplastic enlarged lobular units (HELUs) are important clinically as the earliest histologically identifiable potential precursor of breast cancer. The causes of the hyperplasia are unknown but may include estrogen-simulated growth mediated by estrogen receptor-α, which is highly elevated in HELUs and may be fundamental to their development. The present study used DNA microarray technology and RNA from microdissected pure epithelial cells to examine changes in gene expression and molecular pathways associated with the development of HELUs from TDLUs. The results suggest that HELUs evolve from TDLUs primarily by reactivation of pathways involved in embryonic development and suppression of terminal differentiation. Changes in ERBB genes were particularly prominent, including a uniform switch in ligands for the ERBB1 receptor (14-fold decrease in epidermal growth factor and 10-fold increase in amphiregulin, respectively) in HELUs compared with TDLUs. Epidermal growth factor regulates terminal differentiation in adult breast and amphiregulin is critical to normal embryonic breast development. Because HELUs are such early potential precursors of breast cancer, targeting some of these alterations may be especially promising strategies for breast cancer prevention.
Enlargement of normal terminal duct lobular units (TDLUs) by hyperplastic columnar epithelial cells is one of the most common abnormalities in the adult female human breast.1,2 These hyperplastic enlarged lobular units (HELUs) are often multifocal, bilateral, and up to 100-fold larger (volume and numbers of cells) than the TDLUs they evolve from (Figure 1), representing a major alteration of growth.2 The majority of HELUs are lined by one or two layers of crowded columnar epithelial cells with minimal nuclear atypia, but a substantial minority exhibit more diverse histological features, contributing to the complex terminology that has evolved to describe them.1,2 The HELUs evaluated in this study were the common generally single-layer type with minimal nuclear atypia (see Supplemental Figure S1 at http://ajp.amjpathol.org), which would be referred to as “columnar cell change” using other currently popular terminology.3 We prefer the term HELUs over others because it conveys our viewpoint that epithelial hyperplasia and the resulting enlargement of lobules are the most fundamental characteristics and that the cells may then differentiate or progress in diverse ways depending on additional events (eg, mutations) that follow.
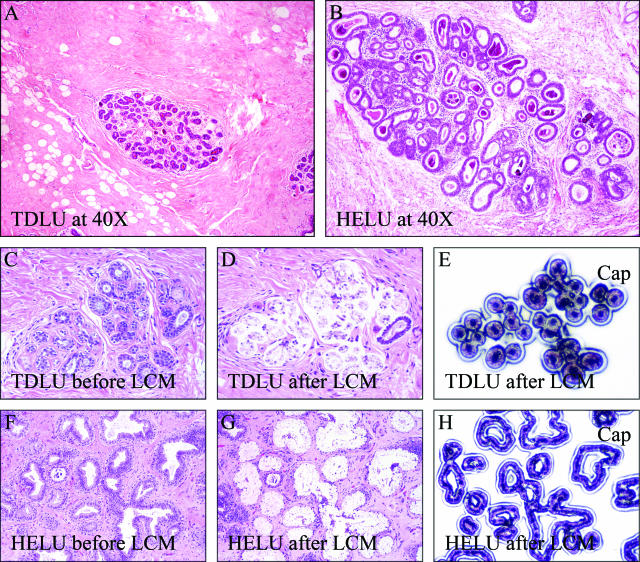
Figure 1.
A and B: HELUs are much larger (up to 100-fold increased volume and number of cells) than the normal TDLUs they evolve from (H&E stain). This study used DNA microarray technology and RNA isolated from nearly pure (>95%) populations of epithelial cells isolated by laser capture microdissection to compare gene expression profiles between paired TDLUs (C–E) and HELUs (F–H). Original magnifications, ×40 (A, B).
Histological, epidemiological, and more recent genetic evidence suggest that HELUs are forerunners of many other abnormalities in the breast, including microcysts, usual ductal hyperplasia, and atypical ductal hyperplasia (ADH).2,4,5 ADH is a well-established potential precursor of breast cancer6,7,8 and, in this sense, HELUs can be viewed as the earliest histologically identifiable lesion with premalignant potential, as suggested at least a century ago.1,9,10
Recent studies from our laboratory demonstrated that the epithelial cells lining HELUs show highly elevated expression of nuclear estrogen receptor-α (ER-α), significantly increased proliferation, and significantly decreased apoptosis compared with adjacent TDLUs.2 Estrogen, by binding and activating ER-α, regulates proliferation and apoptosis in the normal breast,11,12,13 suggesting that elevated ER-α may be a fundamental defect contributing to the development of HELUs, although the underlying reasons for elevated ER-α are unknown, and other mechanisms are almost certainly involved.
This study used DNA microarray technology to learn more about changes in gene expression and molecular pathways associated with the development of HELUs from TDLUs. Because HELUs are thought to be such an early potential precursor of breast cancer, identifying alterations associated with their development may reveal new and especially effective strategies for breast cancer prevention.
Materials and Methods
Tissue Samples
Samples for the microarray studies were derived from eight formalin-fixed paraffin-embedded tissue (FFPET) biopsies containing paired normal TDLUs and well-developed HELUs from noncancerous adult female human breasts obtained within the past 3 years. Populations of nearly pure (>95%) epithelial cells were isolated from the TDLUs and HELUs in each biopsy (∼25,000 cells per sample) by laser capture microdissection (Veritas; Arcturus, Mountain View, CA) (Figure 1). Ten additional paired samples of TDLUs and HELUs from independent FFPET biopsies were prepared in a similar manner for validation studies. Five of these ten samples also contained ADH arising within HELUs, which were also harvested by laser capture microdissection. All samples were anonymous and obtained with institutional review board approval (protocol H-10493).
Microarrays
Total RNA for the microarray experiments was purified from the samples using the Optimum kit (Ambion, Austin, TX), which is based on proteinase K digestion followed by phase separation in a proprietary microfilter cartridge and incubation with DNase to remove contaminating genomic DNA, yielding from 50 to 300 ng per sample. The purified total RNA was then linearly amplified for two rounds and converted to cDNA using the Paradise kit (Arcturus), which is based on an oligo-dT primer and T7 RNA polymerase. The cDNA was then in vitro transcribed to biotin-labeled cRNA with the IVT labeling kit (Affymetrix, Santa Clara, CA).
