Abstract
Clones of CD8+ T cells specific for viral antigens must avoid replicative senescence to maintain continuous production of new effector cells during chronic viral infections. In the present study, we have determined whether this capability may be related to Bmi-1, a transcriptional repressor that is required for the maintenance of hematopoietic stem cells and certain neural stem cells and that mediates its antisenescence function by inhibiting transcription of the Ink4a/Arf tumor suppressor locus. Ligation of the T cell receptor increased the levels of Bmi-1 mRNA and protein in primary CD8+ T cells. The increased expression was reversible upon removal of antigen but could be maintained by using stimulation with the IL-2 receptor. Specific suppression of Bmi-1 by using a lentivirally encoded short hairpin RNA inhibited the proliferation of IL-2-stimulated CTLL-2 cytotoxic T cells and primary CD8+ T cells. Ectopically expressed Bmi-1 enhanced the expansion of primary CD8+ T cells stimulated by IL-2 and IL-7 in vitro and by homeostatic signals in vivo. Taken together, these findings indicate that Bmi-1 is required for CD8+ T cell clonal expansion and is positively regulated by receptors that mediate this response. Therefore, the observation that the ability of the T cell receptor to induce Bmi-1 is maintained in the subset of replication-competent, antigen-experienced CD8+ T cells that do not express the killer cell lectin-like receptor G1 (KLRG1) but is developmentally switched off in the senescent, KLRG1+ subset suggests that Bmi-1 is a molecular determinant of the capacity of a CD8+ T cell clone to persist during chronic viral infections.
Keywords: Ink4a, senescence, IL-2
In the field of CD8+ T cell biology, there is a need to understand how new effector CD8+ T cells are generated during intermittent or persistent microbial infections because two sets of observations suggest that replicative senescence occurs in the terminally differentiated CD8+ T cell. First, certain populations of antigen-experienced CD8+ T cells, such as effector and effector memory cells that are CD62Llow or CD127low, proliferate poorly after adoptive transfer and microbial challenge (1–7). However, this finding may be explained as the result of impaired homing of the transferred cells to appropriate sites in secondary lymphoid organs, so a second set of observations relating to human and murine CD8+ T cells that express killer cell lectin-like receptor G1 (KLRG1) is especially important (8). This marker identifies effector CD8 T cells that have developed during intermittent or persistent viral infections and that proliferate poorly in vivo upon antigenic challenge and in vitro after T cell receptor (TCR) ligation (9–11). The proliferative defect was observed even in the presence of exogenous IL-2 and despite TCR-mediated IFN-γ synthesis, indicating that the senescent phenotype is not caused by an inability to produce IL-2 or by a general defect in TCR signaling, distinguishing the KLRG1+ CD8+ T cell from the “exhausted” CD8+ T cell in which TCR signaling is inhibited by PD-1 (12).
Although senescence in the CD8+ T cell has been ascribed to telomere erosion, T cell activation can induce telomerase activity (13, 14), and replicative senescence is not prevented by expression of ectopic telomerase in human T cells (15). Another cause of senescence may involve the stem cell-associated transcriptional repressor, Bmi-1, and the p16Ink4a/p19Arf tumor suppressor proteins. These two proteins are encoded by the Ink4a/Arf locus and activate the retinoblastoma protein- and p53-dependent pathways of cell cycle arrest, senescence, and apoptosis (16). Transcription of Ink4a/Arf is suppressed by Bmi-1 (17), a member of the Polycomb repressive complex 1 that was discovered as a cooperating oncogene in Eμ-myc transgenic mice (18, 19). Mice deficient in Bmi-1 have impaired self-renewal of hematopoietic stem cells and neural stem cells (20–22), diminished T cell development in the thymus, and decreased numbers of peripheral T and B lymphocytes (23). The few mature lymphocytes that are present in Bmi-1−/− mice have diminished in vitro proliferative responses to mitogenic stimulation, reflecting either an effect of abnormal thymic development or a role for Bmi-1 in the replication of mature T cells. The abnormal lymphocyte phenotype of Bmi-1−/− mice is partially rescued by interruption of the Ink4a/Arf genes (17), with p19Arf appearing to have a more important growth inhibitory role (24). Within the hematopoietic system, Bmi-1 is most highly expressed in hematopoietic stem cells and is down-regulated upon commitment to differentiation to the Gr-1+ granulocytic and Mac-1+ monocytic/macrophage lineages but is maintained in mature splenic B and T lymphocytes (25). The possibility that Bmi-1 may have a role in the clonal expansion of lymphocytes is supported by the finding that ligating the B cell antigen receptor increases the expression of Bmi-1 (26).
These findings suggest that Bmi-1 may be the determinant of the replicative competence of the CD8+ T cell. In the present study, we support this possibility by demonstrating that Bmi-1 is required for optimal proliferation of the CD8+ T cell and that ligation of the TCR causes its expression in naïve and KLRG1− memory cells but not in senescent, KLRG1+ memory cells.
Results
Regulation of Bmi-1 Expression in Naïve CD8+ T Cells by Using Stimuli That Induce Clonal Expansion.
