Abstract
Measures of brain change can be computed from sequential MRI scans, providing valuable information on disease progression, e.g., for patient monitoring and drug trials. Tensor-based morphometry (TBM) creates maps of these brain changes, visualizing the 3D profile and rates of tissue growth or atrophy, but its sensitivity depends on the contrast and geometric stability of the images. A s part of the Alzheimer’s Disease Neuroimaging Initiative (ADNI), 17 normal elderly subjects were scanned twice (at a 2-week interval) with several 3D 1.5 T MRI pulse sequences: high and low flip angle SPGR/FLASH (from which Synthetic T1 images were generated), MP-RAGE, IR-SPGR (N = 10) and MEDIC (N = 7) scans. For each subject and scan type, a 3D deformation map aligned baseline and follow-up scans, computed with a nonlinear, inverse-consistent elastic registration algorithm. Voxelwise statistics, in ICBM stereotaxic space, visualized the profile of mean absolute change and its cross-subject variance; these maps were then compared using permutation testing. Image stability depended on: (1) the pulse sequence; (2) the transmit/receive coil type (birdcage versus phased array); (3) spatial distortion corrections (using MEDIC sequence information); (4) B1-field intensity inhomogeneity correction (using N3). SPGR/FLASH images acquired using a birdcage coil had least overall deviation. N3 correction reduced coil type and pulse sequence differences and improved scan reproducibility, except for Synthetic T1 images (which were intrinsically corrected for B1-inhomogeneity). No strong evidence favored B0 correction. Although SPGR/FLASH images showed least deviation here, pulse sequence selection for the ADNI project was based on multiple additional image analyses, to be reported elsewhere.
Introduction
Serial scanning of the human brain with MRI offers tremendous power to detect the earliest signs of illness, monitor disease progression and resolve drug effects in clinical trials that aim to prevent or slow the rate of brain degeneration. Structural MRI provides high-contrast 3D scans, offering excellent ability to differentiate gray and white matter, CSF and other tissues (including disease-related abnormalities). Moreover, with recent advances in mathematical and computational techniques for nonlinear image registration, researchers can now track local tissue change in the human brain based on serial MRI scans. One such approach is called tensor-based morphometry (TBM), which applies a nonlinear deformation field to the baseline scan to align it with the follow-up scan. Based on local analysis of the applied compression and expansion, rates of brain change can be inferred for specific regions of interest or presented in the form of a map. Tensor-based morphometry has been used to map growth patterns in the developing human brain (Thompson et al., 2000; Chung et al., 2001), degenerative rates in Alzheimer’s disease and other dementias (Fox et al., 1997, 1999, 2000, 2001; O’Brien et al., 2001; Freeborough et al., 1996; Freeborough and Fox, 1997; Studholme et al., 2001) as well as tumor growth and multiple sclerosis lesions (Lemieux et al., 1998; Ge et al., 1999; Rey et al., 2002). In addition, there has been intensive work on the statistical analysis of deformation fields for detecting whether significant changes have occurred (Worsley, 1994, 1999; Thompson et al., 1997; Ashburner et al., 1998; Cao and Worsley, 1999; Gaser et al., 1999; Woods, 2003; Fillard et al., 2005) as well as on the elastic and fluid registration algorithms to compute these deformations (Thompson and Toga, 1996a,b, 2002; Fox and Freeborough, 1997; Studholme et al., 2001; Janke et al., 2001; Crum et al., 2001; Miller et al., 2002; Leow et al., 2005a,b).