Gene expression was measured using U133-X3P Human GeneChip oligo-based microarrays (Affymetrix). These microarrays contain transcripts for an estimated 47,000 genes and 61,359 probe sets (many genes have multiple probe sets). A large majority of the probe sets were constructed to be within 300 bases of the 3′ end of the transcripts to accommodate the relatively fragmented RNA that is typically obtained from FFPET samples. In brief, amplified biotinylated cRNA was further purified with the RNeasy mini kit (Qiagen, Frankfurt, Germany) and fragmented (20 μg of cRNA) in 5× fragmentation buffer (200 mmol/L Tris-acetate, 500 mmol/L KOAc, and 150 mmol/L MgOac at pH 8.1) at a 4:1 volume ratio (cRNA/buffer) for 10 minutes to a range of 35 to 200 bases to optimize hybridization. Fragmented cRNA (20 μg) was then mixed with hybridization controls (bioB, bioC, bioD, and cre) and hybridized to the microarrays for 16 hours. After hybridization, the microarrays were washed, stained, and scanned to generate quantitative digital files. Affymetrix CEL files are available online at Gene Expression Omnibus (accession number GSE7377; http://www.ncbi.nlm.nih.gov/geo/). Measuring global gene expression in FFPET samples using microarray technology is challenging but feasible and accurate.14 The quality of the original RNA was inferred from the 3′/5′ ratios of an internal control gene (GAPDH) and the percent present calls of genes expressed above background in the microarray data, which fell well within satisfactory ranges for FFPET-derived samples as defined by the manufacturers of the kit used to amplify the RNA (Arcturus) and the microarray chip (Affymetrix), respectively (average for TDLUs = 15.9 and 24.5%; average for HELUs = 18.3 and 25.7%).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
qRT-PCR was used to measure the expression of selected genes starting with total RNA template from the original samples to confirm the ability of the microarrays to accurately measure gene expression in FFPET samples and from the independent validation samples to confirm further the biological findings overall. Briefly, first strand cDNA was synthesized from 1 to 5 ng of total RNA using the SuperScript II kit (Invitrogen, Carlsbad, CA). The quantitative PCR reactions were then performed using dual-labeled (5′/6-FAM and 3′/Black Hole Quencher) probes and GeneAmp Fast PCR Master Mix (Applied Biosystems, Foster City, CA) in an ABI 7500 Fast Real Time PCR System instrument (Applied Biosystems) with an initial denaturing cycle at 95°C for 20 seconds followed by 45 alternating production cycles of 95°C for 3 seconds and 60°C for 30 seconds.
Six genes of interest were evaluated in the original samples, including EGFR, EGF, AREG, annexin A8 (ANXA8), prominin 1 (PROM1), and v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (KIT). EGF and AREG were further evaluated in the independent validation samples. Human β2-microglobulin (B2M) was used as the internal reference gene in all analyses. Sequences for the probes and primers (Table 1) were designed using Primer Express Software v2.0 (Applied Biosystems) and synthesized by a commercial vendor (Eurogentec, San Diego CA).
Table 1.
Sequences for Primers and Probes of Genes Evaluated by qRT-PCR
| Gene | Sequence |
|---|---|
| EGFR forward primer | 5′-GCACATTTTGGGAAGTTGCA-3′ |
| EGFR reverse primer | 5′-CTGCTCAAAGGGACAATATTCTTG-3′ |
| EGFR probe | 5′-TCTTCAAACTGTGAAGCATTTACAGAAACGCA-3′ |
| EGF forward primer | 5′-GTGTGCTGGACGCCTGTCT-3′ |
| EGF reverse primer | 5′-CTTACGGAATAGTGGTGGTCATCTT-3′ |
| EGF probe | 5′-TTTGCCCTGACTCTACTCCACCCCCTC-3′ |
| AREG forward primer | 5′-ACCTGGAAGCAGTAACATGCAA-3′ |
| AREG reverse primer | 5′-ACTGTCAATCATGCTGTGAGTTTTC-3′ |
| AREG probe | 5′-CACACCGTTCACCGAAATATTCTTGCTGA-3′ |
| ANX8 forward primer | 5′-CAGGACTGGCCCTCCAAGA-3′ |
| ANX8 reverse primer | 5′-CATCTCATCAGTCCCACGAATC-3′ |
| ANX8 probe | 5′-CTCGCCTGCCGCATACAGATCCTG-3′ |
| PROM1 forward primer | 5′-CCAGCAACGAGTCCTTCCTATAGA-3′ |
| PROM1 reverse primer | 5′-CCATTCCCTGTGCGTTGAA-3′ |
| PROM1 probe | 5′-CAATCACTGAGCACTCTATACCAAAGCGTCAA-3′ |
| KIT forward primer | 5′-TCTTTTCTTTCAACTTGCATCCAA-3′ |
| KIT reverse primer | 5′-AAAGTGTGCTCAGAAAGACAGGATT-3′ |
| KIT probe | 5′-TCCAGGATAGTGGGCACCCCACTG-3′ |
| B2M forward primer | 5′-ATAATTCTACTTTGAGTGCTGTCTCCAT-3′ |
| B2M reverse primer | 5′-TGCCAGCCCTCCTAGAGCTA-3′ |
| B2M probe | 5′-CTGTGGAGCAACCTGCTCAGATACATCAAA-3′ |
Data Analysis
dChip software (http://www.dchip.org) was used for estimating expression and class comparisons. Normalization of raw data were by the invariant set normalization method, and expression was estimated by the perfect match only model. Differentially expressed genes were identified using the t-test, and the empirical false discovery rate was estimated by permutation as previously described.15 More than 70% of the genes measured in this study showed SDs (log2 ratio) <1. With this degree of variability, power = 0.7 to detect significant differences in expression at more than or equal to twofold; P < 0.05 = 0.7; power = 0.7 to detect differences at more than or equal to fourfold; P < 0.001 = 0.7; and power = 0.9 at more than or equal to fourfold; P < 0.05 = 0.9.16
BRB Array Tools software (http://linus.nci.nih.gov/BRB-ArrayTools.html) was used for pathway comparisons using the functional class scoring analysis algorithm, which provides a list of potentially differentially activated pathways (as defined by KEGG and BioCarta) based on having a higher number of differentially expressed genes in the pathways than expected by chance, using LS (mean negative natural logarithm of the P values of the appropriate single gene univariate test) and KS (maximum difference between i/N and pi, where pi is the ith smallest P value of the univariate test) statistics as previously described.17 Probesets were filtered before class comparisons, eliminating genes expressed at or below background, to mitigate multiple comparison errors.
Gene Batch Finder and Gene Ontology (GO) Summary software (http://cgap.nci.nih.gov) were used to identify GO Terms that were significantly different between TDLUs and HELUs, inferred from a list of differentially expressed genes from a supervised comparison using dChip (see Results). Go Terms correspond to biological processes, cellular components, and molecular functions and pathways as defined by the Gene Ontology Consortium (http://www.geneontology.org).
Ingenuity Pathway Analysis software (Ingenuity System, Redwood City, CA) was also used to identify potentially important differences in biological mechanisms and pathways between TDLUs and HELUs, inferred from a list of differentially expressed genes from a supervised comparison using dChip (see Results). This program generates a list of theoretical networks based on algorithms assigning a score based on the number of differentially expressed genes in a dataset that are within annotated pathways and interact in various ways (eg, direct protein binding, activation, and so forth). The higher the score, the more genes and potential interactions there are in the pathways, inferring that they may be important. The networks are unique but not mutually exclusive. Graphical tools allow visualization of potential gene interactions within networks. Typically, the highest scoring networks are merged and visualized together.
