Abstract
The chemotaxis system plays an essential role in swarm cell differentiation and motility. We show in this study that two (Tsr and Tar) of the four chemoreceptors in Escherichia coli can support swarming individually, but sensing their most powerful chemoattractants is not necessary. Conditions that abolish chemotaxis toward serine (presence of serine concentrations that saturate Tsr, or mutations in Tsr that destroy serine binding) have no effect on swarming. Similar results were obtained for the aspartate and maltose chemoreceptor Tar. We also show that although a mutation in the signaling domain of Tsr that inhibits CheA kinase abolishes swarming, nonchemotactic flagellar switch mutants can swarm. Our results suggest that during swarming, the chemoreceptors signal through the chemotaxis pathway and induce swarmer cell differentiation in response to signals other than their known chemoeffectors.
A large number of flagellated eubacteria show a behavior known as “swarming” when propagated on the surface of certain solid media (1). Swarming is defined as an organized surface translocation that depends on extensive flagellation and cell–cell contact (2). Swarmer cells are generally longer and more flagellated than swimmer cells (cells of the same species propagated in liquid media) and move within a milieu of extracellular “slime” surrounding the colony. In some organisms, the swarmer cell state may be associated with pathogenesis (3). Swarming therefore offers a unique opportunity for studying signal transduction mechanisms that operate in bacterial populations growing in close proximity.
Studies in a number of different organisms have established that components of the chemotaxis system play a critical role during swarming (4–7). Although the role of these components during swimming in liquid media has been extensively studied in Escherichia coli and in Salmonella typhimurium (8), their role in swarming is not known. In E. coli and S. typhimurium, defects in all known che genes abolish swarming completely; these mutants are primarily defective in induction of the hyperflagellation response (7). Chemotaxis defects in Serratia marcescens also affect swarmer cell differentiation (unpublished data). Thus, components of the chemotaxis system play a key role in these organisms in transducing “swarm” signals to produce specific changes in gene expression, a function not as yet ascribed to them during the swimming response. Besides understanding this new role for the chemotaxis components, a basic unanswered question is whether chemotaxis, the process by which cells move toward higher concentrations of attractant or away from higher concentrations of repellent in liquid media, is essential for outward migration of a swarmer colony. A brief overview of the known aspects of signaling through the chemotaxis pathway during swimming in liquid media is presented below.
In swimming E. coli and S. typhimurium, the flagellar motors spin counterclockwise (CCW) or clockwise (CW) to produce smooth-swimming or tumbling motion, respectively (9). The chemotaxis system (composed of sensory receptors networking with components of a cytoplasmic phosphorylation cascade) can alter the rotational bias of the motors, allowing the bacteria to move toward attractants and away from repellents. External stimuli are detected by a group of sensory transducers or transmembrane receptors (also referred to as methyl-accepting chemotaxis proteins or MCPs; Tsr, Tar, Trg, Tap in E. coli), which recognize (either directly or via binding proteins) attractants (amino acids, sugars, and oligopeptides), as well as repellents (extreme pH, certain metal ions and hydrophobic amino acids). A signal is initiated when conformational changes induced by ligand binding are conveyed to the cytoplasmic face of the receptors, where they are recognized by an associated CheA/CheW complex. CheA has an autokinase activity that is inhibited by attractant-bound and stimulated by repellent-bound or attractant-free receptors. CheA donates its phosphate to the response regulators CheY and CheB. Phospho-CheY (CheY-P) interacts with the switch complex found at the base of the flagellar motor to generate CW motor rotation. CCW rotation is the default state of the motor. Thus, external chemical signals cause changes in swimming patterns by altering the relative levels of CheY-P. CheZ dephosphorylates CheY-P. CheR and CheB-P add and remove methyl groups to the MCPs, resetting them to a prestimulus level of signaling. Null mutations in the Che proteins alter the CCW/CW bias of the motors. CheA, CheW, CheY, and CheR mutants are smooth-swimming, as are those missing all four membrane transducers; CheZ and CheB mutants are tumbly.
CheA and CheY/CheB belong to a family of two-component, stimulus-response regulators (10). Besides motility, processes regulated by these proteins in diverse bacteria include sporulation, virulence, membrane transport, and intermediary metabolism. Biochemical and genetic experiments suggest that cross-talk between these parallel signal transduction pathways may provide a mechanism for higher-order information processing or a global regulatory network.
To begin understanding the role of the chemotaxis system in swarm cell differentiation, we have investigated whether chemotaxis to the major chemoeffectors of Tsr and Tar (the only two chemoreceptors that supported swarming in E. coli) is involved in swarming. Our results show that neither the presence of saturating chemoeffector concentrations nor of mutations that destroy ligand binding interferes with swarming. However, communication of the chemoreceptors with the CheA kinase is essential. The ability of nonchemotactic flagellar switch mutants to swarm suggests that chemotaxis is not required for swarming.