The Alzheimer’s Disease Neuroimaging Initiative (ADNI; Mueller et al., submitted for publication(a),(b), in press; see http://www.loni.ucla.edu/ADNI and http://ADNI-info.org) is a large multi-site longitudinal MRI and FDG-PET study of 200 elderly controls, 400 mildly cognitively impaired subjects and 200 Alzheimer’s disease subjects. One goal of this project is to develop improved imaging methods to measure longitudinal changes of the brain in normal aging, during the transition to early Alzheimer’s disease, and in Alzheimer’s disease patients. One of our specific aims was to develop a high-resolution 3D T1-weighted MRI scanning protocol that provided both between-vendor and between-site comparability, as well as longitudinal stability. Despite its usefulness for tracking brain change, there is little information regarding the stability and variability of various MR imaging techniques. Most evidence that MRI has good reproducibility comes from studies that have used rigid registration to identify systematic changes in overall brain volume in serial scans (Hajnal et al., 1995a,b; Oatridge et al., 2001; Smith et al., 2002).
Therefore, we performed a series of pilot studies to compare different 3D T1-weighted MRI sequences. Once acquired, these scans were evaluated with a number of different image analysis techniques including: atlas-based measurements of hippocampal volume (Haller et al., 1997; Hsu et al., 2002), the boundary shift integral (Fox and Freeborough, 1997; Fox et al., 2000), voxel-based morphometry using Statistical Parametric Mapping (VBM; Ashburner and Friston, 2000), cortical thickness measures (Fischl and Dale, 2000) and tensor-based morphometry (TBM; Studholme et al., 2001; Leow et al., 2005a,b).
The results provided in this paper concern 3D maps of the stability of different MRI imaging protocols and pre/post-processing techniques, in the context of mapping brain change using nonlinear image registration and TBM. Specifically, our goal was to determine which MR imaging sequences combined with which data correction methods were the most reproducible, and most reliable, resulting in least measurement variability. The foundation of our calibrations was based on the assumption that any serial MRI scan pair in this study should show minimal structural change related to aging or disease, and there should be no consistent change detected in a group of subjects scanned. This is plausible given that the elderly normal subjects were scanned twice using the same protocol, scanner and RF head coil over a very short interval (2 weeks). In individuals, there may still be minor changes due to subject-specific mechanical, circadian or tissue hydration effects on anatomy. There are also (non-pathological) sources of variability due to the interaction of the patient and the sequence/scanner. For example, subject movement is more likely with a longer sequence. Patient positioning is inevitably variable relative to the coil and scanner, which may have different impacts on the images depending on the scanner, sequence and coils. Differences among MRI scanning techniques were assessed by scanning the same subjects with four or five (depending on the MR system) different MRI pulse sequences in the same scanning session (IR-SPGR, MEDIC, high and low flip angle SPGR/FLASH and MP-RAGE). The low flip angle SPGR/FLASH images were not evaluated as an independent image type; they were used along with the high flip SPGR/FLASH scans to generate a Synthetic T1 image. Therefore, any regional structural difference picked up using TBM can be assumed to be random error or artifactual drift, related to geometric distortion of the scanner, uncorrected spatial distortions and variations in imaging signal or contrast-to-noise. Statistical analysis was therefore applied to maps of changes computed using TBM, providing baseline information on MRI imaging reliability, reproducibility and variability.
Methods
Subjects
Seventeen healthy elderly subjects (12 women, 5 men; mean age: 71.1 ± 7.5 years; mean education: 15.7 ± 2.5 years) were scanned twice, at an interval of exactly 2 weeks. Ten were scanned at the Mayo Clinic in Rochester, Minnesota, seven were scanned at the University of California, San Diego, after providing informed consent as directed by the respective Institutional Review Boards. At each acquisition site, multiple sets of 3D image volumes were acquired using various combinations of pulse sequences, including IR-SPGR, MEDIC, high and low flip angle SPGR/FLASH and MP-RAGE (see Methods for descriptions of these sequences). The subjects’ age and gender are shown in Table 1, together with the pulse sequences that were used to scan them. Inclusion criteria required that all subjects be between 55 and 90 years of age, with an informant/caregiver able to provide an independent evaluation of functioning. All enrolled control subjects had Mini-Mental State Exam (MMSE) scores between 28 and 30 and a Clinical Dementia Rating (CDR) of 0, without symptoms of depression, mild cognitive impairment (MCI) or other dementia and no current use of psychoactive medications.