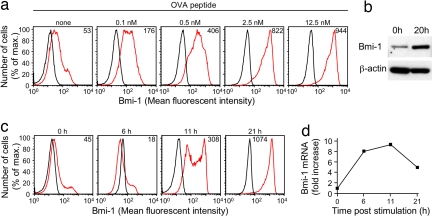
Bmi-1 suppresses the senescence of replicating cells (17), and it was of interest to determine whether receptors that mediate clonal expansion of the CD8+ T cell induce Bmi-1 expression. We cultured purified TCR transgenic OT-I cells (27), which are specific for the ovalbumin-derived peptide OVA257–264 (SIINFEKL) complexed to H-2Kb, with incremental concentrations of OVA peptide for 24 h, and then we stained permeabilized cells with antibody specific for Bmi-1. The intracellular level of Bmi-1, as detected by flow cytometric analysis, increased in a dose-dependent manner, with maximal expression occurring with 2.5 nM peptide (Fig. 1a). The increase in Bmi-1 induced by 2.5 nM peptide also was demonstrated by using Western blot analysis (Fig. 1b). Addition of blocking antibodies to IL-2 and CD25 did not diminish the induction of Bmi-1 (data not shown). A kinetic analysis of TCR-induced Bmi-1 protein (Fig. 1c) and mRNA (Fig. 1d) revealed that maximal protein expression occurred by 21 h of stimulation, whereas the increase in mRNA peaked between 6 and 11 h.
Fig. 1.
TCR signaling and induction of Bmi-1 expression in CD8+ T cells. (a) Naïve OT-I T cells were cultured for 24 h in medium alone or in medium containing incremental concentrations of SIINFEKL peptide and assessed by flow cytometry for Bmi-1 expression after staining with isotype control (black line) or anti-Bmi-1 antibody (red line). (b) Whole-cell lysates of naïve OT-I cells were generated 0 and 20 h after culturing cells in medium containing 2.5 nM SIINFEKL peptide and were assessed by immunoblot for Bmi-1 and β-actin. (c and d) Naïve OT-I cells were cultured in the presence of 2.5 nM SIINFEKL peptide for various lengths of time and then assessed for Bmi-1 expression by flow cytometry after staining with isotype control (black line) or anti-Bmi-1 antibody (red line) (c) and assessed for Bmi-1 mRNA by quantitative RT-PCR (d). The mean fluorescent intensity for specific Bmi-1 staining is represented by the number in each histogram. These experiments were performed multiple times with comparable results.
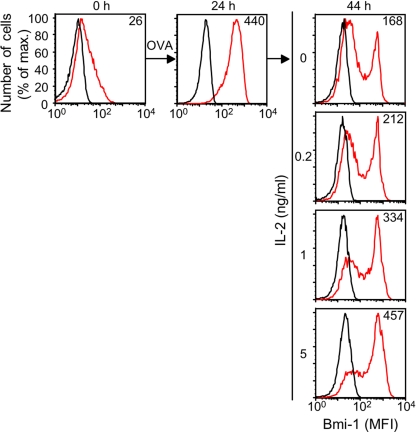
Stimulation of OT-I cells with IL-2 alone did not increase expression of Bmi-1 (data not shown), as would be anticipated because resting CD8+ T cells do not respond to this cytokine. However, addition of incremental concentrations of IL-2 to OT-I cells that had been cultured with 2.5 nM OVA peptide for 24 h maintained Bmi-1 expression during subsequent culture for 20 h in medium lacking antigen, which otherwise decreased in a subpopulation of cells (Fig. 2). Therefore, TCR induces expression of Bmi-1 in the naïve CD8+ T cell, and this increase is maintained by using ligation of the IL-2 receptor, which is consistent with its proliferative effect not requiring repetitive TCR stimulation (28–30).
Fig. 2.
Maintenance of Bmi-1 expression by IL-2. Naïve OT-I cells were cultured for 24 h in medium with 2.5 nM SIINFEKL peptide. The cells were then transferred to fresh medium alone or medium containing incremental concentrations of IL-2 and cultured for an additional 20 h, after which Bmi-1 expression was assessed by staining with isotype control (black line) or anti-Bmi-1 antibody (red line). The mean fluorescent intensity for specific Bmi-1 staining is represented by the number in each histogram. The experiments shown were performed multiple times with comparable results.
Bmi-1 and the Proliferative Capacity of the CD8+ T Cell.