MATERIALS AND METHODS
Bacterial Strains and Plasmids.
Strains and plasmids used in this study are listed in Table 1. pPA56 encodes the cytoplasmic signaling domain of Tsr (amino acids 290–470) (22). pMS103 encodes a similar domain of Tar (amino acids 257–553) fused to a leucine zipper at its N terminus; in pMS105, amino acids 212–553 of Tar are also fused to a leucine zipper, but with an additional flexible linker region in between. The four sites of deamidation/methylation in Tar (QEQE) are mutated to produce QQQQ in these two plasmids (23). All three cytoplasmic fragments are reported to activate CheA kinase and confer a CW rotational bias to the flagella (22, 23).
Table 1.
Strains and plasmids
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| E. coli | ||
| RP437 | Wild type for motility | (11) |
| RP8611 | Δtsr, Δ(tar-tap), Δtrg | (12) |
| RP9352 | Δtsr, Δ(tar-tap), Δtrg, ΔcheZ | (12) |
| RP2867 | Δ(tap-cheB) | (13) |
| RP9535 | ΔcheA | (J. S. Parkinson, Univ. of Utah, Salt Lake City) |
| RP4782 | fliM182 | (14) |
| RP4783 | fliM183 | (14) |
| HCB429 | Δtsr, Δ(tar-tap), Δtrg | (15) |
| S. typhimurium | ||
| SJW1103 | Wild type for motility | (16) |
| KK2051 | cheA∷Tn10 | (17) |
| MY107 | fliG N72Y | (18) |
| SJW2323 | fliG V135F | (18) |
| Plasmids* | ||
| pJC3-Tsr | tsr+ | (J. S. Parkinson) |
| pJC3-Tsr(R64C) | tsr (190 C → T) | This work |
| pJC3-Tsr(T156P) | tsr (466 A → C) | This work |
| pJC3-Tsr(A413V) | tsr (1238 C → T) | This work |
| pNT201-Tar | tar+ | (19) |
| pNT201-Tar(R69K) | tar (205-207 CGT → AAA) | This work |
| pNT201-Tar(T154I) | tar (461 C → T) | This work |
| pMK76-Tar(M76K) | tar (227 T → A) | (20) |
| pAL1-Trg | trg+ | (21) |
| pCG1-Tap | tap+ | This work |
| pPA56 | Cytoplasmic Tsr domain | (ref. 22; see Methods) |
| pMS103 | Cytoplasmic Tar domain | (ref. 23; see Methods) |
| pMS105 | Cytoplasmic Tar domain | (ref. 23; see Methods) |
Genes on most plasmids are expressed from the tac promoter; pMS plasmids use the trc promoter, whereas pMK76 uses the natural Tar promoter. The numbers in parentheses under “relevant genotype” of plasmids refer to the nucleotide positions, starting at the first nucleotide of the ATG codon.
Motility Assays.
Swarming. Media used in swarming assays with E. coli consisted of 0.45% Eiken agar (7) with Eiken broth (3 g/liter meat extract, 10 g/liter peptone, 5 g/liter NaCl), to which 5 g/liter glucose was added after autoclaving. For S. typhimurium, Luria–Bertani broth with 5 g/liter glucose was solidified with 0.6% Difco agar. Where needed, 100 μg/ml ampicillin was added for propagation of plasmids, and 20 μM isopropyl β-d-thiogalactoside (IPTG) was added for inducing protein expression from them. Swarm agar was typically allowed to dry at room temperature for 1 day before use. Swarming efficiency was dramatically improved when cells were inoculated onto the center of swarm plates from 0.3% (swim) agar plates grown overnight at 30°C.
Swimming.
Media used to assay chemotaxis contained 0.3% Difco agar, 10 g/liter Difco tryptone, 5 g/liter NaCl, and where necessary, 100 μg/ml ampicillin and 20 μM IPTG. When required, serine was added as a solution to the media before pouring the plates.
All plates with E. coli were incubated at 30°C, whereas those with S. typhimurium were incubated at 37°C.
The reported smooth-swimming or tumbly phenotypes of all strains used in this study were confirmed by microscopic observation of the bacteria suspended in a drop of liquid. The rotational bias of the switch mutants was determined by tethering cells to a glass surface as described (24) by using cross-reacting antisera raised against S. marcescens flagellin (25). Tethered cells were washed extensively in buffer (10 mM potassium phosphate, pH 7.0/0.1 mM NaCl/0.01 mM EDTA/0.02 mM methionine/0.04 M dl-sodium lactate/100 μg/ml chloramphenicol), observed by videomicroscopy at room temperature, and recorded on videotape.
Flagellar staining of swarmer cells was performed as described (7).
Cloning and Mutagenesis.
Tsr mutants. Plasmid pJC3 contains tsr expressed from a tac promoter. Tsr(R64C), Tsr(T156P), and Tsr(A413V) were obtained by PCR amplification of relevant regions of pJC3 by using mutagenic primers containing a single base change at appropriate positions (Table 1), followed by exchange of restriction fragments between the wild-type and mutant versions of Tsr.