Results
Unsupervised and Supervised Analyses of Gene Expression
Eight paired samples of RNA isolated from microdissected pure epithelial cells of TDLUs and HELUs were evaluated for global gene expression using Affymetrix U133-X3P Human GeneChip microarrays. An unsupervised hierarchical clustering of normalized results using dChip software and 6023 genes filtered from 61,359 total probe sets (filtering criteria: ≥50% present calls; 0.5 < SD/mean <1000 variation across samples; absolute expression >50 U) resulted in near-perfect separation of case-matched TDLUs and HELUs (Figure 2), indicating that there are prominent underlying differences in gene expression associated with, and probably involved in, the development of HELUs from TDLUs.
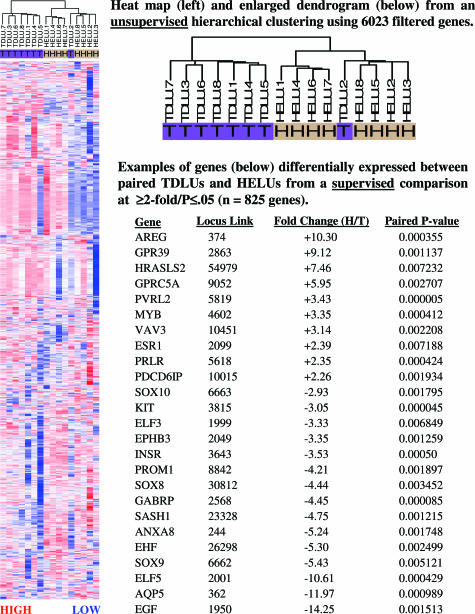
Figure 2.
An unsupervised hierarchical clustering using 6023 filtered genes resulted in near-perfect separation of paired normal TDLUs and HELUs. In a supervised comparison (more than or equal to twofold; P < 0.05), 825 genes showed significantly different levels of expression.
A supervised comparison using dChip software identified 825 genes expressed at more than or equal to twofold different levels (P ≤ 0.05) in HELUs compared with TDLUs (Figure 2 and Supplemental Table S1, which can be viewed at http://ajp.amjpathol.org), ranging from a 17.5-fold increase in epiregulin to a 14.8-fold decrease in odd Oz/ten-m homolog 2. The median false discovery rate for this comparison was 7%, and thus, the 825 genes identified far exceeded the 58 genes (0.07 × 825) expected by chance alone. ER-α expression at the RNA level was up-regulated 2.4-fold (P = 0.007) in HELUs, consistent with our previous study showing highly elevated expression at the protein level.2 ER-β expression was similar in HELUs and TDLUs. Overall, more genes were relatively up-regulated (514 of 825 = 62%) than down-regulated (311 of 825 = 38%).
Pathway Analyses
Three independent analytical strategies were used to identify biological pathways that might be operating differently in TDLUs and HELUs, inferred from differences in expression of individual genes in the pathways (Figure 3).
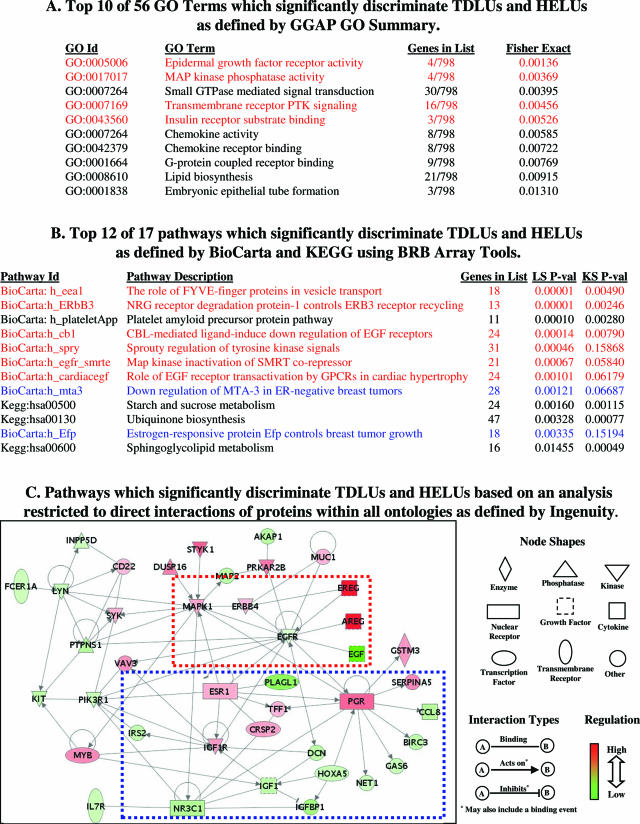
Figure 3.
Selected results from pathway analyses using CGAP GO Summary (A), BRB Array Tools (B), and Ingenuity software (C), highlighting the potential importance of pathways involving ERBB genes (red type or box) and estrogen/ER-regulated genes (blue type or box) in the development of HELUs from TDLUs.
The first strategy used the 825 gene list from the supervised comparison (more than or equal to twofold, P ≤ 0.05) described above and the CGAP GO Summary function to identify 56 GO Terms that were significantly different between TDLUs and HELUs. Four of the top 10 most significant GO Terms involved overlapping pathways in which ERBB genes played a central role. Pathways regulating inflammation/immunity (chemokines, chemokine activity, G-protein chemokine receptors), lipid metabolism, and embryonic development were also prominent.
The second approach used BRB Array Tools pathway analysis software and a list of 26,709 genes filtered from 61,359 total probe sets (filtering criteria: ≥30% present calls; 0.5 < SD/mean <1000 variation across samples; absolute expression >100 U) to identify 17 BioCarta/KEGG-defined pathways that were significantly different between TDLUs and HELUs. Six of the top 12 involved overlapping pathways in which ERBB genes were central. Two of the top 12 involved estrogen/ER-α-regulated pathways. Lipid/carbohydrate metabolism was also prominent.
The third strategy used Ingenuity Pathways Analysis software and the 825 gene list from the supervised comparison (more than or equal to twofold, P ≤ 0.05) described above to identify 43 hypothetical networks of genes that were significantly different between TDLUs and HELUs. The top two (highest scoring) networks were merged and used to create graphic visualizations of potential connections restricted to direct interactions of genes in the networks within all functional ontologies as defined by Ingenuity. This model also emphasized potentially critical roles for ERBB and estrogen/ER-α-regulated genes in the development of HELUs from TDLUs.
Analyses of ERBB Pathway and Estrogen/ER-Regulated Genes
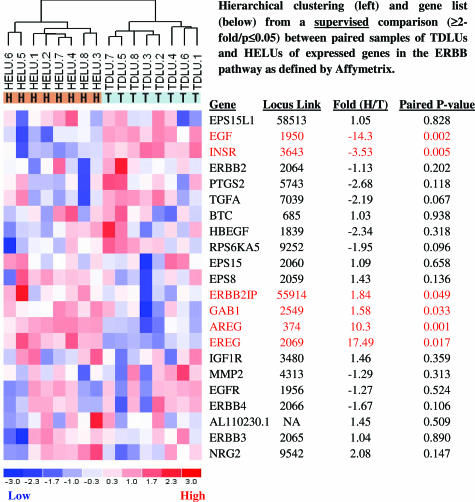
Because previous analyses suggested that changes in the expression of ERBB pathway genes may be especially important in the development of HELUs, dChip software was used to conduct a supervised comparison (more than or equal to twofold, P ≤ 0.05) restricted to the expression of genes (n = 22) on the U133-X3P microarray in the ERBB pathway as defined by Affymetrix (Figure 4). Surprising and interesting differences were observed in the expression of ligands for epidermal growth factor receptor (EGFR)/ERBB1 in HELUs relative to TDLUs, including prominent increases in epiregulin (17.5-fold) and amphiregulin (AREG) (10.3-fold) and a prominent decrease in epidermal growth factor (EGF) (14.3-fold). There was also a substantial decrease (3.5-fold) in insulin receptor (INSR). There were no significant changes in expression of the ERBB receptors, including EGFR/ERBB1.