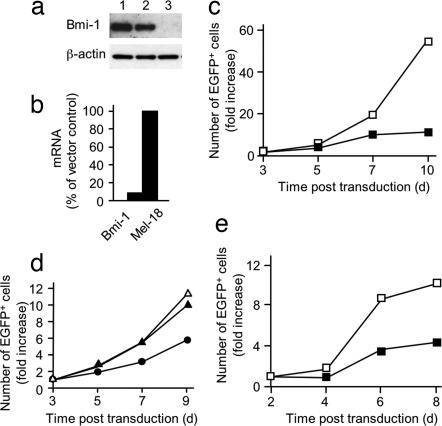
T cells from Bmi-1−/− mice proliferate poorly in vitro, but this defect may be secondary to Bmi-1 deficiency during thymic development (23). To evaluate the effect of depleting Bmi-1 from a Bmi-1-sufficient CD8+ T cell, we first validated the specificity of a lentiviral vector expressing short hairpin RNA (shRNA) by using the murine cytotoxic T cell line CTLL-2. During their IL-2-mediated proliferation, these cells were infected with lentiviral vectors expressing EGFP alone (pll3.7) or together with an shRNA that targets Bmi-1 (pll3.7-shBmi-1). The pll3.7-shBmi-1 vector suppressed both Bmi-1 protein (Fig. 3a) and mRNA (Fig. 3b), but not Mel-18 mRNA (Fig. 3b), and inhibited proliferation of the CTLL-2 cells (Fig. 3c). To exclude off-target effects of this shRNA on proliferation, we first infected replicate samples of CTLL-2 cells with a retroviral vector expressing human CD2 alone or together with a variant Bmi-1 having silent mutations in the shRNA target sequence. The CTLL-2 cells were then transduced with each of the two lentiviral vectors. In contrast to its inhibitory effect on CTLL-2 cells that had been infected with the control retrovirus, the pll3.7-shBmi-1 did not affect the proliferation of the cells expressing the mutant Bmi-1 (Fig. 3d). The rescue of the proliferation of CTLL-2 cells with the mutant Bmi-1 excludes off-target effects of pll3.7-shBmi-1 and validates its use for analysis of primary CD8+ T cells. Therefore, the inhibition of IL-2- and IL-7-mediated expansion of OT-I cells by transduction with pll3.7-shBmi-1 indicates that Bmi-1 expression is required for CD8+ T cell proliferation (Fig. 3e).
Fig. 3.
Inhibition of proliferation of CD8+ T cells by a lentivirus expressing an shRNA specific for Bmi-1. CTLL-2 cells maintained with IL-2 were infected at an moi of 5 with lentiviral vectors expressing EGFP alone (pll3.7) or together with a Bmi-1-specific shRNA (pll3.7-shBmi-1). (a) Whole-cell lysates of CTLL-2 cells (lane 1), cells infected with pll3.7 (lane 2), or cells infected with pll3.7-shBmi-1 (lane 3) were subjected to immunoblot analysis with antibodies against Bmi-1 and β-actin. (b) CTLL-2 cells infected with either the pll3.7 or pll3.7-shBmi-1 lentiviral vector were assessed for Bmi-1 and Mel-18 mRNA by using quantitative RT-PCR. (c) The number of CTLL-2 cells infected with pll3.7 (open squares) or pll3.7-shBmi-1 (filled squares) was measured during continuous culture in the presence of IL-2. (d) CTLL-2 cells were transduced with a retroviral vector expressing human CD2 alone (circles) or together with Bmi-1 containing silent mutations at the shRNA binding site (triangles). Transduced cells were superinfected with either pll3.7 (open triangles) or pll3.7-shBmi-1 (filled triangles and filled circles), and the number of EGFP+, lentivirally transduced cells was assayed periodically during growth in the presence of IL-2. (e) The number of EGFP+ OT-I cells infected with pll3.7 (open squares) or pll3.7-shBmi-1 (filled squares) was measured during continuous culture in the presence of IL-2 and IL-7. The experiments shown were performed multiple times with comparable results.
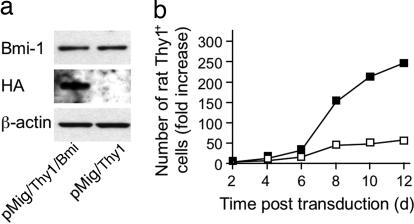
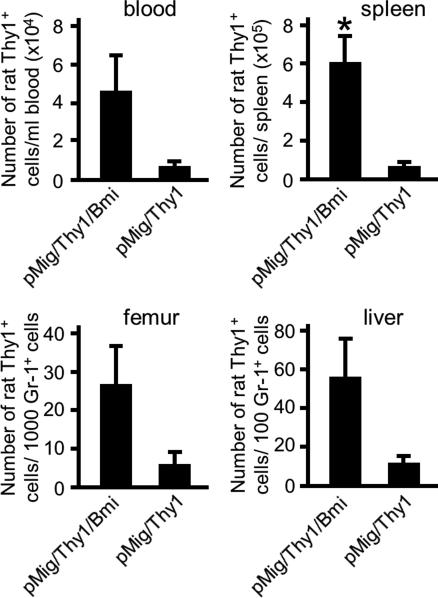
The magnitude of Bmi-1 expression maintained by IL-2 receptor signaling may not be optimal for CD8+ T cell proliferation, as suggested by the occurrence of a subpopulation of OT-I cells with diminished Bmi-1 in the presence of IL-2 (Fig. 2). To examine this possibility, we infected OT-I cells responding to IL-2 and IL-7 with retroviral vectors expressing rat Thy1 alone (pMig/Thy1) or with HA-tagged Bmi-1 (pMig/Thy1/Bmi-1) and measured their expansion during culture with IL-2 and IL-7 over the ensuing 12 d. Although the ectopic Bmi-1 was not overexpressed when cells were assessed on day 4 (Fig. 4a), these transduced OT-I cells expanded more effectively than did the vector control cells (Fig. 4b). We examined the capacity of ectopically expressed Bmi-1 to enhance in vivo expansion of the CD8+ T cell by adoptively transferring OT-I cells that had been transduced with pMig/Thy1 or pMig/Thy1/Bmi-1 into Rag2−/− recipient mice and counting the number of CD8+ T cells from various tissues 215 d later. There was greater expansion of the OT-I cells expressing ectopic Bmi-1 in the blood, spleen, femur, and liver (Fig. 5), indicating that augmented Bmi-1 expression enhances the response of cells to homeostatic stimuli.