Tar mutants.
Plasmid pNT201 contains tar expressed from a tac promoter (19). The R69K, T154I, and M76K mutant variants of tar were created by exchanging restriction fragments harboring these mutations (from pRK69, pTI154, and pMK76, respectively; ref. 20) with pNT201 to replace the wild-type tar gene.
Tap gene cloning.
The tap gene was obtained by PCR amplification from the genomic DNA of RP437. Primers for the 5′ (5′-TCAGGATCCTTACTAACGCGGTCATTGCCGC) and 3′ (5′-GCATGCAAGCTTGCAGAGATGAAGTCATAGCGCC) ends of tap contained a BamHI and a HindIII site, respectively. A BamHI-HindIII fragment of the amplified PCR product was cloned into pJC3 next to the tac promoter, to generate pCG1.
All mutations and regions amplified by PCR were verified by DNA sequencing.
RESULTS
Tsr or Tar Alone Can Support Swarming.
We had shown earlier that E. coli mutants lacking any one of the four inner membrane chemoreceptors (Trg, Tsr, Tar, Tap) swarm normally (7). Although deletion of two transducers (Tar, Tap) had no effect on swarming, deletion of three (Tar, Tap, Tsr) or all four of the transducers abolished swarming (7). These results may either reflect a requirement for a specific subset of transducers, or, because Tsr and Tar are the most abundant transducers (26, 27), may reflect a requirement for a certain threshold number of transducers. Alternatively or in addition, the swarming defects could be due to the predominantly CCW pattern of flagellar rotation in the deletion mutants.
To test whether individual transducers support swarming, we introduced into a strain lacking all four transducers (HCB429) plasmids expressing either Tsr (pJC3), Tar (pNT201), Trg (pAL1), or Tap (pCG1) from IPTG-inducible promoters (Table 1). Only plasmids expressing Tsr and Tar restored swarming to the transducerless strain; Tsr was more effective than Tar (Fig. 1A). Even without added IPTG, basal-level expression of Tsr and Tar proteins was sufficient to support swarming (data not shown). For comparison, the extent of swimming motility (in 0.3% soft agar) in these strains under similar IPTG concentrations is shown in Fig. 1B. The strains expressing Trg and Tap were motile in liquid but did not migrate out significantly in the swim media in spite of expression from an inducible promoter, an observation consistent with that reported by others (28).
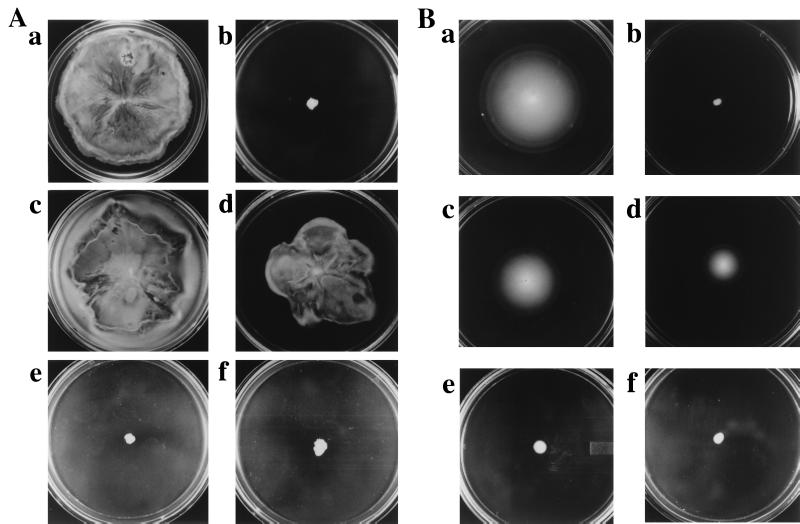
Figure 1.
Tsr or Tar alone supports swarming in E. coli. (A) Swarming motility is observed in wild-type (RP437) bacteria (a), but not in RP437 lacking all four transducers (HCB429) (b). Plasmids expressing Tsr (pJC3) (c) or Tar (pNT201) (d), but not those expressing Trg (pLA1) (e) or Tap (pCG1) (f), restore swarming in HCB429. (B) Swimming motility in the same strains as in A. As described in Methods, 20 μM IPTG was used to induce protein expression from plasmids. Plates in A were incubated for 16 h, whereas those in B were incubated for 12 h at 30°C.
The ability of a single transducer (Tsr or Tar) to support swarming allows us to manipulate them individually to investigate their role in swarming.