Figure 4.
Hierarchical clustering and gene list from a supervised comparison (more than or equal to twofold; P ≤ 0.05) between paired samples of normal TDLUs and HELUs of expressed genes in the ERBB pathway as defined by Affymetrix.
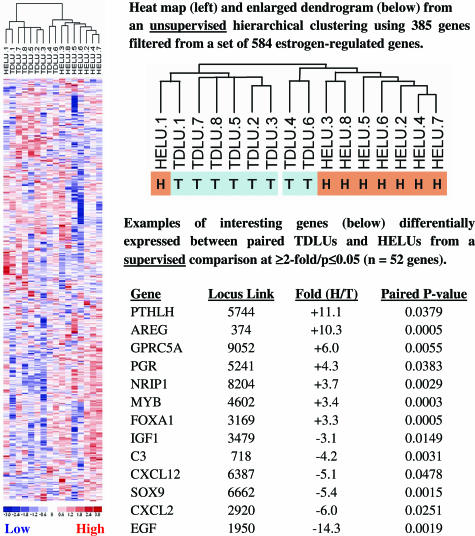
Previous analyses also suggested that estrogen/ER-α-regulated genes may be important in the development of HELUs. An unsupervised hierarchical clustering was conducted using 385 genes filtered (filtering criteria: ≥20% present calls; 0.5 < SD/mean <1000 variation across samples) from a total list of 584 estrogen/ER-α-regulated genes (Figure 5). The gene list (see Supplemental Table S2 at http://ajp.amjpathol.org) was derived from published microarray studies of ER-α-positive breast cancer cell lines stimulated by estrogen.18,19,20,21,22,23,24,25,26,27 There was near-perfect separation of TDLUs and HELUs, indicating that there are substantial underlying differences in estrogen-regulated pathways mediated by ER-α. A supervised comparison (more than or equal to twofold, P ≤ 0.05) identified 52 differentially expressed genes (Figure 5 and Supplemental Table S3, which can be seen at http://ajp.amjpathol.org), including increased AREG (10.3-fold) and decreased EGF (14.3-fold) in HELUs.
Figure 5.
An unsupervised hierarchical clustering using 385 genes filtered from a total list of 584 estrogen-regulated genes resulted in near-perfect separation of paired normal TDLUs and HELUs. In a supervised comparison (more than or equal to twofold; P ≤ 0.05), 52 genes were expressed at significantly different levels in TDLUs and HELUs, including AREG and EGF in a manner consistent with their known regulation by estrogen.
Confirmation and/or Validation by qRT-PCR
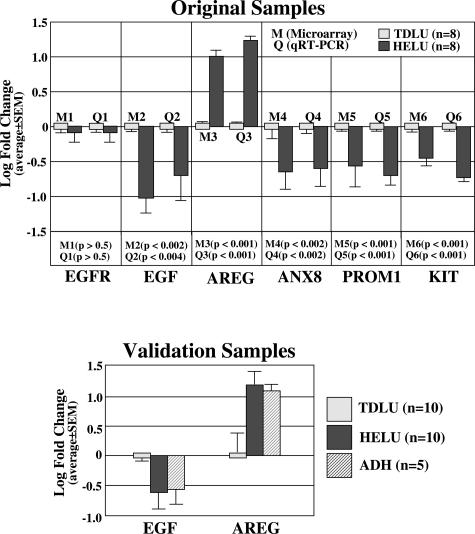
Selected interesting genes (EGFR, EGF, AREG, ANXA8, PROM1, and KIT) were evaluated by qRT-PCR using total RNA from the original eight paired samples of TDLUs and HELUs (Figure 6). The findings were very similar (direction and magnitude) to the original microarray results for each of these six genes, confirming their validity. Additional analyses were performed for EGF and AREG using total RNA isolated from 10 independent paired samples of TDLUs and HELUs. Again, the results were very similar, providing further validation that the switch in these two ligands for EGFR (ie, prominent decreased expression of EGF and increased expression of AREG) is an important biological characteristic of HELUs. Five of the samples also contained areas of ADH arising in the HELUs, and the results were nearly identical in these two compartments.
Figure 6.
qRT-PCR was used to confirm the differential expression of selected interesting genes, including epidermal growth factor receptor (EGFR), EGF, AREG, annexin 8 (ANXA8), prominin 1 (PROM1), and v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (KIT). Changes (direction and magnitude) in all six genes were confirmed using total RNA from the original eight paired samples. AREG and EGF were further evaluated by qRT-PCR using total RNA from 10 additional paired samples of TDLUs and HELUs, with the same results as the original eight samples. Five of these 10 breasts also contained foci of ADH arising within HELUs, and the same patterns of expression in HELUs were observed in ADH.
Discussion
HELUs are a very common and prominent alteration of growth in the adult female human breast. The idea that HELUs might be precursors of breast cancer was proposed as far back as the early 1900s, if not earlier,1,9,10 and it is consistent with more recent studies showing that they are on a histological continuum with breast cancer,2,4,5 that they are risk factors for developing breast cancer,28,29,30 and that they may share genetic alterations (allelic imbalances) with breast cancer, especially when they occur in the same breasts.31,32
The underlying causes of the hyperplasia leading to HELUs are unknown. Some evidence suggests that estrogen may be involved, including the observations that HELUs are more common in premenopausal compared with postmenopausal breasts5 and in cancerous compared with noncancerous breasts5,30 where increased estrogen exposure is such a strong risk factor for developing cancer.33 Recent studies from our laboratory demonstrated that the epithelial cells lining HELUs show highly elevated expression of ER-α, significantly increased proliferation, and significantly decreased apoptosis compared with TDLUs.2 Because estrogen, mediated by ER-α, stimulates proliferation12 and suppresses apoptosis13 in normal cells, elevated ER-α in HELUs may be a fundamental alteration leading to increased growth. A recent study in mice overexpressing ER-α in normal breast epithelium noted the rapid development of hyperplasia that occasionally progressed to cancer,34 supporting the idea that elevated ER-α may be partially responsible for the development of HELUs and their progression to more advanced lesions in humans.
Because HELUs are so common in the population, relatively similar to each other in histological appearance (especially the frequent single-layer type evaluated in this study), and share important biological characteristics (eg, elevated ER-α), their beginnings seem more likely to reflect alterations in development or differentiation than genetic defects per se. Regardless, the end result is increased growth, creating fertile soil for accumulating random genetic defects leading to diversity and progression to more committed breast cancer precursors such as ADH.
