Summary
Numerous cellular processes are regulated by (poly)ubiquitin-mediated signaling events, which involve a covalent modification of the substrate protein by a single ubiquitin or a chain of ubiquitin molecules linked via a specific lysine. Remarkably, the outcome of polyubiquitination is linkage-dependent. For example, Lys48-linked chains are the principal signal for proteasomal degradation, while Lys63-linked chains act as non-proteolytic signals. Despite significant progress in characterization of various cellular pathways involving ubiquitin, understanding of the structural details of polyubiquitin chain recognition by downstream cellular effectors is missing. Here we use NMR to study the interaction of a ubiquitin-interacting motif (UIM) of the proteasomal subunit S5a with di-ubiquitin, the simplest model for polyubiquitin chain, in order to gain insights into the mechanism of polyubiquitin recognition by the proteasome. We have mapped the binding interface and characterized the stoichiometry and the process of UIM binding to Lys48- and Lys63-linked di-ubiquitin chains. Our data provide first direct evidence that UIM binding involves a conformational transition in Lys48-linked di-ubiquitin which opens the hydrophobic interdomain interface. This allows UIM to enter the interface and bind directly to the same ubiquitin hydrophobic-patch surface as utilized in UIM:monoubiquitin complexes. The results indicate that up to two UIM molecules can bind di-ubiquitin, and the binding interface between UIM and ubiquitin units in di-ubiquitin is essentially the same for both Lys48- and Lys63-linked chains. Our data suggest possible structural models for the binding of UIM and of full-length S5a to di-ubiquitin.
Keywords: ubiquitin, polyubiquitin, ubiquitin interacting motif, S5a, chemical shift perturbation mapping, ligand binding
Introduction
The selective degradation of cellular proteins is a precisely regulated process required for various cellular functions. Breakdown of many short-lived enzymes and various regulatory and structural proteins is achieved in eukaryotes via the ubiquitin-proteasomal pathway, in which the attachment of a polyubiquitin (polyUb) chain targets proteins to their degradation by the proteasome 1.
Ubiquitin (Ub) is a highly conserved, 76-residue globular protein with a flexible unstructured C-terminal four-residue segment. In a process of post-translational modification called ubiquitination (e.g. 2) the carboxyl group of the C-terminal glycine of Ub forms an isopeptide bond with the ε-amino of a lysine on a target protein. This modification can result in the attachment of a single Ub or a polyUb chain, where the consecutive Ubs are also linked via isopeptide bonds. Remarkably, the number of ubiquitin moieties in the chain and specific lysine through which they are linked determine the outcome of ubiquitination 3. Thus, while polyUb chains linked via K48 act as a universal recognition signal that targets proteins for proteasomal degradation 4, K63-linked chains act primarily as regulatory rather than proteolytic signals 3, and monoubiquitination is involved in signaling a variety of trafficking events 5. Knowledge of how the polyUb signal is recognized and processed by the proteasome is required for our understanding of the nature of specificity and the mechanisms of regulation in the ubiquitin-proteasomal system.
S5a (Rpn10 in yeast) is an integral subunit of the 19S regulatory complex of the 26S proteasome and is believed to act as one of the polyUb tag receptors, thereby allowing polyubiquitinated proteins to be shuttled to the proteasome for degradation 6; 7. S5a has two ubiquitin-interacting motifs (UIM) which adopt α-helical structures 8 and contain a short stretch of residues of alternating long and short hydrophobic side chains (LALAL motif in UIM-1 and IAYAM in UIM-2) serving as the primary interacting surface with the hydrophobic patch on Ub 6; 8; 9; 10. Several groups have reported binding of UIM-containing peptides to monomeric Ub (monoUb) with relatively low affinity (Kd∼300 μM) 8; 10; 11; 12; 13, although stronger (73 μM) binding was observed for UIM-2 of S5a 14. Interestingly, UIM-2 domains of S5a and Vps27 bind ubiquitin with higher affinity than UIM-1 (5- and 2-fold stronger binding, respectively, in the case of S5a 8 and Vps27 15). A similar difference between the UIMs of S5a was observed in polyUb binding, wherein isolated UIM-2 showed a 10-fold greater affinity 6. The solution structure of a S5a fragment, S5a196–306, encompassing both UIMs, in complex with monoUb 8, shows that each UIM interacts independently with the corresponding Ub molecule. It has been suggested 8 that the hydrophobic contacts between the so-called N-terminal turn in UIM-2 (residues 276–282) and L8, V70, L71 of Ub confer the stronger binding affinity compared to UIM-1.
The fact that in the S5a196–306:monoUb complex UIM-1 and UIM-2 are each bound to a separate Ub molecule suggests that the full length S5a would optimally bind multiple (at least two) Ub monomers in polyUb. In fact, S5a shows strong binding preference toward longer (n ≥ 4) polyUb chains 6; 8; 9. Intriguingly, however, the individual UIMs of S5a also exhibit much higher binding affinity for K48-linked polyUb than for monoUb 6. This suggests that the S5a's binding preference toward polyUb cannot be attributed solely to the presence of two UIMs (note in this regard that Rpn10, the yeast homologue of S5a, contains a single UIM, homologous to UIM-1). This also raises the possibility that polyUb chains might bind UIMs in a different mode compared to monoUb. Detailed understanding of the reasons for selective recognition of polyUb chains by S5a requires knowledge of the structural details of the interaction between polyUb and UIMs, which is currently missing.
Because S5a acts as a proteasomal polyUb-tag receptor, characterization of the interaction of the S5a UIMs with K48-linked polyUb chains could provide clues to the mechanisms of polyUb tag recognition by the proteasome. Specific questions of interest concern comparison of S5a's interaction with monoUb versus polyUb and comparison of the mechanisms of S5a binding to K48-linked and K63-linked polyUb. Intriguingly, the latter chains act primarily as regulatory signals, although they also bind S5a 8; 16 and can signal proteolysis 17. K63- and K48-linked chains adopt distinct conformations under physiological conditions 3; 18. While the former is in extended conformation 18, K48-linked di- and tetra-ubiquitins are predominantly in a “closed” conformation 19; 20; 21, with the hydrophobic patch residues L8, I44, and V70 being sequestered at the Ub/Ub interface. This interface, however, is not rigidly locked, but rather dynamic, such that the functionally important residues (L8, I44, V70) can become exposed and available for interactions with downstream recognition factors as a result of interface opening. Moreover, the linkage via K48 allows di-ubiquitin (Ub2) to form an extended hydrophobic pocket comprising the hydrophobic patches of both Ub units. Reflecting these conformational differences, K63- and K48-linked Ub2 chains bind UBA2 domain of hHR23A in distinct modes: as two independent Ub-like ligand-binding sites 18 and in a sandwich-like fashion 22, respectively. In the latter complex, the UBA2 domain is inserted into the Ub/Ub interface and interacts directly with the hydrophobic patches on both ubiquitins. It is therefore of significant interest to understand if these linkage-dependent mechanisms of binding hold for other ligands, in particular for the UIMs of S5a. Given that the UIMs bind monoUb at its hydrophobic patch, a critical question here is, whether in the case of K48-linked di-ubiquitin and longer chains the UIMs can compete with the Ub:Ub interaction and bind to the hydrophobic patches (buried at the Ub/Ub interface) directly or do they bind at some other sites, perhaps on the outer surface of the closed conformation of the chain.
Here we use NMR to study the interaction of the UIM-2 domain of S5a with di-ubiquitin as the simplest model for polyubiquitin chain, in order to map the binding interface and gain insights into the mechanism of complex formation. We also examine UIM-2 binding to monomeric Ub and to K63-linked Ub2, in order to understand the effect of the chain length and linkage on this interaction.
Results
Mapping the interaction sites between S5a263–307 and monoubiquitin
Binding of the isolated UIM-2 containing peptide, S5a263–307, to monoUb was investigated using the chemical shift perturbation (CSP) mapping approach (e.g. 23). Differences in the resonance frequencies of protein signals detected in the absence and in the presence of a ligand, under identical experimental conditions, indicate a change in the microenvironment of the observed nuclei as a direct or indirect result of interaction with the ligand. The CSPs observed in 15N-labeled monoUb upon addition of S5a263–307 (Fig.1a,e) cluster around residues L8, I44, and V70, consistent with S5a263–307 binding to the hydrophobic patch on Ub surface.