Table 1.
This table summarizes the scans collected from the 17 subjects in this study
| SPGR/FLASH
|
MP-RAGE
|
SYNTH-T1
|
IR-SPGR
|
B0-corrected MEDIC
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Site | Gender | Age | PA | BC | PA | BC | PA | BC | PA | BC | PA | BC |
| 1 | 1 | Female | 71.3 | X | X | X | X | X | X | X | X | ||
| 2 | 1 | Female | 76.9 | X | X | X | X | X | X | X | X | ||
| 3 | 1 | Female | 63.9 | X | X | X | X | X | X | X | X | ||
| 4 | 1 | Female | 73.0 | X | X | X | X | X | X | X | X | ||
| 5 | 1 | Female | 72.8 | X | X | X | X | X | X | X | X | ||
| 6 | 1 | Female | 66.9 | X | X | X | X | X | X | X | X | ||
| 7 | 1 | Male | 66.6 | X | X | X | X | ||||||
| 8 | 2 | Female | 79.6 | X | X | X | X | X | X | X | X | ||
| 9 | 2 | Female | 85.7 | X | X | X | X | X | X | X | X | ||
| 10 | 2 | Female | 62.1 | X | X | X | X | X | X | X | X | ||
| 11 | 2 | Female | 76.7 | X | X | X | X | X | X | X | X | ||
| 12 | 2 | Male | 64.5 | X | X | X | X | X | X | X | X | ||
| 13 | 2 | Male | 66.2 | X | X | X | X | X | X | X | X | ||
| 14 | 2 | Female | 77.8 | X | X | X | X | X | X | X | X | ||
| 15 | 2 | Female | 65.1 | X | X | X | X | X | X | X | X | ||
| 16 | 2 | Male | 59.6 | X | X | X | X | X | X | X | X | ||
| 17 | 2 | Male | 80.0 | X | X | X | X | X | X | X | X | ||
Sites 1 and 2 indicate UCSD and the Mayo Clinic respectively. Note that a Siemens scanner was used at UCSD and a GE scanner was used at Mayo. Therefore, the set of imaging sequences acquired was not identical between the two sites. The major differences were: IR-SPGR was only acquired at Mayo and the MEDIC scans were only acquired at UCSD. The specific scan types acquired for each subject are marked with an “X”. To rank the relative stability of two scan types, we only included subjects with both scan types acquired (for example, when evaluating the effect of B0-correction, only subjects 1–6 from site 1 were analyzed).
Pulse sequences
The following four pulse sequences were used to collect 3D T1-weighted volumes at 1.5 T. All acquisitions used 1.25 – 1.25 mm in-plane spatial resolution and a sufficient number of 1.2 mm thick sagittal slices to completely cover the head.
SPGR/FLASH. The 3D SPGR (spoiled gradient echo) sequence was acquired on a General Electric Healthcare Signa 1.5 T scanner with parameters: TE/TR/flip angle = 4/17/20°. The essentially equivalent 3D FLASH (fast low angle shot) sequence on the Siemens Medical Solutions Symphony 1.5 T scanner used the parameters: TE/TR/flip angle = 4.2/15/20°.
Synthetic T1. A calculated T1 3D volume was obtained by combining two 3D volumetric acquisitions (SPGR for GE and FLASH for Siemens) with different flip angles (5 and 20°; see Deoni et al., 2003; Fischl et al., 2004).
B0-corrected. A MEDIC (multiple-echo data image combination; see Schmid et al., 2005) pulse sequence was acquired on the Siemens Symphony 1.5 T scanner at UCSD with parameters: TE = 2.3, 4.5, 6.6, 8.8 and 11.0 ms, TR = 16 ms. The MEDIC pulse sequence was used to generate images corrected for B0 distortions (see Correction for distortions induced by B0 inhomogeneity (B0 correction)).