Fig. 4.
Enhanced proliferation in vitro of CD8+ T cells expressing ectopic Bmi-1. (a) OT-I cells that had been activated with anti-CD3ε in the presence of IL-2 and IL-7 were transduced with pMig/Thy1 or with pMig/Thy1/Bmi-1 and maintained for 6 d in the presence of IL-2 and IL-7. On day 4 after transduction, cells expressing rat Thy1 were purified, and lysates were subjected to analysis by Western blot with antibodies specific for Bmi-1, HA, and β-actin. (b) In a replicate experiment, OT-I cells were transduced with pMig/Thy1 (open squares) or pMig/Thy1/Bmi-1 (filled squares) and assessed for growth in the presence of IL-2 and IL-7. The experiments shown were performed multiple times with comparable results, except for the Western blot, which was performed once.
Fig. 5.
Enhanced homeostatic expansion of CD8+ T cells expressing ectopic Bmi-1. Rat Thy1+ OT-I cells (4 × 105) that had been transduced with pMig/Thy1 or pMig/Thy1/Bmi-1 during proliferation in the presence of IL-2 and IL-7 were adoptively transferred to Rag2−/− recipients. After 215 d, the number of rat Thy1+ CD8+ T cells in the blood, spleen, femur, and liver was measured, with recovery from the latter two sites being normalized by counting Gr-1+ cells. The means and SE are shown for each determination. The experiment has been repeated with similar results. *, P = 0.043 (paired Student's t test).
Regulation of Bmi-1 Expression in Antigen-Experienced CD8+ T Cells.
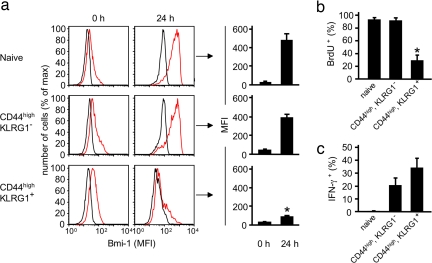
The expression of KLRG1 distinguishes antigen-experienced CD8+ T cells that have lost the potential for clonal expansion from cells that have retained this capability (9, 10). Because Bmi-1 regulates the proliferation of the CD8+ T cell, we evaluated the possibility that loss of its TCR-induced expression may account for the replicative defect of the KLRG1+ subset. We compared the expression of Bmi-1 before and 24 h after TCR ligation in naïve CD44low CD8+ T cells and in the two populations of antigen-experienced CD44high cells that are distinguished by the presence or absence of KLRG1. All cells were purified from the spleens of mice persistently infected with mouse γ-herpesvirus strain 68 (γMHV-68) and were shown to be CD3ε+. The three populations of CD8+ T cells had equivalently low levels of Bmi-1 when assessed immediately after purification, but only the naïve and CD44high KLRG1− cells increased Bmi-1 expression in response to TCR ligation, the KLRG1+ cells having lost this capability (Fig. 6a). This uncoupling of the TCR from Bmi-1 was associated with a decreased capacity for TCR- and IL-2-mediated DNA replication (Fig. 6b), despite the maintenance of TCR-induced IFN-γ production (Fig. 6c), indicating that the senescent phenotype correlates with a selective defect rather than a general defect of TCR signaling. That this defect is responsible for senescence of the KLRG1+ CD8+ T cell is supported by the elevated levels of p16Ink4a and p19Arf mRNA in these cells (Table 1), because these proteins are products of the Ink4a/Arf locus that is transcriptionally repressed by Bmi-1. The increase observed in the KLRG1− memory CD8+ T cells relative to naïve cells may reflect the presence of contaminating KLRG1+ cells. Therefore, whether the TCR is capable of augmenting the expression of Bmi-1 may be the molecular determinant of the replicative potential of the antigen-experienced CD8+ T cell.
Fig. 6.