Figure 1.
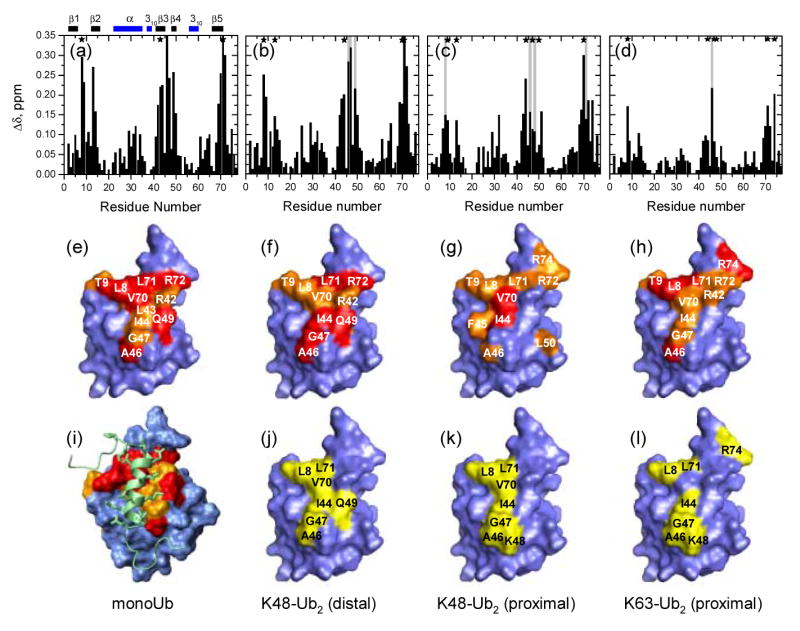
NMR mapping of the S5a263–307-binding surface on monoUb and Ub units in the Ub2 constructs studied here. The top row shows CSPs at saturation as a function of the residue number for the (a) monoUb, (b) distal and (c) proximal Ubs in K48-linked Ub2 and (d) for the proximal Ub in K63-linked Ub2. Panels (e–h) map the sites with significant CSPs on the surface of ubiquitin. The coloring of the CSPs is as follows: (e) Δδ ≥ 0.25 ppm (red), 0.25 > Δδ ≥ 0.15 (orange) (f) Δδ > 0.2 ppm (red), 0.2 ppm > Δδ > 0.15 ppm (orange), (g) Δδ > 0.2 ppm (red), 0.2 ppm > Δδ > 0.125 ppm (orange), (h) Δδ > 0.15 ppm (red), 0.15 ppm > Δδ > 0.075 ppm (orange). Panels (j–l) map the residues that show signal attenuation > 60% (colored yellow) in the ubiquitin units of the corresponding Ub2 constructs. The structure of the monoUb:UIM-2 complex is shown in (i) (PDB code 1YX6 8). Asterisks in (a-d) mark sites showing intermediate exchange (attenuation > 60%) and the vertical grey bars mark sites exhibiting slow exchange. Horizontal bars on the top of panel (a) indicate the elements of secondary structure in ubiquitin, the β-strands are colored red, helices are blue. Most of the molecular images throughout this paper were made in PyMol 45. Panel (i) in this figure as well as Fig.4c were prepared in MolMol 46.
In S5a263–307, the addition of monoUb caused a considerable change in chemical shifts of more than 10 residues, most of them located in the helical region and the remaining few in the region flanking the α-helix (Fig.2a, also Supplementary Material). The magnitude of the perturbations increased with ubiquitin concentration and saturated at [Ub] / [S5a263–307] > 1. The largest CSPs were observed in residues M291, M293, L295, as well as I287 and Q296, indicating that monoUb binding involves the hydrophobic IAYAM motif (287–291) and the C-terminus of the UIM-2's helix. Significant chemical shift changes were also observed in S279-T282 located in the N-terminal turn which has been implicated in the helix stabilization and binding interactions with both monoUb 8 and the UBL domain of hHR23A 12; 24. Interestingly, chemical shifts in the bound state were nearly identical to those observed in UIM-2 complexed with UBL of hHR23A 24, indicating that UIM-2's interactions with UBL and monoUb are, indeed, similar.
Figure 2.

NMR mapping of S5a263–307 residues affected by binding to monomeric Ub and K48-linked Ub2. Panels (a) and (b) depict the CSPs in S5a263–307 at saturation with monoUb and Ub2, respectively, as a function of the residue number. Sites showing significant signal attenuation (> 60%) due to intermediate or slow exchange are marked by the asterisks. Panels (c) and (d) map these CSPs on the surface (c) and the cartoon representation (d) of UIM-2; the coloring is as follows: Δδ > 0.4 ppm (red), 0.4 ppm > Δδ > 0.2 ppm (orange), 0.2 ppm > Δδ > 0.125 ppm (yellow). Horizontal bars on the top of panel (a) indicate the elements of secondary structure in S5a263–307, the helices are blue, the N-terminal turn in S5a263–307 (labeled with “t”) is colored cyan.
The stoichiometry of the S5a263–307:monoUb complex was determined by 1H transverse relaxation measurements. Relaxation time T2 is inversely proportional to the overall tumbling time of a protein, which, in turn, is proportional to protein's molecular weight. Using the 1H T2 values for monoUb (50 ms, molecular weight 8.8 kDa) and Ub2 (26 ms, 17.4 kDa) 18 as “molecular weight markers”, the T2 of 28 ms measured for the S5a263–307:monoUb complex corresponds to ∼16.1 kDa, which indicates a 1:1 stoichiometry (16.5 kDa expected).
The CSP mapping and relaxation data both indicate that monoUb binds a single S5a263–307 molecule and the interaction involves Ub's hydrophobic patch (Fig.1). The same conserved hydrophobic patch on ubiquitin also serves as the binding surface for several Ub-binding domains 25, including UBA 14; 18; 22; 26; 27 and CUE 28; 29; 30 domains, as well as the other Ub moiety in a K48-linked polyUb chain 19. Our surface mapping agrees well with the solution structure of monoUb:UIM-2 complex 8 (Fig. 1i). Note that the signal of K48 was only slightly shifted (Δδ = 0.06 ppm) upon binding, consistent with the observation 8 that this residue is not directly involved in Ub's interaction with the UIMs of S5a.
Mapping the interaction surface between S5a263–307 and Ub2
1. Binding surfaces on Ub2
Using segmentally 15N-labeled K48-linked Ub2 chains synthesized as described in Materials and Methods, we monitored separately perturbations in the distal and proximal units of Ub2 induced by S5a263–307 binding. As evident from Fig.1, the perturbations (CSPs and signal attenuations) observed in both Ubs are clustered around the hydrophobic patch residues L8, I44, and V70, and the perturbed sites are generally consistent with those in monoUb. Moreover, the magnitudes and the directions of the shifts in the signals are different from those associated with the opening of the hydrophobic interface in Ub2 (Supplementary Material). These are important observations, because they indicate, for the first time, that UIM-2 enters the Ub/Ub interface and binds directly to the hydrophobic patch on either Ub domain in K48-linked Ub2.
2. Binding surface on S5a263–307
The CSPs in most residues in S5a263–307 in the presence of K48-linked Ub2 were almost identical to those observed in our titration study with monoUb (Fig.2), both in the magnitude and the direction of the shifts (Supplementary Material). The magnitude of the perturbations increased with Ub2 concentration and saturated at [Ub2] / [S5a263–307] > 1. Noticeably weaker CSPs (accompanied by significant signal attenuation, 75% and 50%, respectively) were observed in I287 and Q296 located at the binding interface between UIM-2 and monoUb, while residues C-terminal to Q296 as well as at the N-terminus of the S5a263–307 construct (T264, S266, Q268) show small but significant resonance shifts in the presence of Ub2, indicative of their possible involvement in the binding. Interestingly, most amides at the N- terminus of S5a263–307 (residues 264–274) show signal attenuation comparable to that in the α-helix and other Ub-binding sites in this construct (Supplementary Material), whereas the C- terminal residues (300–307) show a relative increase in intensity. The latter is consistent with the high flexibility of the C-terminus, which makes these residues practically insensitive to the slower tumbling of the Ub2-bound part of the construct. This is in contrast with monoUb binding where the N- and C-termini both behave as flexible in the UIM-2:monoUb complex, and only the middle part of S5a263–307 (278–295) shows a relative overall decrease in signal intensity reflecting the increased overall tumbling time of the complex (Supplementary Material). All these observations suggest a possible involvement of the N-terminal residues of S5a263–307 in binding to Ub2 (see also below).