MP-RAGE and IR-SPGR. These are magnetization-prepared inversion recovery sequences (i.e., magnetization-prepared rapid gradient echo and inversion-recovery spoiled gradient echo). MP-RAGE scans were acquired on a Siemens Symphony 1.5 T scanner, with parameters: TI/TR/flip angle = 1000/ 2300/8°, MP-RAGE scans were also collected on the GE system using a prototype pulse sequence developed for the study with the following parameters: TI/TR/flip angle = 1000/ 2400/8°. IR-SPGR scans were acquired on a GE Signa 1.5 T scanner with parameters: TI/TR/flip angle = 600/1540/12°. (The value of TR stated here for these pulse sequences is the repetition time for the inversion pulses.)
As noted in Table 1, IR-SPGR scans were collected at the Mayo site only (10 subjects). The inversion recovery techniques have the advantage of providing good contrast between tissues with different T1 relaxation times, thus providing greater gray–white contrast. Advantages of the SPGR/FLASH sequence are greater SNR per unit acquisition time and the fact that a complete set of sequences currently exists for all major MR vendors. This is not the case for magnetization-prepared inversion recovery sequences.
Fig. 1 illustrates these four different MRI sequences in three orthogonal views (axial, coronal and sagittal) for one of the subjects in this study.
Fig. 1.
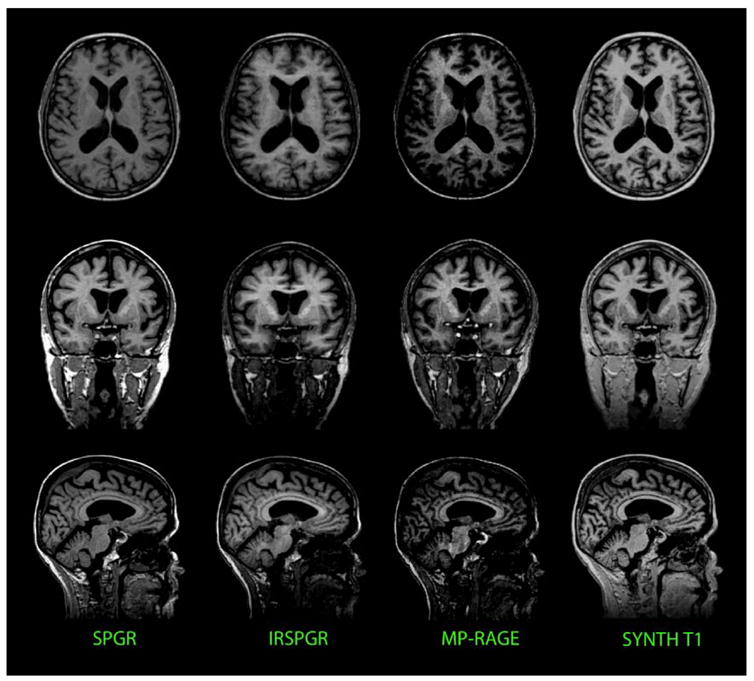
This figure shows the axial, coronal and sagittal views of a subject imaged using four of the five MRI sequences studied in this paper (SPGR, IR-SPGR, MP-RAGE and calculated Synthetic T1).
Several factors can contribute to the degradation of MRI data quality. These include geometric distortion due to gradient nonlinearity, spatially varying tissue contrast due to non-uniform transmit B1-field, geometric distortion due to local B0-field nonuniformity and signal inhomogeneity due to non-uniform B1 sensitivity profiles of some receiver coils (Narayana et al., 1988). Corrections for these are described in the following sections, including some adjustments made to the IR-SPGR sequence.