Saturation of Tsr with Serine Abolishes Chemotaxis but Does Not Affect Swarming.
l-serine, one of the most powerful attractants of E. coli (29), is detected by the periplasmic domain of Tsr (30, 31). Threshold attractant concentrations of serine are estimated to be in the micromolar range (29). At comparable concentrations, Tsr also responds to α-aminoisobutyrate, a nonmetabolizable analog of serine (32). As bacteria grow and metabolize serine in the medium, Tsr detects the presence of the resultant spatial serine gradient, allowing the bacteria to move toward higher serine concentrations. Very high concentrations of serine (in the millimolar range) saturate the receptor and inhibit chemotaxis (32). We have used this observation to ask whether detection of a serine gradient plays a critical role in swarming.
Fig. 2A shows inhibition of the chemotaxis response in HCB429/pJC3 (Tsr) with increasing serine concentrations (0–30 mM), measured by migration of bacteria in soft agar (0.3% “swim” plates). At the same serine concentrations, however, swarming is unaffected (Fig. 2B). Similar results were obtained with α-aminoisobutyrate (not shown). These results indicate that chemotaxis toward serine, the major chemoeffector of Tsr, is unlikely to be required for Tsr-dependent swarming.
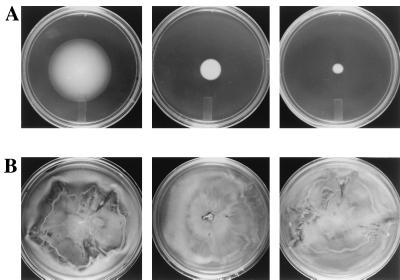
Figure 2.
High serine concentrations inhibit chemotaxis but do not inhibit swarming. Effect of increasing serine concentrations (0, 15, and 30 mM, left to right) on swimming (chemotaxis) (A) and swarming (B) motility of E. coli (HCB429) expressing only Tsr (pJC3). Serine concentrations (30–50 mM) inhibit chemotaxis even when all four transducers are present (not shown). Growth conditions are as in Fig. 1, except all plates were incubated at 30°C for 16 h.
Two Tsr Mutations That Destroy Serine Binding Have No Effect on Swarming.
To test the conclusions of the experiment in Fig. 2 further, we introduced two mutations into the periplasmic domain of Tsr that have been reported to abolish serine binding (R64C and T156P; ref. 31). The mutant plasmids were introduced into HCB429 and tested for swimming and swarming in the presence of increasing serine concentrations (Fig. 3). When compared with wild-type Tsr, bacteria expressing Tsr(R64C) migrated poorly in swim media (Fig. 3A; compare 0 mM serine plates in Figs. 2A and 3A). This is expected, because the mutant Tsr is unable to sense a serine gradient. Also as expected, increasing serine concentrations had no effect on this migration. By contrast, the mutant Tsr supported swarming efficiently (Fig. 3B) and displayed a normal swarmer cell morphology (see Fig. 5b). Similar results were obtained with Tsr(T156P). As indicated in Table 2, both Tsr(R64C) and Tsr(T156P) were more efficient in swarming than wild-type Tsr. These results confirm that chemotaxis toward serine is not required for the ability of Tsr to support swarming.
Figure 3.
Tsr(R64C), a serine-binding mutant, supports swarming. Migration of E. coli (HCB429) expressing Tsr(R64C) in swim (A) and swarm (B) media in the presence of increasing (0–30 mM) serine concentrations. Growth conditions are as in Fig. 2.
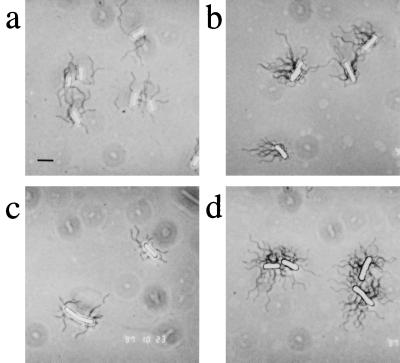
Figure 5.
Flagellation of representative nonchemotactic mutants. Cells grown on swarm media were stained to visualize flagella as described (7). (a) Nonswarming E. coli HCB429 (Fig. 1A, b). (b) Swarming E. coli HCB429/pJC3-Tsr(R64C) (Fig. 3B). (c) Nonswarming E. coli RP2867 (cheBR) (Table 3). (d) Swarming S. typhimurium MY107 (fliG) (Table 3). The morphology of wild-type E. coli (not shown) is indistinguishable from that of the Tsr mutant shown in b. (Bar = 2.8 μm.)
Table 2.