Impaired TCR-mediated induction of Bmi-1 expression in senescent KLRG1+ CD8+ T cells. (a) Naïve CD44low KLRG1− CD8+ T cells and antigen-experienced, CD44high CD8+ T cells that were either KLRG1− or KLRG1+ were purified from the spleens of four mice 8–9 weeks after infection with γMHV-M3-OVA virus and were either immediately assessed for expression of Bmi-1 or stimulated with anti-CD3ε, IL-2, and IL-7 for 24 h. Representative histograms demonstrate staining with isotype control (black lines) or anti-Bmi-1 antibody (red lines) of purified cells of a single mouse. The bar graphs to the right represent the mean ± SE of specific mean fluorescent intensity for Bmi-1 staining of cells from all four mice. (b) Stimulation of cells with anti-CD3ε, IL-2, and IL-7 was continued for an additional 16 h, with BrdU being added during the last 4 h. The bar graphs represent the mean ± SE of the percentage of cells that had incorporated BrdU. (c) Additional replicate samples of cells received Brefeldin A at time 0, when stimulation was initiated, and were then assessed at 3 h for intracellular IFN-γ. The bar graphs represent the mean ± SE of the percentage of cells that had intracellular IFN-γ. *, KLRG-1+ cells are significantly different from naïve and CD44high KLRG-1− cells: P = 0.0036 (a) and P = 0.0015 (b) (paired Student's t test).
Table 1.
p16Ink4a and p19Arf mRNA in memory CD8+ T cells
Data are the fold increase relative to naïve cells and are the mean of three experiments. The percentages of KLRG1+ were calculated by paired Student's t test.
*Significantly different from KLRG1− cells (P = 0.031).
†Significantly different from KLRG1− cells (P = 0.006).
Discussion
The capacity of a CD8+ T cell clone to generate new effector cells during persistent viral infections (31–33) indicates that it must have a stage of antigen-dependent development that maintains replicative function and avoids senescence. Because Bmi-1 mediates this capability in the hematopoietic stem cell (20, 21), we evaluated its role in controlling the replicative potential of the CD8+ T cell in relation to three aspects of clonal expansion: its effect on cellular proliferation, the regulation of its expression by known signals for cellular expansion, and whether its expression is altered in cells with a senescent phenotype.
A lentiviral vector expressing an shRNA for suppressing Bmi-1 expression inhibited the proliferation of CTLL-2 cells responding to IL-2 (Fig. 3). The use of this cell line permitted the introduction of a vector expressing Bmi-1 with silent mutations in the shRNA target sequence. This mutant Bmi-1 rescued cells from growth inhibition, which excluded off-target effects of the shRNA (Fig. 3d). Having determined the specificity of the shRNA-expressing lentiviral vector, we were able to conclude that its inhibition of the proliferation of primary CD8+ T cells was caused specifically by the loss of expression of Bmi-1 (Fig. 3e). Corresponding gain-of-function experiments supported the conclusion that Bmi-1 positively regulates the proliferation of the CD8+ T cell by showing that ectopic Bmi-1 expressed at a level even less than that induced by ligation of the TCR and by maintenance with IL-2 (Figs. 1 and 2) enhanced the expansion of CD8+ T cells responding to IL-2 and IL-7 in vitro (Fig. 4b) and to homeostatic stimuli in vivo (Fig. 5). That a modest increase in Bmi-1 enhances proliferation suggests that small variations in its level caused by physiological signals could affect the proliferative ability of the CD8+ T cell.
Ligation of the TCR selects for clonal expansion of the CD8+ T cell by directly and indirectly mediating entry into cell cycle, the latter through promoting production of and responsiveness to IL-2 and responsiveness to CD70, the ligand for CD27. Therefore, the finding that ligating the TCR induces the expression of Bmi-1 protein (Fig. 1. a–c) and mRNA (Fig. 1d) in naïve and replication-competent, KLRG1−, antigen-experienced CD8+ T cells (Fig. 6a) directly links the regulation of Bmi-1 to clonal expansion. Interestingly, IL-2 maintained the TCR-induced increase in Bmi-1, but ligation of CD27 did not (data not shown), which is consistent with the differing requirements for repetitive TCR signaling in the IL-2 receptor and TCR/CD27 pathways of replication (34). The finding that this critical TCR-mediated function is lost in the KLRG1+, antigen-experienced CD8+ T cells may explain the raised levels of p16Ink4a and p19Arf mRNA and account for the senescent phenotype of these cells. Although falling short of formal proof because we have not yet demonstrated the reversal of senescence by ectopic expression of Bmi-1 in the KLRG1+ cell, these findings are consistent with the uncoupling of Bmi-1 from TCR signaling being the basis for replicative senescence of the CD8+ T cell. Bmi-1 may also suppress senescence by activating telomerase, which has been reported for human mammary epithelial cells but not fibroblasts (35). If Bmi-1 does prove to be the determinant of the replicative competence of the antigen-experienced CD8+ T cell, the original proposal of memory T cells needing to share certain characteristics with stem cells (36) will receive additional experimental support, as it has with the recent proposal of asymmetrical division of the antigen-stimulated CD8+ T cell (37).