Adjustments to the IR-SPGR sequence
During the preparatory phase of the ADNI study, the inversion recovery spoiled gradient echo (IR-SPGR) sequence was evaluated on the GE Healthcare scanners. IR-SPGR is a product pulse sequence, characterized by an initial inversion pulse, followed by a delay equal to the inversion time (TI) and the acquisition of a series of views along the slice-encoded direction in a segmented fashion. Before the start of the preparatory phase, we observed several deficiencies with the use of the product 11.0 M4 and G3 M4 versions of the IR-SPGR pulse sequence for the ADNI study. A discrete ghost artifact signal was observed in the brain, emanating mainly from lipids (e.g., in the scalp). De-selecting the RF-spoiling option removed the discrete artifact. It became clear after the start of the preparatory phase, however, that de-selecting the RF-spoiling option also aggravated artifacts from CSF flow and motion in some subjects. The discrete ghost artifact was addressed by increasing the gradient area of the end-of-sequence spoilers on the readout and slice-encoded axes to 14 mT/m ms. This change was made in the prototype pulse sequences. The use of the increased end-of-sequence spoilers in conjunction with RF spoiling provided good image quality for the remainder of the subjects in the preparatory phase. All the IR-SPGR changes were provided to the manufacturer so that they could incorporate them into future product releases if desired.
Transmit/Receive coil type and B1 RF inhomogeneity correction
In general, smaller coils obtain higher SNR but have a greater B1 inhomogeneity (that is, they have greater spatial inhomogeneity of the RF coil sensitivity profile). Larger coils have more uniform sensitivity profiles, but reduced SNR. On the latest generation of MR scanners, using a uniform RF coil (e.g., body coil) for transmission and a receive-only head coil (or phased array surface coil) for reception, the sensitivity profile of the receive coil(s) can be estimated by simply dividing an image volume obtained with the head coil by a corresponding image volume obtained with the body coil on a voxel-by-voxel basis. Once this sensitivity profile is obtained, all subsequent volumes can be corrected by dividing each voxel’s intensity by the estimated sensitivity value at that location. (This does not work on systems using a combined transmit/receive (T/R) head coil because body coil transmit cannot be used in conjunction with the T/R head coil. Furthermore, even if the two were compatible, the transmit non-uniformity of the T/R head coil also affects the flip angle and hence image contrast.)
Images were acquired using two coil types: birdcage (BC) and phased array (PA). The birdcage design, a combined transmit and receive RF coil, provides a more uniform receive B1-field. The phased array design, on the other hand, provides a higher SNR. (All images acquired using a PA design received a B1 correction as previously described.) We therefore determined if one coil design significantly outperformed the other. This effectively determined whether the boosted SNR increases the stability of the computed maps of brain change and if the B1 correction technique (referred to as B1 in the rest of the paper) was sufficient in removing the RF inhomogeneity.
Spatial distortion due to gradient nonlinearity
A major cause of spatial distortion of anatomical images is gradient nonlinearity: the deviation of the gradient field from an ideal linear function of position. This is particularly prominent on some of the latest generations of MRI systems that have been optimized for gradient amplitude and slew rate at the expense of gradient linearity. A solution to this problem, known as Gradwarp (GW) correction (Hajnal et al., 2001), is described and evaluated by Jovicich et al. (in press). Briefly, gradient nonlinearity is estimated from the geometry of the gradient coil construction. A set of spherical harmonic coefficients is computed uniquely for a particular gradient coil design, which can be used to reverse the nonlinearity embedded in the acquired images. This unwarping matrix is applied after image reconstruction. The effect of GW is examined systematically in other papers (Jovicich et al., in press) and is not reported further here.
Correction for distortions induced by B0 inhomogeneity (B0 correction)
B0 inhomogeneity-induced distortion can result from imperfect magnet shimming or local patient-induced magnetic susceptibility variations and was corrected using an approach described in Jovicich et al. (in press) (see also Jezzard and Balaban, 1995). A MEDIC sequence is used with bipolar gradients, resulting in multiple, high-bandwidth volumes acquired with alternating readout direction (and thus alternating spatial shifts). These echoes are combined in a way that eliminates most of the B0 distortions. No phantom measurements are required, and distortions induced by the subject are corrected. Gray/white contrast was also improved by optimizing the weighting of different echoes, and artifacts due to eye-movements and flow were reduced due to the high bandwidth of each echo. We studied the impact of this B0 correction technique on the maps of change.