Ability of serine and aspartate-binding mutants of Tsr and Tar to support chemotaxis (swimming) and swarming motility
| Strain | Chemotaxis | Swarming |
|---|---|---|
| HCB429 | − | − |
| RP437 | ++++ | ++++ |
| HCB429/pJC3-Tsr | +++ | ++++ |
| /pJC3-Tsr(R64C) | − | +++++ |
| /pJC3-Tsr(T156P) | − | +++++ |
| /pNT201-Tar | ++ | +++ |
| /pNT201-Tar(R69K) | − | +++ |
| /pNT201-Tar(T154I) | − | ++++ |
| /pMK76-Tar(M76K) | ++ | +++ |
Chemotaxis assays were on swim (0.3%) agar plates, whereas swarming assays were on 0.45% agar plates, as described in Methods. Except for pMK76, protein expression from all plasmids was induced with 20 μM IPTG. Results are from several independent experiments. In any one experiment, the diameter of the swim/swarm colony after 16 h at 30°C was measured, and the results from four plates were averaged. Chemotaxis/swarming phenotype: −, no migration; +++, migration of ≈6 cm; ++++, migration of ≈8 cm (i.e., entire petri dish); +++++, migration of ≈8 cm after 12 h.
Mutations in Tar That Destroy Aspartate or Maltose Binding Do Not Affect Swarming.
Unlike Tsr, Tar responds to two chemoeffectors—aspartate and maltose. Because swarm media contains significant levels of aspartate, we tested the effect of two mutations in Tar (R69K and T154I) that destroy aspartate binding (20). We also tested the effect of a mutation (R76K) that destroys maltose binding via maltose binding protein (20). Although none of the mutants affected swarming, one of the two aspartate-binding mutants [Tar(T154I)] showed a better swarming response than wild-type Tar (Fig. 4). The properties of all mutants are summarized in Table 2. All Tar mutants had a normal swarmer cell morphology (not shown).
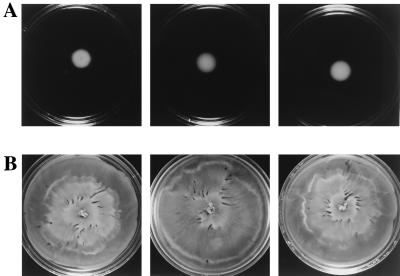
Figure 4.
Tar(T154I), an aspartate-binding mutant, supports swarming. Swarming motility in HCB429 expressing wild-type (A) or mutant (B) Tar(T154I) proteins. Growth conditions are as in Fig. 2.
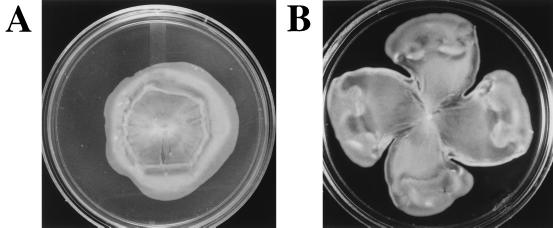
We note that ligand-binding mutants of both Tsr (Fig. 3) and Tar (Fig. 4) lend distinctive patterns to the swarm colony, compared with those generated by their wild-type counterparts.
A Cytoplasmic Signaling Mutant of Tsr Inhibits Swarming.
A single amino acid change (A413V) in the cytoplasmic signaling domain of Tsr results in a transducer with a locked CCW output (33). Studies with truncated signaling domains carrying the A413V mutation have shown that the mutant fragments block CW flagellar rotation in vivo and inhibit CheA autophosphorylation in vitro (22). Tsr(A413V) should therefore effectively behave like a CheA mutation and be defective in swarming. The result of introduction of a plasmid carrying Tsr(A413V) into a transducerless strain (HCB429) and a wild-type strain (RP437) is shown in Table 3. Although the presence of the mutant transducer alone failed to support swarming, its presence in cells with all four wild-type transducers also blocked swarming. The latter observation is consistent with the reported dominant effect of this mutation (for chemotaxis) over both its wild-type counterpart and over heterologous transducers (33).
Table 3.
Flagellar rotational bias and swarming
| Strain | Reported/observed bias | Swarming |
|---|---|---|
| HCB429 | CCW (12) | − |
| HCB429/pJC3-AV413 | CCW (33) | − |
| HCB429/pPA56 | CCW/CW (22) | − |
| RP8611 | CCW (22) | − |
| RP8611/pMS103 | CCW/CW (23) | − |
| RP8611/pMS105 | CCW/CW (23) | − |
| RP437/pJC3-AV413 | CCW (33) | −/+ |
| RP2867 (cheBR) | CCW/CW (34) | − |
| RP9352 (MCP−cheZ) | CCW/CW (22) | − |
| RP437 | CCW/CW (13); | ++++ |
| RP4782 (fliM) | CCW (14); | ++ |
| RP4783 (fliM) | CCW (14); | + |
| SJW1103 | CCW/CW (18); | ++++ |
| MY107 (fliG) | CCW (18); | +++ |
| SJW2323 (fliG) | CW (18); | ++ |
| CCW↔CW |
The flagellar rotational bias of tethered cells (displayed in the histograms) was measured in the absence of chemotactic stimuli by observing 20 rotating cells for 60 sec each, as described in Methods. Cells were classified into five categories (from left to right): exclusively CCW, CCW biased with some reversals, frequent reversals with no bias, CW biased with some reversals, and exclusively CW. The height of the bars corresponds to the percentage of cells in each category. These rotational profiles were quite reproducible. The rotational bias of an E. coli cheA strain (RP9535) was CCW with cell distribution in each category (left to right) being 17, 1, 0, 0, 2; for an S. typhimurium cheA strain (KK2051; ref. 17) it was 17, 0, 0, 1, 2. These biases were very similar to those of the CCW switch mutants. Swarming ability in strains with plasmids was tested over a range (0–1,000 μM) of IPTG concentrations. Even without added IPTG, there is a basal level of gene expression. The negative results in strains with most plasmids were seen over the entire range of IPTG concentrations tested. In RP437/pJC3-AV413, the −/+ indicates either no swarming or 1-2 cm migration in different experiments with no added IPTG; swarming was completely blocked with even 10 μM IPTG. ++++, ≈8 cm; +++, ≈6 cm; ++, ≈4 cm; +, ≈2 cm after 16 h incubation.