This study used DNA microarray technology to learn more about changes in gene expression and molecular pathways associated with the development of HELUs from TDLUs. Several analyses identified pathways involving ERBB genes as important. A particularly interesting finding involved a switch in ligands for ERBB1/EGFR, including a uniform down-regulation of EGF (14-fold) and transforming growth factor-α (twofold) and up-regulation of AREG (10-fold) and epiregulin (17-fold) in HELUs compared with TDLUs. The changes in EGF and AREG were especially large in both absolute and relative terms. In contrast, expression of all ERBB receptors (including ERBB1/EGFR) did not change significantly. Recent studies have shown that estrogen, mediated by ER-α, can induce AREG35,36 and suppress EGF21 in normal and malignant breast epithelial cells. Thus, estrogen may be partially responsible for the switch in these ligands, particularly in the setting of the highly elevated ER-α observed in HELUs.2
There is ample evidence in breast and other types of cells that activation of ERBB1/EGFR by different ligands may have different functional consequences. For example, AREG is essential for ductal morphogenesis in the embryonic mouse mammary gland, whereas EGF is important for alveolar development in the adult.37,38 In monolayer cultures of human kidney Madin-Darby canine kidney cells expressing EGFR, AREG induces an epithelial-to-mesenchymal transformation, whereas EGF has no apparent effect.39 In three-dimensional collagen gels, EGF also induces Madin-Darby canine kidney cells to form well-organized gland-like spheres and ducts, whereas AREG results in the formation of disorganized rudimentary structures,39 suggesting that AREG disrupts differentiation in this setting. Collectively, this evidence suggests that the development of HELUs from TDLUs may involve reactivation of embryonic developmental pathways and/or inhibition of terminal differentiation partially mediated by up-regulation of AREG and down-regulation of EGF.
AREG has also been shown to enhance the tumorigenicity of certain human breast cancer cell lines40 and to stimulate proliferation in the human premalignant cell line MCF10A.41 Baseline expression of AREG is very low in MCF10A but increases dramatically when the cells are transfected with ras and c-neu oncogenes, which also enhances their ability to progress to cancer in xenografts.41 Thus, AREG may help promote tumor progression, consistent with our observation that the up-regulation of AREG observed in HELUs was also present in ADH that evolved from the HELUs. These observations suggest that pharmacological targeting of AREG may be an effective strategy for breast cancer prevention in high-risk patients with ADH.
Alterations of genes typically associated with inflammation and immunity may also be important in the development of HELUs. Several chemokines were uniformly down-regulated, including chemokine (C-X-C motif) ligand 5 (CXCL5, 7-fold), CXCL2 (6.0-fold), CXCL3 (5.8-fold), CXCL12 (5.1-fold), chemokine (C-C motif) ligand 28 (CCL28, 4.5-fold), CCL8 (3.4-fold), chemokine (C-X3-C motif) ligand 1 (CX3CL1, 3.0-fold), and CXCL14 (2.1-fold). Most are potent attractants and activators of inflammatory cells, including neutrophils (CXCL5, CXCL12, and CXCL14), monocytes (CCL8 and CXCL14), and lymphocytes (CCL28 and CCL8).42 CXCL14 has also been implicated in the differentiation of monocytes to macrophages.43 The change in CXCL12 was particularly large in both absolute and relative levels of expression. ER-α was recently shown to activate the transcription of CXCL12 in breast cancer cell lines, resulting in increased proliferation,44 but it seems unlikely to be stimulating growth in HELUs because it was down-regulated in this setting. CXCL12 was also recently shown to be highly expressed in the myoepithelial cells of noninvasive breast cancer compared with normal TDLUs,45 but our assessment of HELUs was restricted to epithelial cells, preventing comparison.
Several other genes usually associated with inflammation and immunity were also down-regulated in HELUs, including monocyte to macrophage differentiation-associated gene (MMD, 2.2-fold) and the transcription factors nuclear factor of κ light polypeptide gene enhancer in B-cells 1 (NFKB1, 2.1-fold) and nuclear factor κB inhibitor ζ (NFKBIZ, 3.7-fold). The latter two also suppress apoptosis in certain cells,46,47 although they are unlikely to be involved in the suppression of apoptosis observed in HELUs2 because both were down-regulated. Killer cell lectin-like receptor subfamily K member 1 (KLRK1) and T-cell receptor α constant (TRAC) were also decreased in HELUs (3.2- and 3.8-fold, respectively), which is surprising because the former is a receptor on natural killer cells mediating cytotoxicity,48 and the latter is a component of the T-cell receptor.49
It is possible that these and other genes mentioned above were measured in inflammatory cells contaminating the samples in our study. However, we think this is unlikely to be a major factor because the samples showed no obvious inflammation histologically, they were carefully microdissected to be composed of nearly pure populations of epithelial cells, and the expression of many of the genes in question was too high to be easily explained by contamination (eg, NFKBIZ and TRAC). Perhaps some of these genes are expressed in breast epithelium as well, which would be interesting. The cell source, however, is not important in the sense that they were still expressed at significantly reduced levels in HELUs compared with normal TDLUs.
Inflammatory cells play critical roles in the normal breast. For example, macrophages are essential during the involution of lactating breast tissue by inducing apoptosis of epithelial cells and participating in their phagocytic clearance.50,51 Normal luminal epithelial cells also participate in this process as cannibalistic phagocytes,52 and perhaps they express inflammation-associated genes to accomplish this. Similar processes may be involved in the remodeling of TDLUs during the menstrual cycle, although this has not been formally studied to our knowledge. Macrophages are also critical for development and differentiation in the embryonic mammary gland.53 Our results suggest that suppression of inflammation and immunity may be important in the development of HELUs, perhaps by reducing cell turnover and/or inhibiting terminal differentiation during normal tissue remodeling.
Many other genes involved in development and differentiation also showed altered expression in HELUs. For example, several epithelial-specific Ets transcription factors54 were uniformly down-regulated, including E74-like factor 5 (ELF5, 10.6-fold), Ets homologous factor (EHF, 5.3-fold), E74-like factor 3 (ELF3, 3.3-fold), and Ets variant gene 6 (ETV6, also known as the TEL oncogene; 3.1-fold). Other developmentally important transcription factors showing decreased expression included sex determining region Y box 9 (SOX9, 5.4-fold), SOX8 (4.4-fold), SOX10 (2.9-fold), mastermind-like 2 (MAML2, 3.6-fold), and forkhead box C1 (FOXC1, 10.1-fold).55,56,57,58,59 PROM1 (CD133), KIT, and ANXA8 were also significantly down-regulated in HELUs (4.2-, 3.1-, and 5.2-fold, respectively). PROM1 and KIT are expressed in several types of stem cells,60,61 KIT may play a role in maintaining normal differentiation in the adult breast,62 and ANXA8 is important during ductal morphogenesis in the embryonic breast.63 ANXA8 was also recently reported as being elevated in the myoepithelial cells of ADH (which evolve from HELUs),64 but our study was restricted to epithelial cells, barring comparisons. Transforming growth factor-β1 suppresses proliferation in ER-α-positive/proliferating breast epithelium, which may represent an important progenitor cell population.65 We recently reported a sixfold increase in ER-α-positive/proliferating cells in HELUs compared with TDLUs.2 However, there were no changes in the expression of relevant transforming growth factor-β pathway genes at the RNA level in this study (data not shown), but it would be necessary to assess protein at the individual cellular level to exclude their involvement completely. Collectively, these data also suggest that delays in development and suppression of terminal differentiation are important in the evolution of HELUs from TDLUs.