Non-parametric non-uniform intensity normalization (N3 correction)
The non-parametric non-uniform intensity normalization, commonly referred to as N3, was first proposed by Sled et al. (1998) as a novel approach to correcting for intensity nonuniformity in MRI. The software is publicly available at http://www.bic.mni.mcgill.ca/software/N3/. The correction is based on a non-parametric framework and thus operates without the presence of a statistical model for tissue classification. This method is independent of different pulse sequences and somewhat insensitive to pathological data that might otherwise violate model assumptions. To eliminate the dependence of the field estimate on anatomy, an iterative approach is employed to estimate both the multiplicative bias field and the sharpness of the histogram of the tissue intensities. N3 correction (MNI N3, version 1.02) was applied after aligning data to ICBM space, using 200 iterations and specifying the ICBM space brain mask as the region of interest. We also determined how N3 interacted with the abovementioned B1 correction technique, as both adjust for intensity inhomogeneity.
Tensor-based morphometry (TBM) based on 3D nonlinear deformation
The baseline scans for all subjects were first aligned to the ICBM-53 average brain template using a 9-parameter linear transformation, driven by a mutual information cost function (Collins et al., 1994). The follow-up scans were then registered to the baseline scan using a second 9-parameter linear transformation—this 9 degree-of-freedom registration should correct to a first approximation (linear) voxel size drifts so that TBM will not be assessing any global variability in voxel sizes. This transformation was followed by a high-dimensional nonlinear registration using a mutual information-based inverse-consistent algorithm (Leow et al., 2005a,b). After this step, deformation fields were obtained by registering follow-up scans (source) to baseline scans (target). The Jacobian determinant operator was then applied to the forward deformation field to show regions of tissue expansion (Jacobian >1) or contraction (Jacobian <1), as in prior TBM and voxel compression mapping studies (Fox and Freedborough, 1997; Thompson et al., 2000; Ashburner et al., 1998, 2003; Studholme et al., 2001; Leow et al., 2005a,b). This map of local tissue change can then be color-coded and overlaid on the baseline image as illustrated in Fig. 2. For more fair comparison of regions with tissue loss or expansion, we take the natural logarithm of the Jacobian determinant values (denoted by log J) in this paper (see Ashburner et al., 1998; Cachier and Rey, 2000; Woods, 2003; Leow et al., 2005a,b for a discussion of why the Jacobians are typically logged before statistical analysis).
Fig. 2.
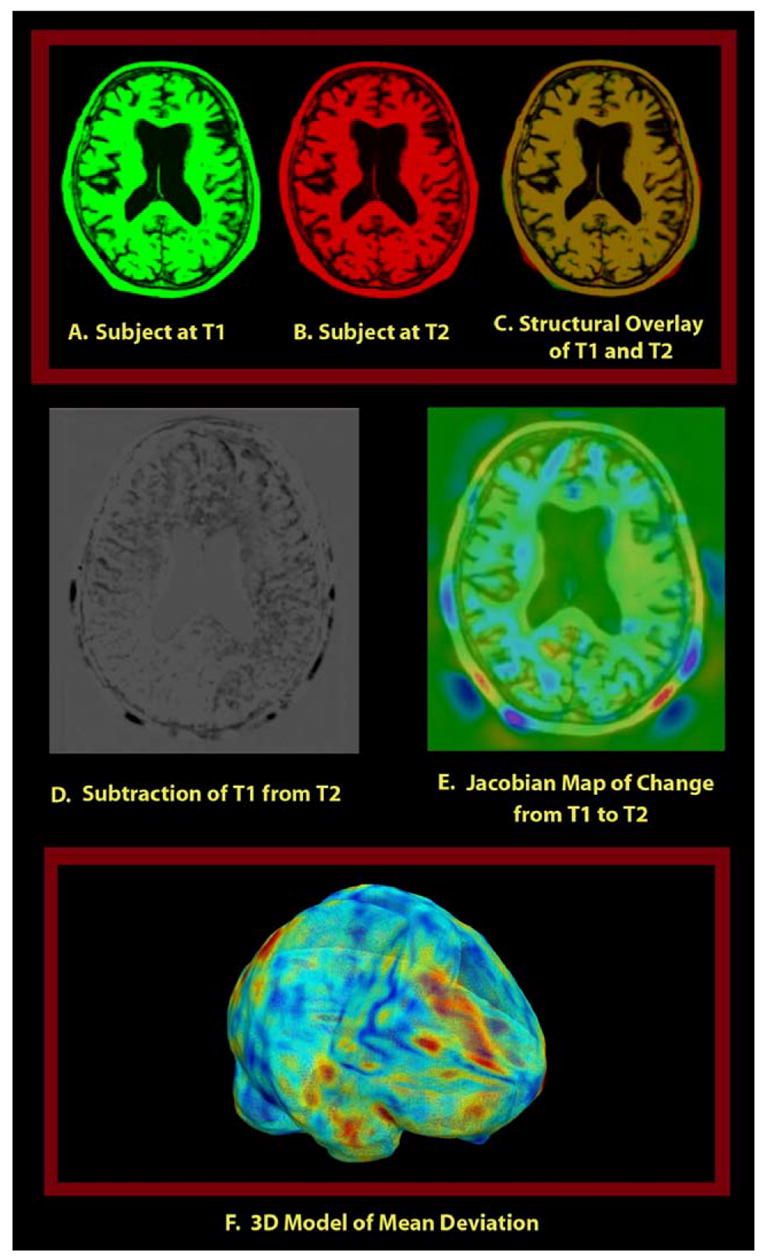