We do not know the signals that lead to the senescent stage of CD8+ T cell development. However, there must be a subset of antigen-experienced cells that escapes these signals and maintains TCR-induced Bmi-1 expression. Because the senescent KLRG1+ CD8+ T cell is highly differentiated and effector differentiation is IL-2-dependent, it may be that the self-renewing subset of cells avoids replicative senescence by proliferating via an IL-2-independent pathway, such as that mediated by ligation of the TCR and CD27 (34).
Materials and Methods
Mice.
C57BL/6 mice were purchased from Charles River U.K. Ltd. (Margate, U.K.). OT-I RAG−/− mice and RAG2−/− mice were generous gifts from C. Reis e Sousa (Cancer Research UK London Research Institute, London, U.K.) and A. Betz (Laboratory of Molecular Biology, Cambridge, U.K.), respectively. Mice were housed under specific pathogen-free conditions at the University of Cambridge. For all animals, housing and procedures were carried out in accordance to the U.K. Home Office guidelines.
Cell Lines and Culture Conditions.
Culture conditions and treatment of the murine B cell lymphoma line, A20 (BALB/cAnN, H-2d), and the human embryonic kidney cell line, HEK293, have been described (34). The murine cytotoxic T cell line CTLL-2 was a generous gift from J. S. Hill Gaston (Department of Medicine Addenbrooke's Hospital, University of Cambridge, U.K.) and was cultured in complete RPMI medium 1640 supplemented with 125 international units IL-2. The murine fibroblast cell line NIH 3T3 was purchased from American Type Culture Collection (Manassas, VA) and cultured in complete DMEM (34).
Purification and Stimulation of Primary CD8+ T Cells.
Naïve CD8+ OT-I T cells were purified as described (34) or by negative selection with anti-biotin microbeads (Miltenyi Biotec, Auburn, CA) after binding of biotinylated antibodies against murine CD11c (N418), CD11b (M1/70), Gr-1 (RB6-8C5), MHC Class II (M5/114.15.2), CD49b (Dx-5) (eBioscience, San Diego, CA), and NK1.1 (PK136) (BD Biosciences, San Jose, CA). Cells were stimulated with anti-CD3ε (clone 145-2C11) (eBioscience), either plate-bound (0.5 μg/ml), in the presence of Fc receptor-expressing A20 cells (34), in the presence of primary B cells (1 μg/ml), or with SIINFEKL peptide (Peptides International, Louisville, KY) (34) in the presence of mouse IL-7 (2–5 ng/ml) and IL-2 (2–5 ng/ml) (R & D Systems, Minneapolis, MN). To some cultures anti-IL-2 (clone JS6–1A12), anti-CD25 (clone PC61), and anti-CD70 (clone FR70) (eBioscience) were added.
Viruses.
The retroviral vector pMig and the pCL-ECO packaging construct have been described previously (38). Full-length Bmi-1 cDNA was fused with a C-terminal HA-tag and inserted into the pMig retroviral vector. For shRNA rescue experiments, Bmi-1 coding sequence was mutated at positions 458, 461, 464, 467, 470, 473, and 476 using the Gene Tailor site-directed mutagenesis system (Invitrogen, Carlsbad, CA). In the original pMig construct, EGFP served as a reporter gene. In other cases, cDNA of rat Thy1 or a truncated human CD2 (39) was inserted into pMig as alternative reporter genes. Retroviruses were produced in HEK293 cells as previously described (38). The lentiviral plasmid pll.3.7 (40) was a generous gift from Christopher P. Dillon (Massachusetts Institute of Technology, Cambridge, MA). The sequences for siRNA against Bmi-1 were obtained from Ambion (Foster City, CA) and cloned as shRNA with a loop sequence from Brummelkamp et al. (41) into pll3.7. Lentiviral production and titration were performed as described (40). Primary CD8+ T cells were transduced 24 h after activation in the presence of IL-2, IL-7, and 6 μg/ml polybrene (hexadimethrine bromide; Sigma, St. Louis, MO), and IL-7 remained absent from the culture medium when CTLL-2 cells were transduced at any time.
γMHV Infection.
The γMHV-M3-OVA virus was a generous gift from Philip Stevenson (University of Cambridge). C57BL/6 mice were infected intranasally with 2 × 104 pfu of γMHV-M3-OVA. Splenocytes were depleted of B cells (Miltenyi Biotec), and CD8+ CD44low, CD44high KLRG1−, and CD44high KLRG1+ were sorted (MoFlo; Dako, Glostrup, Denmark) after staining with appropriate monoclonal antibodies.
Assays.
Intracellular staining of Bmi-1 was performed by using the Cytofix/Cytoperm kit (BD Biosciences) and anti-Bmi-1 monoclonal antibody (1.T.21) (Abcam, Cambridge, MA) followed by anti-mouse IgG1, (A85-1) or (X56) (BD Biosciences). Staining for intracellular IFN-γ (34) was performed after stimulation with anti-CD3ε in the presence of primary B cells for 3 h. For BrdU staining, the BrdU Flow kit (BD Biosciences) was used according to the manufacturer's instructions. The antibodies used were anti-CD8a (5H10; Caltag Laboratories, Carlsbad, CA, and 53-6.7; eBioscience), anti-CD3ε (145-2C11), anti-KLRG1 (2F1), anti-CD44 (IM7), anti-Thy1.1 (HIS51; eBioscience), anti-Gr-1 (RB6-8C5), and anti-CD2 (S5.2; BD Biosciences).