In summary, HELUs are a common abnormality of growth in the breast and the first histologically identifiable potential precursor of breast cancer. To the extent that function can be inferred from static measurements of gene expression, our data suggest that the evolution of HELUs from TDLUs involves alterations leading to reactivation of embryonic development and/or suppression of terminal differentiation. Alterations of pathways involving ERBB genes were especially prominent, including a switch from adult to embryonic ligands for EGFR/ERBB1 (EGF and AREG, respectively). Suppression of many genes usually associated with inflammation and immunity also seem to be important, perhaps by delaying cell turnover during normal tissue remodeling. Finally, several of the most interesting changes involve estrogen-regulated genes, and the effects of these changes may be greatly augmented by the highly elevated expression of ER-α observed in HELUs. Elevated expression of ER-α may be a fundamental defect responsible for widespread hyperplasia leading to HELUs, setting the stage for further progression to more committed precursors of breast cancer.
Supplementary Material
Footnotes
Address reprint requests to D. Craig Allred, M.D., Department of Pathology and Immunology, Washington University School of Medicine, 660 S. Euclid Ave., Box 8118, St. Louis, MO 63110. E-mail: dcallred@path.wustl.edu.
Supported by the National Institutes of Health (National Cancer Institute grant U01-CA84243) and AstraZeneca Pharmaceuticals.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Nasser SM. Columnar cell lesions: current classification and controversies. Semin Diagn Pathol. 2004;21:18–24. doi: 10.1053/j.semdp.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Lee S, Mohsin SK, Mao S, Hilsenbeck SG, Medina D, Allred DC. Hormones, receptors, and growth in hyperplastic enlarged lobular units: early potential precursors of breast cancer. Breast Cancer Res. 2005;8:R6. doi: 10.1186/bcr1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitt SJ, Vincent-Salomon A. Columnar cell lesions of the breast. Adv Anatomic Pathol. 2003;10:113–124. doi: 10.1097/00125480-200305000-00001. [DOI] [PubMed] [Google Scholar]
- Wellings RR, Jensen HM. On the origin and progression of ductal carcinoma in the human breast. J Natl Cancer Inst. 1973;50:1111–1118. doi: 10.1093/jnci/50.5.1111. [DOI] [PubMed] [Google Scholar]
- Wellings SR, Jensen HM, Marcum RG. An atlas of subgross pathology of the human breast with special reference to possible precancerous lesions. J Natl Cancer Inst. 1975;55:231–273. [PubMed] [Google Scholar]
- Allred DC, Hilsenbeck SG, Mohsin SK. Biologic features of human premalignant breast disease. Harris JR, Lippman ME, Morrow M, Osborne CK, editors. Philadelphia: Lippincott Williams & Wilkins,; Diseases of the Breast. 2004:pp 512–513. [Google Scholar]
- Page DL, Dupont WD. Anatomic indicators (histologic and cytologic) of increased breast cancer risk. Breast Cancer Res Treat. 1993;28:157–166. doi: 10.1007/BF00666428. [DOI] [PubMed] [Google Scholar]
- Page DL, Dupont WD, Rogers LW, Rados MS. Atypical hyperplastic lesions of the female breast. A long-term follow-up study. Cancer. 1985;55:2698–2708. doi: 10.1002/1097-0142(19850601)55:11<2698::aid-cncr2820551127>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Bloodgood JC. Senile parenchymatous hypertrophy of female breast. Its relation to cyst formation and carcinoma. Surg Gynecol Obstet. 1906;3:721–730. [Google Scholar]
- Cheatle GL. Cysts, and primary cancer in cysts, of the breast. Br J Surg. 1920;8:149–166. [Google Scholar]
- Fuqua SAW, Schiff R. The biology of estrogen receptors. Harris JR, Lippman ME, Morrow M, Osborne CK, editors. Philadelphia: Lippincott Williams & Wilkins,; Diseases of the Breast. 2004:pp 585–602. [Google Scholar]
- Anderson E, Clarke RB. Steroid receptors and cell cycle in normal mammary epithelium. J Mammary Gland Biol Neoplasia. 2004;9:3–13. doi: 10.1023/B:JOMG.0000023584.01750.16. [DOI] [PubMed] [Google Scholar]
- Gompel A, Somai S, Chaouat M, Kazem A, Kloosterboer HJ, Beusman I, Forgez P, Mimoun M, Rostene W. Hormonal regulation of apoptosis in breast cells and tissues. Steroids. 2000;65:593–598. doi: 10.1016/s0039-128x(00)00172-0. [DOI] [PubMed] [Google Scholar]
- Scicchitano MS, Dalmas DA, Bertiaux MA, Anderson SM, Turner LR, Thomas RA, Mirable R, Boyce RW. Preliminary comparison of quantity, quality, and microarray performance of RNA extracted from formalin-fixed, paraffin-embedded, and unfixed frozen tissue samples. J Histochem Cytochem. 2006;54:1229–1237. doi: 10.1369/jhc.6A6999.2006. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C, Li J, Bumgarner RE. Sample size for detecting differentially expressed genes in microarray experiments. BMC Genomics. 2004;5:87–93. doi: 10.1186/1471-2164-5-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlidis P, Qin J, Arango V, Mann JJ, Sibille E. Using the gene ontology for microarray data mining: a comparison of methods and application to age effects in human prefrontal cortex. Neurochem Res. 2004;29:1213–1222. doi: 10.1023/b:nere.0000023608.29741.45. [DOI] [PubMed] [Google Scholar]
- Bouras T, Southey MC, Chang AC, Reddel RR, Willhite D, Glynne R, Henderson MA, Armes JE, Venter DJ. Stanniocalcin 2 is an estrogen-responsive gene coexpressed with the estrogen receptor in human breast cancer. Cancer Res. 2002;62:1289–1295. [PubMed] [Google Scholar]
- Bourdeau V, Deschenes J, Metivier R, Nagai Y, Nguyen D, Bretschneider N, Gannon F, White JH, Mader S. Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol Endocrinol. 2004;18:1411–1427. doi: 10.1210/me.2003-0441. [DOI] [PubMed] [Google Scholar]
- Cicatiello L, Scafoglio C, Altucci L, Cancemi M, Natoli G, Facchiano A, Iazzetti G, Calogero R, Biglia N, De Bortoli M, Sfiligoi C, Sismondi P, Bresciani F, Weisz A. A genomic view of estrogen actions in human breast cancer cells by expression profiling of the hormone-responsive transcriptome. J Mol Endocrinol. 2004;32:719–775. doi: 10.1677/jme.0.0320719. [DOI] [PubMed] [Google Scholar]