The fact that the CSPs are seen on the same “face” of UIM as in the titration with monoUb, indicates that there is no secondary binding site on S5a263–307 for the other ubiquitin moiety in Ub2. Combined with the mapping data for Ub domains (Fig.1), these results suggest that at least two UIM-2 molecules are bound to Ub2.
Ubiquitin units in di-ubiquitin chains can bind two UIM-2 moieties independently
To verify the above prediction, the stoichiometry of the S5a263–307:Ub2 complex was determined using 1H transverse (T2) and 15N longitudinal (T1) relaxation time measurements. A comparison of the measured 1H T2s with those for Ub2 (26 ms, 17.4 kDa) and Ub4 (13 ms, 34.4 kDa) 18 was used to estimate the molecular weight of the S5a263–307:Ub2 complex. The 1H T2 of 13.6 ms observed independently for two K48-linked Ub2 samples (Ub2-D and Ub2-P), as well as for K63-linked Ub2 (Ub2-P), in the presence of saturating amounts of S5a263–307, corresponds to 32.9 kDa, in good agreement with 32.8 kDa expected for a 2:1 S5a263–307: Ub2 complex. In order to further characterize the binding event, a series of 15N T1 relaxation measurements were performed with increasing S5a263–307: Ub2 molar ratio for K48-linked Ub2 (Table 1). At [S5a263–307]/[Ub2] = 0.5, the 15N T1 averaged over residues belonging to Ub core was 796 ± 25 ms (mean ± std), corresponding to a molecular weight of ∼22 kDa, smaller than 25.1 kDa expected for a 1:1 S5a263–307:Ub2 complex (see the calibration curve in Fig.4d). At a 1:1 molar ratio, the average 15N T1 was 909 ± 73 ms, which corresponds to a molecular weight range of 22–27 kDa, consistent with the molecular weight expected for a 1:1 complex. At [S5a263–307]/[Ub2] =2, the 15N T1 was 1022 ± 130 ms, corresponding to a molecular weight range of 24–32 kDa, comparable with 32.8 kDa expected for a 2:1 complex. The results of all these independent measurements thus indicate that a single K48-linked Ub2 chain can bind up to two UIMs.
Table 1. Using 15N relaxation time T1 to verify the stoichiometry of the S5a263–307:Ub2 complex at various titration points.
| [S5a]/[Ub2] | 15N T1 (ms)a | Molecular weight range (kDa) corresponding to the 15N T1 valueb | Molecular weight (kDa) expected for this stoichiometry |
|---|---|---|---|
| 0.5 | 796 ± 25 | 20–22 | < 25.1 |
| 1 | 909 ± 73 | 22–27 | 25.1 |
| 2 | 1022 ± 130 | 24–32 | 32.8 |
15N longitudinal relaxation time (mean ± standard deviation over the core residues) in K48-linked Ub2 (Ub2-P) in the presence of the specified molar amounts of S5a263–307.
The molecular weight was estimated based on the measured 15N T1 value using the calibration curve in Fig.4d.
Figure 4.
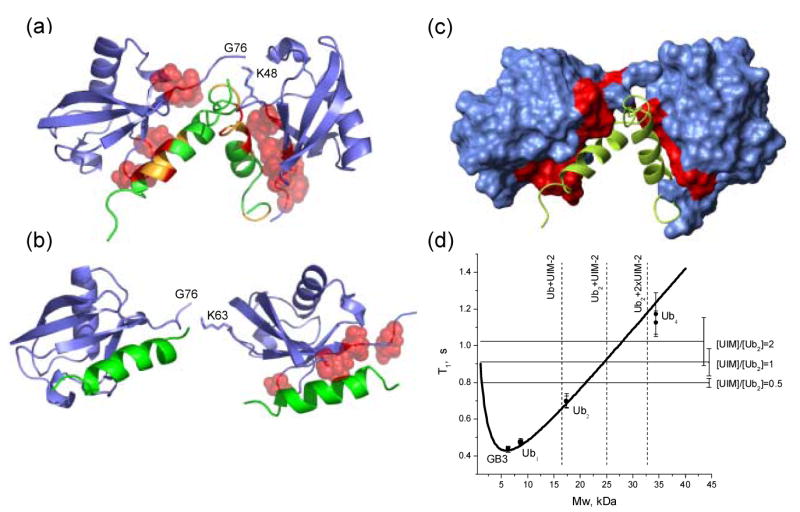
Models of two UIM-2 helices bound to (a) K48-linked Ub2 and (b) K63-linked Ub2. Side chains of Ub2 residues with Δδ > 0.2 ppm are represented by red spheres, while CSPs on UIM-2 (in panel (a)) are colored as follows: Δδ ≥ 0.4 ppm (red), 0.4 ppm > Δδ > 0.125 ppm (orange). These structures were obtained assuming that each UIM-2 binds to the corresponding Ub unit in the chain in the same way/orientation as to monoUb (PDB code 1YX6 8), and using the structures of K48-linked Ub2 in its complex with UBA2 (PDB: 1ZO6 22) and of free K63-linked Ub218 as structural models for the open conformation of the corresponding chain. The resulting structures were obtained by direct superimposition of ubiquitin atoms in the UIM:Ub complex with those for each Ub unit in Ub2. (c) The same complex as in panel (a), with Ub2 shown in surface representation, painted red for those sites on both Ub units that show significant CSPs and/or signal attenuations (see Fig.1). (d) NMR characterization of the stoichiometry of Ub2:S5a263–307 binding using 15N longitudinal relaxation rate measurements. The “molecular weight calibration” curve representing the molecular mass dependence of 15N T1 in proteins was calibrated as described in 22. The horizontal lines correspond to 15N T1 values measured in Ub2-P at various titration steps, as indicated (see also Table 1); the error bars represent standard deviations in T1 over core residues in Ub. The dashed vertical lines are “molecular weight markers” indicating the expected mass of the corresponding constructs.
S5a263–307 shows higher affinity for K48-linked Ub2 compared to monoUb
In addition to the strong similarity in the magnitude of the CSPs in UIM-2 bound to monoUb and Ub2, slow exchange phenomenon was observed in S5a263–307 both in the presence of monoUb and Ub2. Thus, residues M291, M293, L295, and Q296 located at the C-terminus of the helix, as well as I287 (of the IAYAM motif) clearly showed a slow-exchange behavior when titrated with monoUb. A similar picture was observed upon addition of K48-linked Ub2, where signals from the same residues, M291, M293, L295, I287, as well as M281 (N-terminal turn) showed strong signal attenuation from the beginning of titration and attenuated significantly (>80%) already in the presence of 0.2 molar equivalent of Ub2; these signals reappeared at a different position in the spectrum at the end of the titration (Supplementary Material).
There are, however, differences in the behavior of S5a263–307 signals upon binding to monoUb and Ub2. Thus, S280, M281, Q292, and S294 exhibit a typical fast exchange behavior upon addition of monoUb, but are in slow (S280, M281) or intermediate (Q292, S294) exchange in the presence of Ub2. Intriguingly, significant signal attenuation (>90%) was detected in the N-terminal residues T265, I266, and E270 in S5a263–307 upon binding to Ub2, while no such attenuation was observed in its complex with monoUb. Together with the abovementioned CSPs in this part of the construct, these data indicate that the N-terminus of S5a263–307 might be involved in interaction with Ub2. The greater number of sites showing slow or intermediate exchange (reflecting slower off-rates, hence higher binding affinity) suggests stronger S5a263–307 binding to Ub2 than to monoUb. In the UIM-2:monoUb complex, the C-terminus of the UIM helix containing M291 and M293 contacts I44, G47, and V70 of Ub while L295 is in contact with A46 and G47 8. Residues S280 and M281 are located in the N-terminal turn that caps the hydrophobic Ub surface formed by L8, V70, L71. Interestingly, all these Ub sites in both domains in Ub2 also show slow or intermediate exchange behavior (Supplementary Material).