For quantitative RT-PCR analysis primers and TaqMan probes for detection of murine Bmi-1, CD3ε, and Mel-18 mRNA were purchased from Applied Biosystems (Foster City, CA). The sequences of the primers and probes for p16Ink4a and p19Arf used were as follows: p19Arf, 5′-gccgcaccggaatcct-3′ (sense); p19Arf, 5′-ttgagcagaagagctgctacgt-3′ (antisense); p19Arf, 5′-accaggtgatgatgatgggcaacgt-3′ (probe); p16Ink4a, 5′-cgtaccccgattcaggtgat-3′ (sense); p16Ink4a, 5′-gggtcctcgcagttcgaa-3′ (antisense); and p16Ink4a, 5′-cgttcacgtagcagctcttctgctcaact-3′ (probe).
The antibodies used for immunoblot analysis of Bmi-1 expression were mouse anti-Bmi-1 antibody [1.T.21] (Abcam), goat anti-mouse IgG1-HRP (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-β-actin (Abcam), goat anti-rabbit IgG-HRP (Santa Cruz Biotechnology), and mouse anti-MonoHA 1.1 (Covance, Princeton, NJ).
Acknowledgments
We thank Dr. Christopher M. Smith for infection of mice with γMHV, Prof. Philip G. Stevenson for γMHV-M3-OVA virus, and all members of the D.T.F. laboratory for helpful discussions. This work was supported by grants from the Wellcome Trust, National Institutes of Health, Medical Research Council, and Raymond and Beverly Sackler Studentship.
Abbreviations
- γMHV-68
mouse γ-herpesvirus strain 68
- KLRG1
killer cell lectin-like receptor G1
- TCR
T cell receptor
- shRNA
short hairpin RNA.
Footnotes
The authors declare no conflict of interest.
References
- 1.Wherry EJ, Teichgraber V, Becker TC, Masopust D, Kaech SM, Antia R, von Andrian UH, Ahmed R. Nat Immunol. 2003;4:225–234. doi: 10.1038/ni889. [DOI] [PubMed] [Google Scholar]
- 2.Roberts AD, Ely KH, Woodland DL. J Exp Med. 2005;202:123–133. doi: 10.1084/jem.20050137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bachmann MF, Wolint P, Schwarz K, Jager P, Oxenius A. J Immunol. 2005;175:4686–4696. doi: 10.4049/jimmunol.175.7.4686. [DOI] [PubMed] [Google Scholar]
- 4.Bouneaud C, Garcia Z, Kourilsky P, Pannetier C. J Exp Med. 2005;201:579–590. doi: 10.1084/jem.20040876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaech SM, Tan JT, Wherry EJ, Konieczny BT, Surh CD, Ahmed R. Nat Immunol. 2003;4:1191–1198. doi: 10.1038/ni1009. [DOI] [PubMed] [Google Scholar]
- 6.Stock AT, Jones CM, Heath WR, Carbone FR. J Immunol. 2006;177:1411–1415. doi: 10.4049/jimmunol.177.3.1411. [DOI] [PubMed] [Google Scholar]
- 7.Pahl-Seibert MF, Juelch M, Podlech J, Thomas D, Deegen P, Reddehase MJ, Holtappels R. J Virol. 2005;79:5400–5413. doi: 10.1128/JVI.79.9.5400-5413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ortega Soto E, Pecht I. J Immunol. 1988;141:4324–4332. [PubMed] [Google Scholar]
- 9.Voehringer D, Blaser C, Brawand P, Raulet DH, Hanke T, Pircher H. J Immunol. 2001;167:4838–4843. doi: 10.4049/jimmunol.167.9.4838. [DOI] [PubMed] [Google Scholar]
- 10.Voehringer D, Koschella M, Pircher H. Blood. 2002;100:3698–3702. doi: 10.1182/blood-2002-02-0657. [DOI] [PubMed] [Google Scholar]
- 11.Thimme R, Appay V, Koschella M, Panther E, Roth E, Hislop AD, Rickinson AB, Rowland-Jones SL, Blum HE, Pircher H. J Virol. 2005;79:12112–12116. doi: 10.1128/JVI.79.18.12112-12116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barber DL, Wherry EJ, Masopust D, Zhu B, Allison JP, Sharpe AH, Freeman GJ, Ahmed R. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 13.Maini MK, Soares MV, Zilch CF, Akbar AN, Beverley PC. J Immunol. 1999;162:4521–4526. [PubMed] [Google Scholar]
- 14.Hathcock KS, Kaech SM, Ahmed R, Hodes RJ. J Immunol. 2003;170:147–152. doi: 10.4049/jimmunol.170.1.147. [DOI] [PubMed] [Google Scholar]
- 15.Migliaccio M, Amacker M, Just T, Reichenbach P, Valmori D, Cerottini JC, Romero P, Nabholz M. J Immunol. 2000;165:4978–4984. doi: 10.4049/jimmunol.165.9.4978. [DOI] [PubMed] [Google Scholar]