- DeNardo DG, Kim HT, Hilsenbeck S, Cuba V, Tsimelzon A, Brown PH. Global gene expression analysis of estrogen receptor transcription factor cross talk in breast cancer: identification of estrogen-induced/activator protein-1-dependent genes. Mol Endocrinol. 2005;19:362–378. doi: 10.1210/me.2004-0267. [DOI] [PubMed] [Google Scholar]
- Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144:4562–4574. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- Inoue A, Yoshida N, Omoto Y, Oguchi S, Yamori T, Kiyama R, Hayashi S. Development of cDNA microarray for expression profiling of estrogen-responsive genes. J Mol Endocrinol. 2002;29:175–192. doi: 10.1677/jme.0.0290175. [DOI] [PubMed] [Google Scholar]
- Lin CY, Strom A, Vega VB, Kong SL, Yeo AL, Thomsen JS, Chan WC, Doray B, Bangarusamy DK, Ramasamy A, Vergara LA, Tang S, Chong A, Bajic VB, Miller LD, Gustafsson JA, Liu ET. Discovery of estrogen receptor alpha target genes and response elements in breast tumor cells. Genome Biol. 2004;5:R66. doi: 10.1186/gb-2004-5-9-r66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tynan SH, Lundeen SG, Allan GF. Cell type-specific bidirectional regulation of the glucocorticoid-induced leucine zipper (GILZ) gene by estrogen. J Steroid Biochem Mol Biol. 2004;91:225–239. doi: 10.1016/j.jsbmb.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Wang DY, Fulthorpe R, Liss SN, Edwards EA. Identification of estrogen-responsive genes by complementary deoxyribonucleic acid microarray and characterization of a novel early estrogen-induced gene: EEIG1. Mol Endocrinol. 2004;18:402–411. doi: 10.1210/me.2003-0202. [DOI] [PubMed] [Google Scholar]
- Weisz A, Basile W, Scafoglio C, Altucci L, Bresciani F, Facchiano A, Sismondi P, Cicatiello L, De Bortoli M. Molecular identification of ERalpha-positive breast cancer cells by the expression profile of an intrinsic set of estrogen regulated genes. J Cell Physiol. 2004;200:440–450. doi: 10.1002/jcp.20039. [DOI] [PubMed] [Google Scholar]
- McLaren BK, Gobbi H, Schuyler PA, Olson SJ, Parl FF, Dupont WD, Page DL. Immunohistochemical expression of estrogen receptor in enlarged lobular units with columnar alteration in benign breast biopsies: a nested case-control study. Am J Surg Pathol. 2005;29:105–108. doi: 10.1097/01.pas.0000146013.76881.d9. [DOI] [PubMed] [Google Scholar]
- Page DL, Dupont WD, Rogers LW. Breast cancer risk of lobular-based hyperplasia after biopsy: “ductal” pattern lesions. Cancer Detect Prev. 1986;9:441–448. [PubMed] [Google Scholar]
- Shaaban AM, Sloane JP, West CR, Moore FR, Jarvis C, Williams EM, Foster CS. Histopathologic types of benign breast lesions and the risk of breast cancer: case-control study. Am J Surg Pathol. 2002;26:421–430. doi: 10.1097/00000478-200204000-00003. [DOI] [PubMed] [Google Scholar]
- Moinfar F, Man Y-G, Bratthauer GL, Ratschek M, Tavassoli FA. Genetic abnormalities in mammary intraepithelial neoplasia-flat type (“clinging ductal carcinoma in situ”): a simulator of normal mammary epithelium. Cancer. 2000;88:2072–2081. [PubMed] [Google Scholar]
- Simpson PT, Gale T, Reis-Filho JS, Jones C, Parry S, Sloane JP, Hanby A, Pinder SE, Lee AH, Humphreys S, Ellis IO, Lakhani SR. Columnar cell lesions of the breast: the missing link in breast cancer progression? A morphological and molecular analysis. Am J Surg Pathol. 2005;29:734–746. doi: 10.1097/01.pas.0000157295.93914.3b. [DOI] [PubMed] [Google Scholar]
- Martin AM, Weber BL. Genetic and hormonal risk factors in breast cancer. J Natl Cancer Inst. 2000;92:1126–1135. doi: 10.1093/jnci/92.14.1126. [DOI] [PubMed] [Google Scholar]
- Frech MS, Halama ED, Tilli MT, Singh B, Gunther EJ, Chodosh LA, Flaws JA, Furth PA. Deregulated estrogen receptor alpha expression in mammary epithelial cells of transgeneic mice results in the development of ductal carcinoma in situ. Cancer Res. 2005;65:681–685. [PMC free article] [PubMed] [Google Scholar]
- Martínez-Lacaci I, Saceda M, Plowman GD, Johnson GR, Normanno N, Salomon DS, Dickson RB. Estrogen and phorbol esters regulate amphiregulin expression by two separate mechanisms in human breast cancer cell lines. Endocrinology. 1995;136:3983–3992. doi: 10.1210/endo.136.9.7649107. [DOI] [PubMed] [Google Scholar]
- Wilson CL, Sims AH, Howell A, Miller CJ, Clarke RB. Effects of oestrogen on gene expression in epithelium and stroma of normal human breast tissue. Endocr Relat Cancer. 2006;13:617–628. doi: 10.1677/erc.1.01165. [DOI] [PubMed] [Google Scholar]
- Luetteke NC, Qiu TH, Fenton SE, Troyer KL, Riedel RF, Chang A, Lee DC. Targeted inactivation of the EGF and amphiregulin genes reveals distinct roles for EGF receptor ligands in mouse mammary gland development. Development. 1999;126:2739–2750. doi: 10.1242/dev.126.12.2739. [DOI] [PubMed] [Google Scholar]
- Troyer KL, Lee DC. Regulation of mouse mammary gland development and tumorigenesis by the ERBB signaling network. J Mammary Gland Biol Neoplasia. 2001;6:7–21. doi: 10.1023/a:1009560330359. [DOI] [PubMed] [Google Scholar]
- Chung E, Graves-Deal R, Franklin JL, Coffey RJ. Differential effects of amphiregulin and TGF-alpha on the morphology of MDCK cells. Exp Cell Res. 2005;309:149–160. doi: 10.1016/j.yexcr.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Ma L, Gauville C, Berthois Y, Millot G, Johnson GR, Calvo F. Antisense expression for amphiregulin suppresses tumorigenicity of a transformed human breast epithelial cell line. Oncogene. 1999;18:6513–6520. doi: 10.1038/sj.onc.1203042. [DOI] [PubMed] [Google Scholar]
- Normanno N, Selvam MP, Qi CF, Saeki T, Johnson G, Kim N, Ciardiello F, Shoyab M, Plowman G, Brandt R, Todaro G, Salomon DS. Amphiregulin as an autocrine growth factor for c-Ha-ras- and c-erbB-2-transformed human mammary epithelial cells. Proc Natl Acad Sci USA. 1994;91:2790–2794. doi: 10.1073/pnas.91.7.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann MF, Kopf M, Marsland BJ. Chemokines: more than just road signs. Nat Rev Immunol. 2006;6:159–164. doi: 10.1038/nri1776. [DOI] [PubMed] [Google Scholar]