A gel mobility assay of S5a263–307 binding to monoUb and Ub2 provides a qualitative glimpse into preferential binding of S5a263–307 to Ub2. As shown in Figure 3a, in a mixture of Ub2 and S5a263–307 (the rightmost lane on the gel), a significant portion of S5a263–307 migrates with Ub2 as evident from the decrease in intensity of the S5a263–307 band in the presence of Ub2.
Figure 3.
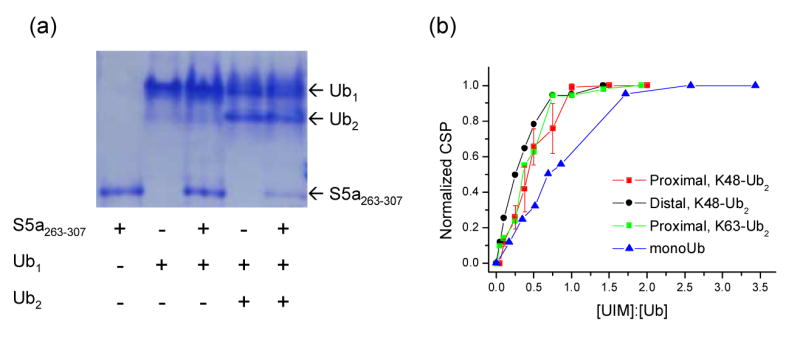
Comparison of S5a263–307 binding to monoUb and Ub2. (a) Gel mobility assay comparing binding of S5a263–307 to monoUb (lane 3) and Ub2 (lane 5). A non-reducing, non-denaturing polyacrylamide gel was loaded with equimolar amount of the proteins (2 nmoles each). (b) NMR titration curves presenting normalized chemical shift perturbations (averaged over Ub residues H68, L69, V70) for monoUb, K48- and K63-linked Ub2 (as indicated) plotted as a function of the S5a263–307:Ub molar ratio, i.e. the number of S5a263–307 molecules per Ub unit. The normalization was achieved by dividing all CSPs for a given residue by its CSP at the endpoint of the titration. The error bars in (b) represent the standard deviation (among selected residues) for each averaged CSP value in the proximal Ub. The molar ratios shown on the abscissa axis in (b) were normalized according to the number of available binding sites. Accurate fitting of these titration curves to simple models (e.g. two independent binding sites model) was not possible, likely due to the complex nature of S5a263–307 binding to Ub2. However, from the fact that the Ub2 CSPs are close to saturation (>90%) at [S5a263–307]/[Ub] ∼ 1, when the molar concentrations of Ub2 and S5a263–307, respectively, are 0.45 and 0.9 mM (proximal Ub titration), 0.44 and 0.88 mM (distal Ub), and 0.37 and 0.74 mM (proximal Ub in K63-Ub2), we estimate that the microscopic Kd is of the order of or less than 10 μM, assuming that Ub2 contains two independent and equivalent binding sites for S5a263–307. For comparison, the level of saturation (∼60%) observed in monoUb at a similar point in the titration ([S5a263–307]/[Ub] =0.86, the molar concentrations 0.395 and 0.339 mM) corresponds to Kd ∼70 μM, consistent with the literature data 14.
In the UIM-2:monoUb structure, the UIM helix is positioned parallel to and in direct contact with the β5 strand of Ub. Therefore we used titration data for residues H68, L69, V70 in this strand to monitor and compare UIM binding to Ub units in all the constructs considered here. Figure 3b illustrates an increase and subsequent saturation of the CSPs in all Ub units upon addition of S5a263–307, which clearly indicates the complex formation. When plotted against the number of S5a263–307 molecules per Ub unit, [S5a263–307]/[Ub], the CSPs in all Ub2 constructs studied here saturate at [S5a263–307]/[Ub] ∼1 (Fig.3b). This is consistent with the binding stoichiometry of one S5a263–307 per Ub unit. The fact that the CSPs in monoUb saturate at a higher [S5a263–307]/[Ub] ratio (Fig.3b) suggests a higher apparent affinity of S5a263–307 for individual Ub units in Ub2 compared to monoUb. This is likely due to the effect of local Ub concentration: one could anticipate that, when S5a263–307 dissociates from a Ub unit, it has a higher probability to bind to another Ub in the same molecule rather than to a Ub in a different chain. Even stronger binding could then be expected in the case of tetraUb and longer chains. This is in agreement with the well known binding preference of S5a for longer chains 8; 31, where the literature data suggest possible binding cooperativity in addition to the increase in the number of Ubs due to chain elongation.
Does UIM-2 differentiate between the distal and the proximal domains in K48-linked Ub2?
Despite the overall similarity between the two ubiquitin units in terms of the binding surface (mapped by the CSPs and signal attenuations), the CSPs observed in the distal domain are overall larger than in the proximal (Fig.1). Larger CSPs at saturation could indicate stronger binding, although the precise relationship between the side-chain conformations and the backbone NH chemical shifts in proteins is not fully understood. On the other hand, as evident from Fig.3b, the averaged normalized titration curves for the two domains are not very different from each other. Although the CSPs in the proximal Ub show a slower increase (compared to the distal Ub) upon addition of S5a263–307 (Fig.3b), which could indicate a somewhat weaker UIM-2 binding, accurate quantification of this observation in terms of the difference in the Kd values turned out problematic due to the site-so-site variations in the titration curves likely reflecting the complexity of the binding and the conformational changes in Ub2 occurring upon addition of the ligand.
A detailed analysis revealed further differences between UIM-2 binding to the proximal and distal Ubs in K48-linked Ub2. Thus, the CSP pattern in the distal Ub is very similar to that in monoUb. This is not surprising, given that the only differences between the distal Ub and the D77 Ub mutant used in our monoUb binding studies include a K48C mutation in the former and modifications of the C-terminus (not involved in UIM-2 binding): the G76-K48(proximal) isopeptide bond in the former and the D77 extension in the latter. The CSP pattern in the proximal Ub is somewhat different. Here the largest CSPs were observed in I44, L69, V70, located in the hydrophobic patch, as well as in R72, R74 at the flexible C-terminus. The differences between the distal and the proximal Ub in the β3-β4 region (residues 45–51), evident from Fig.1, suggest that the involvement of K48 of the proximal Ub in the isopeptide linkage is the likely reason for the observed differences in the S5a263–307-induced perturbations in these domains. Although K48 in monoUb does not directly interact with UIM-2 (see above), this modification could sterically affect UIM's access to and local interaction with the neighboring surface residues in the proximal Ub. Note in this regard that in the proximal Ub, the K48 signal itself shows a stronger CSP (Δδ = 0.11 ppm) than in the distal Ub and a slow-exchange behavior.
It should be mentioned, however, that not all sites in the proximal Ub show a somewhat weaker S5a263–307 binding compared to their counterparts in the distal Ub. Thus, L8 and L71, located at the opening of the Ub/Ub interface, show slow exchange in the proximal but not in the distal Ub, indicative of stronger UIM binding to these particular sites. In addition, V70 in the proximal Ub has a stronger CSP than in both monoUb and the distal Ub (Fig.1, also Supplementary Material). These observations emphasize the complexity of the UIM interaction with Ub2, as well as the ability of NMR titration experiments to provide a detailed site-specific picture of a binding event.
S5a263–307 binding to K63-linked Ub2
In order to compare binding of UIM-2 to polyUb chains of different linkages, we examined S5a263–307 binding to K63-linked Ub2. One of the goals was to test whether the sequestration of the hydrophobic patches in the closed conformation of K48-linked Ub2 has any hindering effect on UIM's binding to Ub2. Because the closed Ub/Ub interface is not formed in K63-linked Ub2 (hence no hindering expected), this chain serves as a control for such study. Another goal was to examine if the involvement of K48 in the isopeptide linker has any effect on the UIM binding properties of the proximal Ub in K48-linked Ub2. In the K63-linked Ub2, K48 is not modified, and K63 is located far away from the UIM-binding sites, hence one could expect that the proximal domain binds UIM similarly to the distal Ub in K63- or K48-linked chain. We also wanted to test whether the extended hydrophobic binding pocket in Ub222, which is formed due to the G76-K48 linkage and allows Ub2 to bind UBA domains in a sandwich-like fashion 22; 32, plays any role in the S5a263–307 binding. If this was true, one could expect that the absence of such a pocket in K63-linked Ub2 would result in weaker S5a263–307 binding to this chain.