Illustration of the approach employed in this paper where a color map of the Jacobian determinant (a measure of expansion or compression of time) is generated by registering follow-up scan (source image) to baseline (target image) and computing the Jacobian of the forward mapping (baseline to follow-up). This map is overlaid on the baseline structural image to visualize voxelwise local tissue change. Red colors denote regions where local expansion is detected, blue colors denote compression, and green denotes regions with no detected change. Notice that the 3D local tissue change, encoded using the Jacobian map, could not be easily visualized by inspecting the subtraction map due to difficulties in reconstructing 3D spatial relationships from 2D slices.
Statistical testing on the mean log Jacobian
Two different tests of stability can be constructed. Firstly, we would like to test if the mean log J is zero, that is, from a statistical standpoint, a particular pulse sequence type should have a zero group mean log J at any point inside the brain within 2 weeks. Thus, one scan type would be considered inappropriate for TBM purposes if this particular scan type has a statistically non-zero mean value as it would indicate a ‘‘geometric drift’’ or changing spatial miscalibration over time. Before we go on and discuss how to conduct statistical testing on the mean, we have to first differentiate two different concepts of mean: the mean log J at the region-wise level (average log J value inside the brain as a whole for each individual) versus the mean at the voxelwise level (given observations from multiple subjects).
In Leow et al. (2005a,b), we showed (using the Kullback–Leibler divergence on material density functions) that the log Jacobian of any non-trivial, smooth, bijective (e.g., fixed or sliding boundary condition) deformation mapping plotted on the target domain has a negative mean value:
| (1) |
Although in our case the brain boundary does not stay fixed from the baseline to the follow-up scans and thus the deformation mapping is not globally volume-preserving for the region inside the brain, it is reasonable to assume that the boundaries remain very similar. Moreover, since all baseline scans have been 9-parameter registered by 9-parameter transformation to the ICBM space, the region defined by the ICBM brain space has a (almost) fixed boundary condition over a 2-week interval, and the region-wise mean log J value (inside the whole brain) is negative when averaged across the brain.
| (2) |
Here, volbrain denotes the brain volume defined in the ICBM space. In contrast, the concept of voxelwise mean log Jacobian is easier to understand and of greater importance as localized group mean tissue changes are ultimately what brain imagers seek. However, when conducting corrections on the localized tissue change map for multiple comparisons, we have to consider the spatial properties of the log Jacobian maps, and thus both voxelwise and region-wise mean log Jacobian concepts are important. For an inverse-consistent image registration algorithm, the voxelwise mean (across subjects) of a log Jacobian map should be zero under the null hypothesis, but the region-wise mean should be negative.