We conclude that although the serine-binding ability of Tsr is not required, its CheA kinase-mediated activity is essential for swarming. Results in the next section show that a CCW bias by itself does not inhibit swarming.
Flagellar Rotation Bias and Swarming.
The results in Figs. 1–4 along with previous results (7) show that the chemotaxis system is required for swarming. Evidence against the argument that the swarming defects in che mutants are simply the result of an altered flagellar rotation bias was provided by our earlier observation that a strain with a cheBcheR deletion (RP 2867), which has a near-normal CW/CCW motor rotation bias (34), also fails to swarm (ref. 7; see also Table 3). Additional experiments to test this idea are shown in Table 3. A strain lacking all MCPs as well as CheZ (RP9352) also shows both CW and CCW flagellar rotation (22), yet fails to swarm. We next tested a CW signaling fragment of Tsr (pPA56), whose expression in a transducerless strain is reported to shift the initial CCW bias of the strain to a CW mode (22). Controlled expression of this fragment at varying inducer concentrations produces cells with different levels of CW/CCW bias. However, swarming was not observed in this strain at any inducer concentration. Two similar signaling fragments of Tar (pMS103 and pMS105; ref. 23) also failed to rescue swarming. These results suggest that it is a defect in signaling through the chemotaxis system in the presence of membrane-bound transducers, rather than the flagellar rotational bias, that is primarily responsible for lack of swarming in the che and the MCP mutants. The swarming defect in all the mutants described in Table 3 appears to be at the level of hyperflagellation. This was confirmed by flagellar staining (Fig. 5 a–c) and flagellin reporter gene assays (data not shown).
The experiments above do not address the issue of whether an extreme flagellar rotational bias (CCW or CW) in itself would also inhibit swarming. To determine this we examined mutations in the “switch” components found at the base of the motor (9). We tested the ability of FliM mutants of E. coli and FliG mutants of S. typhimurium (both components of the flagellar switch mechanism), which have a reported CCW or CW bias. These mutants have an intact signal transduction system, yet are nonchemotactic. The data in Table 3 show that the CCW-biased mutants in fliM (RP4782 and RP4783) and fliG (MY107) can swarm, although not as well as their wild-type counterparts (RP437 and SJW1103, respectively). Tethering assays showed that the rotational bias of these switch mutants was predominantly exclusively CCW, similar to that of nonswarming cheA null mutants of these strains (Table 3). The CW fliG mutant (SJW2323) swarmed less efficiently than the CCW fliG mutant. The rotational bias of this mutant was determined to be predominantly CW with some reversals. All switch mutants were able to differentiate into swarmer cells (Fig. 5d shows morphology of MY107). The initial kinetics of swarming appeared to be similar in all strains; the smaller diameter of the colony appears to be related to a gradual slowing down in rate of cell movement (the data in Table 3 were recorded after 16 h).
We conclude that an extreme CCW bias in the rotational state of the motor does not interfere significantly with swarming motility. A predominantly CW bias with some reversals also allows swarming; it is possible that an extreme CW bias may not. The ability of the nonchemotactic switch mutants to swarm lends further support to our findings that chemotaxis is not required for swarming. However, an intact chemotaxis system is essential.
DISCUSSION
The central question related to swarm cell differentiation is the nature of the swarm signal(s) and the manner in which it is transduced. Although it is not yet known what the signal is in organisms like E. coli and S. typhimurium, it is clear that components of the chemotaxis system play a critical role in inducing the hyperflagellation response in swarmer cells of these species.