The CSPs in the proximal Ub in K63-linked Ub2 clearly demonstrate that S5a263–307 binding occurs at the same hydrophobic Ub surface as in monoUb and K48-linked Ub2. The NMR titration curve is also similar to that for the K48-linked Ub2 (Fig.3b); the magnitude of the perturbations increases with S5a263–307 concentration and saturates at [S5a263–307]/[Ub2] ∼ 2 (or [S5a263–307]/[Ub] ∼ 1). The CSP pattern (Fig.1d) and the directions of the signal shifts (Supplementary Material) here are similar to those in monoUb and in the distal Ub of K48-linked Ub2, suggesting a similar mode of the UIM:Ub interaction. Similar sites in K63-linked Ub2 show signal attenuations upon addition of S5a263–307; the gradual shifts of the corresponding signals accompanied by their broadening (Supplementary Material) indicate intermediate exchange, also observed in the K48-linked Ub2. Also the averaged normalized titration curve for the β5 strand (Fig.3b) is strikingly similar to that for the proximal as well as for the distal Ub in K48-linked Ub2. All these data point to the conclusion that, despite the differences in local perturbations, the involvement of K48 in the isopeptide linkage does not significantly affect the affinity of UIM-2 binding to the proximal Ub. On the other hand, however, the overall magnitude of the CSPs is markedly smaller even compared to the proximal Ub of K48-linked Ub2. The reasons for this are not yet clear and could be related to the linkage-dependence of Ub2's conformation.
Discussion
General comments on ligand binding to polyubiquitin
Ligand binding to polyUb could be complicated by the presence of the intramolecular interaction between Ub monomers in the chain 19. Thus the predominant (85% populated) conformation of K48-linked Ub2 in solution at neutral pH is characterized by a close contact between the two ubiquitins 19; 33, which bind to each other via their hydrophobic patches. In this intramolecular complex, the functionally important residues L8-I44-V70 are buried at the Ub/Ub interface. The same contacts are present in K48-linked tetraUb 19; 20; 21. The opening of the Ub/Ub interface is essential for polyUb's binding to UIM or any other ligand targeting the hydrophobic patch on Ub. Indeed, locking the chain in the closed conformation would greatly reduce ligand's access to the ligand-binding sites on ubiquitins, and could render polyUb binding-incompetent 34. One could then expect that the predominantly closed conformation of Ub2 and the necessity for a ligand (here UIM-2) to compete with the Ub:Ub interaction might control its binding to the chain. Thus, ligand binding to polyUb would depend on several factors, including (i) the strength of the ligand:Ub versus Ub:Ub interaction, (ii) the entropic contributions including the hydrophobic effect and the cost of ligand's immobilization (see e.g. 35), and (iii) the rate of interface opening, which depends on the height of the corresponding activation barrier between the closed and open states of Ub2.
A Model for UIM-2 Binding to K48-linked Ub2
1. A mechanistic perspective
A plausible binding mechanism emerging from the behavior observed in this study involves opening of the interdomain interface in Ub2, thus providing a window of opportunity for a ligand to access the hydrophobic patches on Ub units. Because the open conformation is only weakly populated (∼15%) at neutral pH, the rate of interconversion between the closed and open states of Ub2 could be the limiting factor controlling this binding event. The finding that the Ub domains in K48-linked Ub2 bind S5a263–307 with at least comparable strength to that of a Ub unit in the K63-linked Ub2 suggests that the opening/closing dynamics in the former are sufficiently fast, such that the UIM's access to the hydrophobic patches on Ub units in this chain is not significantly hindered. This is in agreement with the NMR-based estimate that the interconversion occurs on a ∼10 ns time scale 33, i.e. is fast compared to UIM's on-rate, assuming that the latter is controlled by molecular diffusion.
The question however remains, if UIM-2 plays merely a passive role “waiting” for Ub2 to open, or can UIM:Ub2 interaction induce the interface opening? Answering this question is important for the understanding of the mechanism of ligand binding to polyUb.
In the closed conformation of K48-linked Ub2 (PDB codes 2AAR, 2BFG) the side chains of L8, T9, and L71 on either Ub unit are oriented such that they form an almost continuous hydrophobic strip along the outer edge of the Ub/Ub interface, extended by A46 of the other Ub across the interface (Supplementary Material). These residues could serve as the ligand-recognition region on the surface of Ub2, i.e. be the first hydrophobic sites to get in contact with UIM-2. Because these sites are located at the outer edge of the Ub/Ub interface, their interaction with the ligand could promote further opening of the interface, which could subsequently lead to full access to the hydrophobic patch residues on both Ub units. In support of the role of the abovementioned residues, consider their perturbations in the presence of a 0.5 molar equivalent of S5a263–307 (i.e. [S5a263–307]/[Ub2] = 0.5). At this point in the titration, in the proximal Ub residue L8 shows the largest CSP and its signal is attenuated by >70%, while A46 is even more attenuated (∼90%). In the distal Ub, L8 and A46 have the largest CSPs, and the most attenuated residues are A46, G47, Q49 (>80%) and L8 (∼75%).
Reflecting the two-fold symmetry of the closed conformation of K48-linked Ub2, L8, T9, V70, L71 of the proximal Ub are in close contact with A46, G47, Q49 of the distal, and vice versa. Most of these residues show intermediate or slow exchange in the presence of S5a263–307 (Fig.1), thus indicating strong interaction of these sites with the ligand. Interestingly, however, the slow exchange observed in L8, L71 of the proximal and G47 of the distal Ub is in contrast with the intermediate exchange in their counterparts on the other domain. A possible mechanistic explanation for this observation is that the abovementioned residues are located on the opposite end from the isopeptide linkage acting as a hinge, whereas L8, L71 of the distal and G47 of the proximal Ub are positioned in close proximity to the hinge region, thus these latter sites could be less accessible to the ligand, at least during the initial steps of the interface opening. Distinct ligand interactions with the same residues in the two Ub units suggested by this mechanistic Ub2 model are in agreement with the finding 36 that L8A mutations of Ub pairs in K48-linked Ub4 have distinct effects on its binding to the proteasome. Considering tetraubiquitin's structure as a dimer of Ub2's 21, the L8A mutations that had the strongest effect on the binding were in the Ub units that are analogous to the proximal Ub in Ub2. Note that the mechanism of interface opening implied here follows from the comparison of the Ub2 structures in the closed state (PDB codes 1AAR, 2BFG) with the Ub2:UBA complex (PDB code 1ZO6) and, independently, from the analysis of equilibrium dynamics in K48-linked Ub2 from NMR relaxation data33.
The total buried surface area between UIM-2 and monoUb is 1,197 Å2, compared to ∼1,500–1,750 A2 between Ub units in Ub2. This suggests that the formation of a 1:1 S5a263–307:Ub2 complex using the same S5a263–307:Ub contacts as in the case of monoUb and thus exposing the hydrophobic surface of one of the two Ub units, is not entropically favorable. In the case of two S5a263–307 molecules bound to Ub2, the total buried area is expected to be about 2,240 A2, which would make the formation of such a complex more favorable with regard to the hydrophobic effect. This gain, however, could be offset by the entropy loss due to the immobilization of an additional UIM moiety. Our NMR titration data presented above indicate that, in fact, S5a263–307 binding to either Ub unit in Ub2 is tighter than to monoUb. This is in agreement with the published pull-down data indicating that S5a, as well as the individual UIMs, bind polyUb more efficiently than monoUb 6; 8.
Because two UIM molecules ultimately bind to Ub2, the complex must remain open or continue opening after binding of the first UIM, rather than wrapping around it to form a Ub-UIM-Ub sandwich. This conclusion is supported by our NMR mapping data. Indeed, the Ub2-binding surface on S5a263–307 involves the same “face” of the UIM helix as in binding to monoUb, and no additional perturbations were observed on the opposite “face” of UIM-2 (Fig.2) thus excluding the sandwich model. In addition, the S5a263–307-binding surfaces on both K48- and K63-linked Ub2 constructs (Fig.1) agree well with that on monoUb (Fig.1a,e) and with the published structure of the UIM-2:monoUb complex 8. Moreover, our data provide no indication of any interaction between the two S5a263–307 molecules in the fully loaded, 2:1 S5a263–307:Ub2 complex, thus rendering unlikely a more complex model where the two UIMs, bound to the individual Ub domains, interact with each other (e.g. sandwiched within the Ub2's hydrophobic pocket). Note that sedimentation velocity measurements, as well as NMR studies showed no interaction between S5a UIMs 8; 15. It seems then reasonable to model the formation of the UIM:Ub2 complex as a two-step process (possibly cooperative) in which the binding of one UIM shifts the equilibrium towards an open conformation of Ub2, thus causing a conformational change in the chain which, in turn, exposes the hydrophobic patch on the other domain and makes it available for binding to a second UIM.