- Shurin GV, Ferris RL, Tourkova IL, Perez L, Lokshin A, Balkir L, Collins B, Chatta GS, Shurin MR. Loss of new chemokine CXCL14 in tumor tissue is associated with low infiltration by dendritic cells (DC), while restoration of human CXCL14 expression in tumor cells causes attraction of DC both in vitro and in vivo. J Immunol. 2005;174:5490–5498. doi: 10.4049/jimmunol.174.9.5490. [DOI] [PubMed] [Google Scholar]
- Hall JM, Korach KS. Stromal cell-derived factor 1, a novel target of estrogen receptor action, mediates the mitogenic effects of estradiol in ovarian and breast cancer cells. Mol Endocrinol. 2003;17:792–803. doi: 10.1210/me.2002-0438. [DOI] [PubMed] [Google Scholar]
- Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, Porter D, Hu M, Chin L, Richardson A, Schnitt S, Sellers WR, Polyak K. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- Karin M, Cao Y, Greten FR, Li ZW. NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2:301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- Rayet B, Gelinas C. Aberrant rel/nfkb genes and activity in human cancer. Oncogene. 1999;18:6938–6947. doi: 10.1038/sj.onc.1203221. [DOI] [PubMed] [Google Scholar]
- Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol. 2003;3:781–790. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- Koop BF, Rowen L, Wang K, Kuo CL, Seto D, Lenstra JA, Howard S, Shan W, Deshpande P, Hood L. The human T-cell receptor TCRAC/TCRDC (C alpha/C delta) region: organization, sequence, and evolution of 97.6 kb of DNA. Genomics. 1994;19:478–493. doi: 10.1006/geno.1994.1097. [DOI] [PubMed] [Google Scholar]
- Clarkson RW, Wayland MT, Lee J, Freeman T, Watson CJ. Gene expression profiling of mammary gland development reveals putative roles for death receptors and immune mediators in post-lactational regression. Breast Cancer Res. 2004;6:R92–R109. doi: 10.1186/bcr754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein T, Morris JS, Davies CR, Weber-Hall SJ, Duffy MA, Heath VJ, Bell AK, Ferrier RK, Sandilands GP, Gusterson BA. Involution of the mouse mammary gland is associated with an immune cascade and an acute-phase response, involving LBP, CD14 and STAT3. Breast Cancer Res. 2004;6:R75–R91. doi: 10.1186/bcr753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks J, Geske FJ, Lehman L, Fadok VA. Do inflammatory cells participate in mammary gland involution? J Mammary Gland Biol Neoplasia. 2002;7:163–176. doi: 10.1023/a:1020351919634. [DOI] [PubMed] [Google Scholar]
- Sternlicht MD. Key stages in mammary gland development: the cues that regulate ductal branching morphogenesis. Breast Cancer Res. 2006;8:201. doi: 10.1186/bcr1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharrocks AD. The ET:S-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- Bannykh SI, Stolt CC, Kim J, Perry A, Wegner M. Oligodendroglial-specific transcriptional factor SOX10 is ubiquitously expressed in human gliomas. J Neurooncol. 2006;76:115–127. doi: 10.1007/s11060-005-5533-x. [DOI] [PubMed] [Google Scholar]
- Coffer PJ, Burgering BM. Forkhead-box transcription factors and their role in the immune system. Nat Rev Immunol. 2004;4:889–899. doi: 10.1038/nri1488. [DOI] [PubMed] [Google Scholar]
- Drivdahl R, Haugk KH, Sprenger CC, Nelson PS, Tennant MK, Plymate SR. Suppression of growth and tumorigenicity in the prostate tumor cell line M12 by overexpression of the transcription factor SOX9. Oncogene. 2004;23:4584–4593. doi: 10.1038/sj.onc.1207603. [DOI] [PubMed] [Google Scholar]
- Enlund F, Behboudi A, Andren Y, Oberg C, Lendahl U, Mark J, Stenman G. Altered Notch signaling resulting from expression of a WAMTP1-MAML2 gene fusion in mucoepidermoid carcinomas and benign Warthin’s tumors. Exp Cell Res. 2004;292:21–28. doi: 10.1016/j.yexcr.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Kim JH, Do HJ, Yang HM, Oh JH, Choi SJ, Kim DK, Cha KY, Chung HM. Overexpression of SOX9 in mouse embryonic stem cells directs the immediate chondrogenic commitment. Exp Mol Med. 2005;37:261–268. doi: 10.1038/emm.2005.35. [DOI] [PubMed] [Google Scholar]
- Kania G, Corbeil D, Fuchs J, Tarasov KV, Blyszczuk P, Huttner WB, Boheler KR, Wobus AM. Somatic stem cell marker prominin-1/CD133 is expressed in embryonic stem cell-derived progenitors. Stem Cells. 2005;23:791–804. doi: 10.1634/stemcells.2004-0232. [DOI] [PubMed] [Google Scholar]
- Miettinen M, Lasota J. KIT (CD117): a review on expression in normal and neoplastic tissues, and mutations and their clinicopathologic correlation. Appl Immunohistochem Mol Morphol. 2005;13:205–220. doi: 10.1097/01.pai.0000173054.83414.22. [DOI] [PubMed] [Google Scholar]
- Ulivi P, Zoli W, Medri L, Amadori D, Saragoni L, Barbanti F, Calistri D, Silvestrini R. c-kit and SCF expression in normal and tumor breast tissue. Breast Cancer Res Treat. 2004;83:33–42. doi: 10.1023/B:BREA.0000010694.35023.9e. [DOI] [PubMed] [Google Scholar]
- Morris JS, Stein T, Pringle MA, Davies CR, Weber-Hall S, Ferrier RK, Bell AK, Heath VJ, Gusterson BA. Involvement of axonal guidance proteins and their signaling partners in the developing mouse mammary gland. J Cell Physiol. 2006;206:16–24. doi: 10.1002/jcp.20427. [DOI] [PubMed] [Google Scholar]
- Stein T, Price KN, Morris JS, Heath VJ, Ferrier RK, Bell AK, Pringle MA, Villadsen R, Petersen OW, Sauter G, Bryson G, Mallon EA, Gusterson BA. Annexin A8 is up-regulated during mouse mammary gland involution and predicts poor survival in breast cancer. Clin Cancer Res. 2005;11:6872–6879. doi: 10.1158/1078-0432.CCR-05-0547. [DOI] [PubMed] [Google Scholar]
- Ewan KBR, Oketch-Rabah HA, Ravani SA, Shyamala G, Moses HL, Barcellos-Hoff MH. TGF-β1 restrains proliferation of estrogen receptor-α-positive mammary epithelial cells. Am J Pathol. 2005;167:409–417. doi: 10.1016/s0002-9440(10)62985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.