To get a handle on the nature of the signal detected by the chemotaxis system, we have begun analysis of the transmembrane chemoreceptors. When tested individually, only Tsr and Tar were able to complement a receptorless strain for swarming (Fig. 1). A strong conclusion from our studies is that although Tsr or Tar is required for swarming, sensing their most powerful attractants is not needed (Figs. 1–4 and Table 2). What then are the receptors responding to? Although Tsr is the primary chemoreceptor for serine, it binds to other amino acids with lower affinity, is a receptor for repellents l-leucine, indole, and low external pH values, and is also a thermo-receptor (35). However, a receptor saturated by one attractant shows no response when exposed to another attractant that shares the same binding site (36). High serine concentrations are also known to block the thermosensing response of Tsr (37). Because swarming is unaffected under these conditions (Fig. 2), it is unlikely that Tsr functions through thermosensing. A similar argument may be made for responding to lower-affinity attractants. In addition, we have observed that the repellent response of wild-type Tsr to leucine is completely inhibited at high serine concentrations [as assayed by the chemical-in-plug method described by Tso and Adler (38)], and the repellent response toward acetate is significantly diminished (data not shown).
If a common mechanism is to be evoked for Tar or Tsr in swarming, it is unlikely that it would involve repellent sensing. First, these two receptors have been reported to exhibit opposing responses to pH (ref. 39; and unpublished results with acetate). Second, our observation that nonchemotactic switch mutants in FliM and FliG (which have an intact signaling system but a defective motor switch) can swarm (Table 3) argues against a role for chemotaxis, negative or positive, in swarming. We note that a similar conclusion was reached in earlier results with P. mirabilis (40).
The demonstration that the CCW mutants in FliM and FliG (predominantly exclusively CCW; Table 3) can differentiate into swarmer cells (Fig. 5) as well as swarm (Table 3), although not as robustly as wild-type bacteria, suggests that the ability to rotate flagella in both directions is important but not critical for swarming motility. Thus, the extreme CCW flagellar rotation bias of che A, W, R, and Y mutants cannot in itself be responsible for their swarming defects. The CW FliG mutant could also differentiate into swarmer cells and swarm, but less efficiently than the CCW FliG mutant (Table 3). Because the rotational bias of this mutant was not exclusively CW, it is possible that an extreme CW bias might not support swarming motility. Results with Rhodospirillum centenum are consistent with these conclusions, in that mutations in the che genes that are CCW biased do not interfere with swarming, whereas a CW-biased che mutant fails to swarm (41). In this organism, the chemotaxis components appear not to be involved in swarm cell differentiation. Our results suggest that in E. coli and S. typhimurium, it is not chemotactic behavior, but rather a lack of signaling through the chemotaxis components, that inhibits swarming in the che mutants (7).
The data presented in this paper compel us to consider the possibility that the sensory transducers Tsr and Tar may detect hitherto unknown signals that are still processed through the chemotaxis system. These signals might be chemical or physicochemical in nature. Examples of the latter may include membrane alterations induced in response to either the viscosity of the extracellular slime (increased viscosity is known to induce hyperflagellation in several bacteria; ref. 1) or the process of cell elongation, or to cell–cell interactions within the closely packed colony (42).
Acknowledgments
We thank Howard Berg, Jerry Hazelbauer, Bob Macnab, Mike Manson, Sandy Parkinson, and Michael Surette for gifts of strains and plasmids, Howard Berg for helpful comments on the manuscript, and Catherine Gordon for isolating the tap gene from E. coli.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviations: IPTG, isopropyl β-d-thiogalactoside; CCW, counter-clockwise; CW, clockwise.
References
- 1.Harshey R M. Mol Microbiol. 1994;13:389–394. doi: 10.1111/j.1365-2958.1994.tb00433.x. [DOI] [PubMed] [Google Scholar]
- 2.Henrichsen J. Bacteriol Rev. 1972;36:478–503. doi: 10.1128/br.36.4.478-503.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Otteman K M, Miller J F. Mol Microbiol. 1997;24:1109–1117. doi: 10.1046/j.1365-2958.1997.4281787.x. [DOI] [PubMed] [Google Scholar]
- 4.Sar N L, McCarter L, Simon M, Silverman M. J Bacteriol. 1990;172:334–341. doi: 10.1128/jb.172.1.334-341.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belas R, Erskine D, Flaherty D. J Bacteriol. 1991;173:6279–6288. doi: 10.1128/jb.173.19.6279-6288.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Rear J, Alberti L, Harshey R M. J Bacteriol. 1992;174:6125–6173. doi: 10.1128/jb.174.19.6125-6137.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harshey R M, Matsuyama T. Proc Natl Acad Sci USA. 1994;91:8631–8635. doi: 10.1073/pnas.91.18.8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stock J B, Surette M G. In: Escherichia coli and Salmonella, Cellular and Molecular Biology. Neidhardt F C, editor. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1103–1129. [Google Scholar]