2. Structural perspective
Based on the data presented above, we have modeled the complexes of UIM-2 with K48-and K63-linked Ub2 (Fig.4). The Ub2 structure in the complex with the hHR23A UBA2 domain (PDB code 1ZO6, 22) was used as a structural model for the open conformation of K48-linked Ub2, while the (extended) conformation of free K63-linked Ub2 in solution18 was used in Fig.4b The UIM-2:monoUb structure 8 was used to model the UIM-2/Ub contact for both Ub units in each chain. It should be mentioned that, based on the NMR analysis of the equilibrium dynamics in K48-linked Ub2, the relative orientation of the two Ub domains in the open state of free Ub2 at pH6.8 is similar to that in the Ub2:UBA2 complex 33; 37, therefore we used this structure as a model for the open state of Ub2. The structure model shown in Fig.4a,c illustrates how two UIMs can be accommodated in the hydrophobic pocket of K48-linked Ub2. The steric clashes involving flexible regions flanking the UIMs may be relieved by further opening of Ub2 (which seems plausible due to the flexibility of the Ub-Ub linker) as well as by structural rearrangement of the clashing flexible fragments. No steric clashes are seen in the K63-Ub2:UIM complex, in which the Ub units are expected to bind two UIM-2 molecules independently, in analogy to that observed in the chain's binding to UBA2 domains18.
The similarity in UIM-2 binding between K48- and K63-linked Ub2s is in contrast to their interaction with hHR23A UBA2. This is in agreement with the SPR data 16 indicating that, unlike the UBA2 domain, S5a does not discriminate between K48- and K63-linked Ub4. It is important to emphasize here that the binding preference toward a particular chain linkage depends on at least two factors: (1) the Ub-binding properties of a ligand and (2) the ability (dictated by the linkage) of a given polyUb chain to adopt a specific conformation necessary to bind this ligand in the energetically most favorable way. For example, the three-helix bundle structure of UBA2 allows it to interact simultaneously with both ubiquitins in the K48- but not in the K63-linked Ub2, where the conformational properties of the chain do not allow a similar, high-affinity binding mode. This, however, cannot be generalized for all UBAs, which were shown to exhibit diverse chain-linkage preferences 16. The nondiscriminatory binding of polyUb chains by UIM-2 of S5a could be a result of UIM-2 being a single, one-sided Ub-binding helix. It is possible that other UIMs are linkage selective, for example, those from Eps15 and Hrs, predicted to exhibit double-sided binding 38.
Putative models for the Ub2 complex with full-length S5a
As mentioned in the Introduction, strong binding preference of S5a and the individual UIMs toward polyUb might imply that the chains bind UIMs in a different mode compared to monoUb. However, this possibility is not supported by the data presented here which suggest that the mode of UIM-2 binding to each Ub unit in Ub2 is similar to that in the UIM-2:monoUb complex (Figs 1, 2). Detailed structural characterization of these complexes is required in order to fully address this issue. Although at this stage we cannot completely rule out a possibility that longer chains (n ≥ 4) can bind UIM-2 in a different mode, the observation that K48-linked tetra-Ub behaves in solution as a dimer of Ub2's 19; 21 renders the above possibility less likely.
The finding that each Ub moiety in K48-linked Ub2 binds a single UIM, combined with the fact that each UIM in S5a binds a monoUb, suggests a 1:1 stoichiometry model for the Ub2's complex with the full length S5a. In fact, this model is supported by our NMR relaxation measurements for the complex of K48-linked Ub2 with a tandem-UIM construct, S5a201–310 (generously provided by Dr. John P. O'Bryan, University of Illinois, Chicago). Here 1H T2 was 14.3 ms (at the 1:1 molar ratio), and 15N T1 was 1103 ± 112 ms and 1076.5 ± 156 ms at [S5a]/[Ub2] = 0.8 and 1.5, respectively, which corresponds to the molecular weight range of 26–33 kDa, in agreement with 30 kDa expected for a 1:1 complex.
These data suggest a putative model of the complex of full-length S5a and K48-linked Ub2 (schematically shown in Fig.5), in which both UIMs of S5a are bound, each to a separate Ub unit in the chain. This model is based on the similarity of the intermolecular UIM-2:Ub contacts observed in S5a263–307 binding to monoUb and Ub2, and on the assumption that the same should apply to UIM-1. Because UIM-2 binds mono/polyUb stronger than UIM-1 does 6; 8, it is natural to assume that its binding preferences could dictate the structure of the S5a:Ub2 complex. However, since an isolated UIM-2 does not show strong preference for a particular Ub unit in Ub2, both possible scenarios are considered here, i.e. with UIM-2 bound to the distal or to the proximal Ub, and UIM-1 bound to the other Ub unit in Ub2 (Fig.5). Unlike UIM-2, where the Ub-interacting IAYAM sequence is located in the middle of a relatively short α-helix, a similar LALAL motif in UIM-1 is positioned at the N-terminus of a very long α-helix (9 turns compared to 4 turns in UIM-2). This could potentially cause steric clashes between the helices as well as with the Ub-Ub linker. Our preliminary modeling (Supplementary Material) suggests that the more likely scenario is the one shown in Fig.5b, in which UIM-2 is bound to the proximal Ub, because this arrangement could avoid clashes between UIM-1 and UIM-2 near the Ub-Ub linker and, in addition, would allow the flexible part of S5a (between the UIMs) to extend away from this region (Fig.5b). Atomic-resolution structural studies are required to determine the mode of binding between the full length S5a and K48-linked Ub2 and longer chains, and to verify if specific UIMs bind to each particular ubiquitin in the chain.
Figure 5.

Schematic representation of the putative models for the complex of K48-linked Ub2 and the tandem-UIM fragment of S5a. Panels (a) and (b) show two possible ways the S5a UIMs can contact individual Ub units in Ub2. The linker connecting UIMs in S5a is flexible in solution 8. Preliminary structural models of the complex obtained by superimposition of the structures of the corresponding UIM/monoUb complexes can be found in Supplementary Material.
Conclusions
Here we used NMR to map the interacting surfaces involved in binding of monoUb, K48- and K63-linked di-ubiquitin chains to a UIM-2-containing construct of S5a. Our data suggest that these di-ubiquitin chains can bind two UIM-2 moieties per chain and with higher affinity than monoUb. Our mapping of the binding interface between UIM-2 and the individual ubiquitins in these chains suggests that, from the structural perspective, these contacts are similar to that between UIM-2 and monomeric Ub. Moreover, these studies provide first experimental evidence that UIM-2 does enter the interdomain interface in K48-linked Ub2 and binds directly to the hydrophobic patches on ubiquitins, thus confirming that the Ub/Ub interface is not rigidly locked.
Differences in the exchange regimes (on/off-rates) and in the magnitudes and directions of chemical shift perturbations between the two ubiquitins in K48-linked Ub2 indicate a complex mode of binding. This complexity can putatively be attributed to the conformational transition accompanying the opening of the Ub/Ub interface in K48-linked chains and to the way the two ubiquitins are linked. Our results suggest that UIM-2 (and possibly the full-length S5a) bind to K48-linked Ub2 in a mode distinct from the sandwich-like binding observed in the Ub2:UBA2 complex.
Based on our data, we proposed putative structural models of how di-ubiquitin might interact with UIM-2 and the full-length S5a containing two UIMs. Because di-ubiquitin is a structural element in the conformation of polyubiqutin chain, these models provides insights into the possible interactions of polyubiquitin with the proteasome. The linkage-dependent conformation and flexibility of polyubiquitin chains are expected to play a key role in their recognition and specific binding in the proteolytic pathway.