- 16.Sherr CJ. Cell. 2004;116:235–246. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- 17.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 18.Haupt Y, Alexander WS, Barri G, Klinken SP, Adams JM. Cell. 1991;65:753–763. doi: 10.1016/0092-8674(91)90383-a. [DOI] [PubMed] [Google Scholar]
- 19.van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der Gulden H, Berns A. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- 20.Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 21.Lessard J, Sauvageau G. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 22.Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Lugt NM, Domen J, Linders K, van Roon M, Robanus-Maandag E, te Riele H, van der Valk M, Deschamps J, Sofroniew M, van Lohuizen M, et al. Genes Dev. 1994;8:757–769. doi: 10.1101/gad.8.7.757. [DOI] [PubMed] [Google Scholar]
- 24.Bruggeman SW, Valk-Lingbeek ME, van der Stoop PP, Jacobs JJ, Kieboom K, Tanger E, Hulsman D, Leung C, Arsenijevic Y, Marino S, van Lohuizen M. Genes Dev. 2005;19:1438–1443. doi: 10.1101/gad.1299305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwama A, Oguro H, Negishi M, Kato Y, Morita Y, Tsukui H, Ema H, Kamijo T, Katoh-Fukui Y, Koseki H, et al. Immunity. 2004;21:843–851. doi: 10.1016/j.immuni.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 26.Hasegawa M, Tetsu O, Kanno R, Inoue H, Ishihara H, Kamiyasu M, Taniguchi M, Kanno M. Mol Immunol. 1998;35:559–563. doi: 10.1016/s0161-5890(98)00048-0. [DOI] [PubMed] [Google Scholar]
- 27.Hogquist KA, Jameson SC, Heath WR, Howard JL, Bevan MJ, Carbone FR. Cell. 1994;76:17–27. doi: 10.1016/0092-8674(94)90169-4. [DOI] [PubMed] [Google Scholar]
- 28.Kaech SM, Ahmed R. Nat Immunol. 2001;2:415–422. doi: 10.1038/87720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Stipdonk MJ, Lemmens EE, Schoenberger SP. Nat Immunol. 2001;2:423–429. doi: 10.1038/87730. [DOI] [PubMed] [Google Scholar]
- 30.Wong P, Pamer EG. J Immunol. 2001;166:5864–5868. doi: 10.4049/jimmunol.166.10.5864. [DOI] [PubMed] [Google Scholar]
- 31.Khan N, Shariff N, Cobbold M, Bruton R, Ainsworth JA, Sinclair AJ, Nayak L, Moss PA. J Immunol. 2002;169:1984–1992. doi: 10.4049/jimmunol.169.4.1984. [DOI] [PubMed] [Google Scholar]
- 32.Weekes MP, Carmichael AJ, Wills MR, Mynard K, Sissons JG. J Immunol. 1999;162:7569–7577. [PubMed] [Google Scholar]
- 33.Cohen GB, Islam SA, Noble MS, Lau C, Brander C, Altfeld MA, Rosenberg ES, Schmitz JE, Cameron TO, Kalams SA. Virology. 2002;304:474–484. doi: 10.1006/viro.2002.1743. [DOI] [PubMed] [Google Scholar]
- 34.Carr JM, Carrasco MJ, Thaventhiran JE, Bambrough PJ, Kraman M, Edwards AD, Al-Shamkhani A, Fearon DT. Proc Natl Acad Sci USA. 2006;103:19454–19459. doi: 10.1073/pnas.0609706104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dimri GP, Martinez JL, Jacobs JJ, Keblusek P, Itahana K, Van Lohuizen M, Campisi J, Wazer DE, Band V. Cancer Res. 2002;62:4736–4745. [PubMed] [Google Scholar]
- 36.Fearon DT, Manders P, Wagner SD. Science. 2001;293:248–250. doi: 10.1126/science.1062589. [DOI] [PubMed] [Google Scholar]
- 37.Chang JT, Palanivel VR, Kinjyo I, Schambach F, Intlekofer AM, Banerjee A, Longworth SA, Vinup KE, Mrass P, Oliaro J, et al. Science. 2007;315:1687–1691. doi: 10.1126/science.1139393. [DOI] [PubMed] [Google Scholar]
- 38.Manders PM, Hunter PJ, Telaranta AI, Carr JM, Marshall JL, Carrasco M, Murakami Y, Palmowski MJ, Cerundolo V, Kaech SM, et al. Proc Natl Acad Sci USA. 2005;102:7418–7425. doi: 10.1073/pnas.0501585102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riviere I, Sunshine MJ, Littman DR. Immunity. 1998;9:217–228. doi: 10.1016/s1074-7613(00)80604-4. [DOI] [PubMed] [Google Scholar]
- 40.Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Ihrig MM, McManus MT, Gertler FB, et al. Nat Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- 41.Brummelkamp TR, Bernards R, Agami R. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]