- 9.Macnab R M. In: Escherichia coli and Salmonella, Cellular and Molecular Biology. Neidhardt F C, editor. Vol. 1. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 123–145. [Google Scholar]
- 10.Hoch J A, Silhavy T J, editors. Two-Component Signal Transduction. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 11.Parkinson J S. J Bacteriol. 1978;135:45–53. doi: 10.1128/jb.135.1.45-53.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J D, Parkinson J S. Proc Natl Acad Sci USA. 1989;86:8703–8707. doi: 10.1073/pnas.86.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parkinson J S, Houts S E. J Bacteriol. 1982;151:106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parkinson J S. J Bacteriol. 1976;126:758–770. doi: 10.1128/jb.126.2.758-770.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wolfe A F, Conley M P, Kramer T J, Berg H C. J Bacteriol. 1987;169:1878–1885. doi: 10.1128/jb.169.5.1878-1885.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamaguchi S, Fujita H, Sugata K, Taira T, Iino T. J Gen Microbiol. 1984;130:255–265. doi: 10.1099/00221287-130-2-255. [DOI] [PubMed] [Google Scholar]
- 17.Kutsukake K, Ohya Y, Iino T. J Bacteriol. 1990;172:741–747. doi: 10.1128/jb.172.2.741-747.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamaguchi S, Aizawa S, Kiharu M, Isomura M, Jones C J, Macnab R M. J Bacteriol. 1986;168:1172–1179. doi: 10.1128/jb.168.3.1172-1179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Borkovich K A, Kaplan N, Hess J F, Simon M I. Proc Natl Acad Sci USA. 1989;86:1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gardina P, Conway C, Kossman M, Manson M. J Bacteriol. 1992;174:1528–1536. doi: 10.1128/jb.174.5.1528-1536.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee F, Lebert M R, Lilly A A, Hazelbauer G L. Proc Natl Acad Sci USA. 1995;92:3391–3395. doi: 10.1073/pnas.92.8.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ames P, Parkinson J S. J Bacteriol. 1994;176:6340–6348. doi: 10.1128/jb.176.20.6340-6348.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Surette M G, Stock J B. J Biol Chem. 1996;271:17966–17973. doi: 10.1074/jbc.271.30.17966. [DOI] [PubMed] [Google Scholar]
- 24.Manson M D, Tedesco P M, Berg H C. J Mol Biol. 1980;138:541–561. doi: 10.1016/s0022-2836(80)80017-9. [DOI] [PubMed] [Google Scholar]
- 25.Alberti L, Harshey R M. J Bacteriol. 1992;172:4322–4328. doi: 10.1128/jb.172.8.4322-4328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hazelbauer G L, Engstrom P. Nature (London) 1980;283:98–100. doi: 10.1038/283098a0. [DOI] [PubMed] [Google Scholar]
- 27.Slocum M K, Parkinson J S. J Bacteriol. 1983;155:565–577. doi: 10.1128/jb.155.2.565-577.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng X, Baumgartner J W, Hazelbauer G L. J Bacteriol. 1997;179:6714–6720. doi: 10.1128/jb.179.21.6714-6720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adler J. Science. 1969;166:1588–1597. doi: 10.1126/science.166.3913.1588. [DOI] [PubMed] [Google Scholar]
- 30.Hedblom M L, Adler J. J Bacteriol. 1980;144:1048–1060. doi: 10.1128/jb.144.3.1048-1060.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee L, Mizuno T, Imae Y. J Bacteriol. 1988;170:4769–4774. doi: 10.1128/jb.170.10.4769-4774.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mesibov R, Adler J. J Bacteriol. 1972;112:315–326. doi: 10.1128/jb.112.1.315-326.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ames P, Parkinson J S. Cell. 1988;55:817–826. doi: 10.1016/0092-8674(88)90137-7. [DOI] [PubMed] [Google Scholar]
- 34.Segall J E, Manson M D, Berg H B. Nature (London) 1982;296:855–857. doi: 10.1038/296855a0. [DOI] [PubMed] [Google Scholar]
- 35.Manson M D. Adv Microbial Physiol. 1992;33:277–346. doi: 10.1016/s0065-2911(08)60219-2. [DOI] [PubMed] [Google Scholar]
- 36.Adler J. Annu Rev Biochem. 1975;44:341–356. doi: 10.1146/annurev.bi.44.070175.002013. [DOI] [PubMed] [Google Scholar]
- 37.Maeda K, Imae Y. Proc Natl Acad Sci USA. 1979;76:91–95. doi: 10.1073/pnas.76.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tso W-W, Adler J. J Bacteriol. 1974;118:560–576. doi: 10.1128/jb.118.2.560-576.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krikos A, Conley P M, Boyd A, Berg H C, Simon M I. Proc Natl Acad Sci USA. 1985;82:1326–1330. doi: 10.1073/pnas.82.5.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams F D, Anderson D M, Hoffman P S, Schwarzhoff R H, Leonard S. J Bacteriol. 1976;127:237–248. doi: 10.1128/jb.127.1.237-248.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang Z-J, Gest H, Bauer C. J Bacteriol. 1997;179:5720–5727. doi: 10.1128/jb.179.18.5720-5727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sogaard-Andersen L, Kaiser D. Proc Natl Acad Sci USA. 1996;93:2675–2679. doi: 10.1073/pnas.93.7.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]