Materials and Methods
Biochemicals were from Sigma unless otherwise stated. E1 was from BostonBiochem, Inc. C170S-E225K, Ub (K48C), Ub (K48R), Ub (K63R), and Ub (D77) were expressed and purified as described in 39. Yeast Mms2 and Ubc13 were expressed and purified as in previous studies 17; 40. For 15N-labeled Ub (D77), Ub (K48C) and S5a263–307, Escherichia coli cells were grown in minimal media with 15NH4Cl as the sole source of nitrogen. Ub2 chains 15N-enriched at the distal or proximal (with respect to the free C-terminus) Ub unit were assembled from 15N-labeled and unlabeled recombinant monomers as described in 18; 19 (see also 17; 41). Throughout the text these constructs are referred to as Ub2-D and Ub2-P, respectively. The correct chain assembly was confirmed by the observed chemical shift perturbations in the C-terminal residues of Ub2-D and in K48 (or K63) and adjacent residues in Ub2-P when compared with monoUb.
The UIM-2 construct for these studies contained residues 263–307 of the S5a subunit of the 26S human proteasome (SWS P55036) and, in addition, an N-terminal His tag MG(H)12SSGHIEGRH. The plasmid was a generous gift from Dr. Patrick Young (Stockholm University). Its expression and purification was performed as described in 6.
NMR spectroscopy
NMR samples of the proteins under study were prepared in a 20 mM sodium phosphate buffer (pH 6.8) containing 7% D2O and 0.02% (w/v) NaN3.
All NMR studies were performed on a Bruker Avance 600 spectrometer at 25 °C. 1H-15N HSQC spectra and 2D planes for 15N relaxation experiments were acquired with spectral widths of 7.2 kHz and 2 kHz in the 1H and 15N dimensions, respectively. Typically, 128 or 256 t1 increments, each consisting of 1024 complex points, were collected for each 2D plane. The 15N relaxation studies included measurements of the longitudinal 15N relaxation (T1) using standard pulse sequences described elsewhere 42. NMR signal assignments for monoUb and Ub2 at pH 6.8 were taken from previous studies 18. NMR signal assignments for S5a263–307 were derived using a combination of 1H-15N HSQC, homonuclear TOCSY measurements, and the literature data 24 as a starting point. NMR data were processed with XWINNMR and then analyzed using XEASY 43 and CARA 44 and in-house software. The signals in slow exchange were assigned based on a combination of 2D TOCSY and 1H-15N HSQC spectra, with the aid (for S5a263–307) of published assignments for UIM-2 complex with hHR23A UBL 24. In ubiquitins, the peaks in slow exchange were isolated and had relatively small Δδ, such that their assignment was straightforward.
NMR Titration Studies of Ubx/S5a263–307 Binding
Mapping of the binding interface was achieved in a series of NMR titration experiments, in which 2D 1H-15N HSQC spectra of a 15N-labeled species of interest (e.g. Ubx, see below) were recorded as a function of the increasing amount of the unlabeled binding partner (S5a263–307). To map the binding surface on the binding partner (S5a263–307), a similar assay was performed, this time adding unlabeled Ubx to 15N-labeled S5a263–307.
Specifically, to determine the binding surface on the distal or proximal Ub in Ub2, 0.8–1.1 mM 15N-labeled K48-linked Ub2 samples (Ub2-P or Ub2-D, respectively) were titrated with increasing amounts of unlabeled S5a263–307 (from a concentrated stock solution). Titration for the proximal and distal Ub continued up to S5a263–307:Ub2 molar ratio of 4 and 3, respectively. Similar studies were performed on 1 mM 15N-labeled K63-linked Ub2 samples up to a S5a263–307:Ub2 molar ratio of 3.9. In a reverse titration experiment, a 0.5 mM 15N-labeled S5a263–307 sample was titrated with increasing amounts of unlabeled K48-linked Ub2, up to [Ub2]/[S5a263–307] = 2. In a control experiment, increasing amounts of unlabeled S5a263–307 were titrated into a 0.6 mM 15N-labeled monoUb sample up to a S5a263–307:Ub molar ratio of 3.4:1. For the reverse titration, a 0.7 mM 15N-labeled S5a263–307 sample was titrated with monoUb up to a molar ratio of 2:1. Some of these titration experiments were repeated to assure reproducibility of the measurements.
Binding was monitored through accompanying changes in the signals in the 1H-15N HSQC spectra. The shifts in peak positions were quantified using combined amide chemical shift perturbation (CSP) calculated as Δδ= [(ΔδH)2+(ΔδN/5)2]1/2, where ΔδH and ΔδN are the observed chemical shift changes (for 1H and 15N, respectively). To monitor site-specific changes in NMR signal intensities in the course of binding, the spectra obtained upon titration of S5a263–307 (or Ubx) were uniformly scaled to compensate for the higher molecular weight of the complex. The signal attenuation for each residue was then calculated as the ratio of peak intensities in HSQC spectra of the free and bound protein. The stoichiometry of binding was determined from NMR relaxation measurements as described in the Results section. The titration curves representing CSP dependence on the protein and ligand concentrations were fitted to the appropriate stoichiometry/binding models as detailed in 18; 22 using in-house software.
Based on the observed changes in NMR signal positions and intensities, the following classification of the exchange regimes was used here. The resonances that shift during titration abut show little or no signal attenuation are considered in fast exchange on the NMR chemical shift time scale; those that experience significant attenuation (≥60%) and broadening and a noticeable shift are in the intermediate exchange, whereas the presence of both strong attenuation of the original (almost unshifted) resonance and the subsequent appearance of a second (bound) peak with an increasing intensity upon titration was assigned to slow exchange regime.
Supplementary Material
Acknowledgments
Supported by NIH grant GM065334 to DF. We thank Dr. Patrick Young for providing us with S5a clones, Dr. John P. O'Bryan for the tandem-UIM S5a construct, and Drs. Kylie Walters and Ranjani Varadan for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. A Multiubiquitin Chain Is Confined to Specific Lysine in a Targeted Short-Lived Protein. Science. 1989;243:1576–1583. doi: 10.1126/science.2538923. [DOI] [PubMed] [Google Scholar]
- 2.Hershko A, Ciechanover A. The ubiquitin system. Annual Review of Biochemistry. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 3.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–6. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Pickart CM. Ubiquitin in chains. Trends in Biochemical Sciences. 2000;25:544–548. doi: 10.1016/s0968-0004(00)01681-9. [DOI] [PubMed] [Google Scholar]
- 5.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annual Review of Cell and Developmental Biology. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 6.Young P, Deveraux Q, Beal RE, Pickart CM, Rechsteiner M. Characterization of two polyubiquitin binding sites in the 26 S protease subunit 5a. Journal of Biological Chemistry. 1998;273:5461–5467. doi: 10.1074/jbc.273.10.5461. [DOI] [PubMed] [Google Scholar]
- 7.Hiyama H, Yokoi M, Masutani C, Sugasawa K, Maekawa T, Tanaka K, Hoeijmakers JHJ, Hanaoka F. Interaction of hHR23 with S5a - The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. Journal of Biological Chemistry. 1999;274:28019–28025. doi: 10.1074/jbc.274.39.28019. [DOI] [PubMed] [Google Scholar]
- 8.Wang QH, Young P, Walters KJ. Structure of S5a Bound to Monoubiquitin Provides a Model for Polyubiquitin Recognition. J Mol Biol. 2005;348:727–739. doi: 10.1016/j.jmb.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Beal R, Deveraux Q, Xia G, Rechsteiner M, Pickart C. Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. PNAS. 1996;93:861–866. doi: 10.1073/pnas.93.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fisher RD, Wang B, Alam SL, Higginson DS, Robinson H, Sundquist WI, Hill CP. Structure and ubiquitin binding of the ubiquitin-interacting motif. Journal of Biological Chemistry. 2003;278:28976–28984. doi: 10.1074/jbc.M302596200. [DOI] [PubMed] [Google Scholar]
- 11.Shekhtman A, Cowburn D. A ubiquitin-interacting motif from Hrs binds to and occludes the ubiquitin surface necessary for polyubiquitination in monoubiquitinated proteins. Biochemical and Biophysical Research Communications. 2002;296:1222–1227. doi: 10.1016/s0006-291x(02)02006-5. [DOI] [PubMed] [Google Scholar]
- 12.Fujiwara K, Tenno T, Sugasawa K, Jee JG, Ohki I, Kojima C, Tochio H, Hiroaki H, Hanaoka F, Shirakawa M. Structure of the ubiquitin-interacting motif of S5a bound to the ubiquitin-like domain of HR23B. Journal of Biological Chemistry. 2004;279:4760–4767. doi: 10.1074/jbc.M309448200. [DOI] [PubMed] [Google Scholar]
- 13.Raiborg C, Bache KG, Gillooly DJ, Madshush IH, Stang E, Stenmark H. Hrs sorts ubiquitinated proteins into clathrin-coated microdomains of early endosomes. Nature Cell Biology. 2002;4:394–398. doi: 10.1038/ncb791. [DOI] [PubMed] [Google Scholar]
- 14.Ryu KS, Lee KJ, Bae SH, Kim BK, Kim KA, Choi BS. Binding surface mapping of intra- and interdomain interactions among hHR23B, ubiquitin, and polyubiquitin binding site 2 of S5a. Journal of Biological Chemistry. 2003;278:36621–36627. doi: 10.1074/jbc.M304628200. [DOI] [PubMed] [Google Scholar]
- 15.Swanson KA, Kang RS, Stamenova SD, Hicke L, Radhakrishnan I. Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation. Embo Journal. 2003;22:4597–4606. doi: 10.1093/emboj/cdg471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–14. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 17.Hofmann RM, Pickart CM. In vitro assembly and recognition of Lys-63 polyubiquitin chains. J Biol Chem. 2001;276:27936–43. doi: 10.1074/jbc.M103378200. [DOI] [PubMed] [Google Scholar]
- 18.Varadan R, Assfalg N, Haririnia A, Raasi S, Pickart C, Fushman D. Solution conformation of Lys(63)-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. Journal of Biological Chemistry. 2004;279:7055–7063. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- 19.Varadan R, Walker O, Pickart C, Fushman D. Structural properties of polyubiquitin chains in solution. Journal of Molecular Biology. 2002;324:637–647. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- 20.Tenno T, Fujiwara K, Tochio H, Iwai K, Morita EH, Hayashi H, Murata S, Hiroaki H, Sato M, Tanaka K, Shirakawa M. Structural basis for distinct roles of Lys63- and Lys48-linked polyubiquitin chains. Genes Cells. 2004;9:865–75. doi: 10.1111/j.1365-2443.2004.00780.x. [DOI] [PubMed] [Google Scholar]
- 21.Eddins MJ, Varadan R, Fushman D, Pickart CM, Wolberger C. Crystal Structure and Solution NMR Studies of Lys48-linked Tetraubiquitin at Neutral pH. J Mol Biol. 2006 doi: 10.1016/j.jmb.2006.12.065. [DOI] [PubMed] [Google Scholar]
- 22.Varadan R, Assfalg M, Raasi S, Pickart C, Fushman D. Structural Determinants for Selective Recognition of a Lys48-Linked Polyubiquitin Chain by a UBA Domain. Mol Cell. 2005;18:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 23.Zuiderweg ER. Mapping protein-protein interactions in solution by NMR spectroscopy. Biochemistry. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 24.Mueller TD, Feigon J. Structural determinants for the binding of ubiquitin-like domains to the proteasome. Embo Journal. 2003;22:4634–4645. doi: 10.1093/emboj/cdg467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–72. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mueller TD, Kamionka M, Feigon J. Specificity of the interaction between UBA domains and ubiquitin. J Biol Chem. 2004;279:11926–36. doi: 10.1074/jbc.M312865200. [DOI] [PubMed] [Google Scholar]
- 27.Swanson KA, Hicke L, Radhakrishnan I. Structural basis for monoubiquitin recognition by the Ede1 UBA domain. J Mol Biol. 2006;358:713–24. doi: 10.1016/j.jmb.2006.02.059. [DOI] [PubMed] [Google Scholar]
- 28.Prag G, Misra S, Jones EA, Ghirlando R, Davies BA, Horazdovsky BF, Hurley JH. Mechanism of ubiquitin recognition by the CUE domain of Vps9p. Cell. 2003;113:609–620. doi: 10.1016/s0092-8674(03)00364-7. [DOI] [PubMed] [Google Scholar]
- 29.Kang RS, Daniels CM, Francis SA, Shih SC, Salerno WJ, Hicke L, Radhakrishnan I. Solution structure of a CUE-ubiquitin complex reveals a conserved mode of ubiquitin binding. Cell. 2003;113:621–630. doi: 10.1016/s0092-8674(03)00362-3. [DOI] [PubMed] [Google Scholar]
- 30.Donaldson KM, Yin HW, Gekakis N, Supek F, Joazeiro CAP. Ubiquitin signals protein trafficking via interaction with a novel ubiquitin binding domain in the membrane fusion regulator, Vps9p. Current Biology. 2003;13:258–262. doi: 10.1016/s0960-9822(03)00043-5. [DOI] [PubMed] [Google Scholar]
- 31.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 32.Trempe JF, Brown NR, Lowe ED, Gordon C, Campbell ID, Noble ME, Endicott JA. Mechanism of Lys48-linked polyubiquitin chain recognition by the Mud1 UBA domain. Embo J. 2005;24:3178–89. doi: 10.1038/sj.emboj.7600797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ryabov Y, Fushman D. Interdomain mobility in di-ubiquitin revealed by NMR. Proteins. 2006;63:787–96. doi: 10.1002/prot.20917. [DOI] [PubMed] [Google Scholar]
- 34.Dickinson B, Varadan R, Fushman D. Effects of cyclization on conformational dynamics and binding properties of Lys48-linked di-ubiquitin. Protein Sci. 2007;16 doi: 10.1110/ps.062508007. XXX, in press (published online Jan 22, 2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finkelstein AV, Janin J. The price of lost freedom: entropy of biomolecular complex formation. Protein Engineering. 1989;3:1–3. doi: 10.1093/protein/3.1.1. [DOI] [PubMed] [Google Scholar]
- 36.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. Embo Journal. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ryabov Y, Fushman D. A Model of Interdomain Mobility in a Multi-Domain Protein. J Amer Chem Soc. 2007;129 doi: 10.1021/ja067667r. XXX (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirano S, Kawasaki M, Ura H, Kato R, Raiborg C, Stenmark H, Wakatsuki S. Double-sided ubiquitin binding of Hrs-UIM in endosomal protein sorting. Nat Struct Mol Biol. 2006;13:272–7. doi: 10.1038/nsmb1051. [DOI] [PubMed] [Google Scholar]
- 39.Haldeman MT, Xia G, Kasperek EM, Pickart CM. Structure and function of ubiquitin conjugating enzyme E2-25K: The tail is a core-dependent activity element. Biochemistry. 1997;36:10526–10537. doi: 10.1021/bi970750u. [DOI] [PubMed] [Google Scholar]
- 40.Raasi S, Pickart CM. Rad23 ubiquitin-associated domains (UBA) inhibit 26 S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. Journal of Biological Chemistry. 2003;278:8951–8959. doi: 10.1074/jbc.m212841200. [DOI] [PubMed] [Google Scholar]
- 41.Piotrowski J, Beal R, Hoffman L, Wilkinson KD, Cohen RE, Pickart CM. Inhibition of the 26 S proteasome by polyubiquitin chains synthesized to have defined lengths. Journal of Biological Chemistry. 1997;272:23712–23721. doi: 10.1074/jbc.272.38.23712. [DOI] [PubMed] [Google Scholar]
- 42.Fushman D, Cahill S, Cowburn D. The main-chain dynamics of the dynamin pleckstrin homology (PH) domain in solution: Analysis of N-15 relaxation with monomer/dimer equilibration. Journal of Molecular Biology. 1997;266:173–194. doi: 10.1006/jmbi.1996.0771. [DOI] [PubMed] [Google Scholar]
- 43.Bartels C, Xia TH, Billeter M, Guntert P, Wuthrich K. The Program Xeasy for Computer-Supported Nmr Spectral-Analysis of Biological Macromolecules. Journal of Biomolecular Nmr. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 44.Keller R. The Computer Aided Resonance Assignment Tutorial. CANTINA Verlag; 2004. [Google Scholar]
- 45.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos: 2002. [Google Scholar]
- 46.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