Let us first discuss the statistical testing on the voxelwise mean (please refer to the Correction for multiple comparison for multiple comparisons section for discussions of the region-wise mean). Since we have a log J map from each of the n control subjects for each scan type, whose log J values at voxel x will be denoted as log J1(x), log J2(x),..., log Jn(x) in the rest of the paper, a voxelwise standard t test can thus be conducted on the n observations, allowing us to test the validity of the zero-mean hypothesis at that voxel. The following voxelwise T statistic can then be compared to a two-tailed Student’s t distribution with n − 1 degrees of freedom to test the above null hypothesis:
| (3) |
where
We reject the null hypothesis if the magnitude of the T value calculated above exceeds a pre-set threshold based on a suitable confidence interval. Notice the voxelwise variance of log J provides us with a way to assess the repeatability of a scan type, i.e., measuring the voxelwise spread of the given multiple observations (higher variance implying poorer repeatability). In the Results section, we plot the log J variance maps to visualize the repeatability of the scans. However, as will be discussed, the concept of repeatability does not translate directly into the concept of performance.
Statistical testing on the deviation of log Jacobian maps
The above t test at a given voxel, if significant, implies that there is a bias, or geometric drift over time, in the spatial accuracy of a scan type at that voxel. Now, let us consider the second type of statistical testing: assessing the performance. For an ideal scanner, no mean structural change should be detected within 2 weeks, so any deviation of the Jacobian map from one should be considered error. Thus, the best scan type should have log J values closest to 0 (in the sequel, we will interchangeably use the two terms: better performance/lower deviation). Mathematically speaking, testing the performance is to consider the deviation map dev of the logged Jacobian away from zero, defined at each voxel as
| (4) |
For two different sequences A and B in any subject, we define the voxelwise score or gain of sequence A over sequence B in this subject (denoted by image data, SA,B) as
| (5) |
Again, we are given n observations at each voxel: , so we can compare the performance of sequence A and B at each voxel by considering the distributions of the n observations, using similar methods to those previously described.
Visually, the performance of a sequence can be assessed by inspecting the estimated mean deviation map. This is defined for sequence A as follows
| (6) |
To statistically compare the performance of two scan types, we again rely on the standard t test on the mean of S. To construct a suitable null hypothesis, the following relation should hold, assuming sequence A outperforms B
| (7) |
Thus, the null hypothesis in this case would be testing if the mean score is zero
| (8) |
To determine the ranking of A and B, we have to consider one-sided alternative hypotheses. For example, when testing if sequence A outperforms B, we use the following alternative hypothesis
| (9) |
The voxelwise T statistic, defined as
| (10) |
where
thus follows the Student’s T distribution with n − 1 degrees of freedom under the null hypothesis and can be used to determine the P value at each voxel. If the alternative hypothesis is accepted, sequence A outperforms B at point x. Similarly, the hypothesis that B outperforms A can be tested by switching the sign in the alternative hypothesis. We rank A and B equally if the null hypothesis is not rejected for either test.
Correction for multiple comparisons
To determine the overall effects of different pulse sequences (and image corrections) on both the mean and the deviation of log Jacobian maps throughout the brain, we need to adjust for multiple comparisons. The above analyses are conducted voxel by voxel, so this results in statistical parametric maps, i.e., maps of statistics.
Two types of permutation tests (see Bullmore et al., 1999; Nichols and Holmes, 2001) were applied. The first type, the percentage test, uses an ROI that defines brain voxels in standard ICBM space and mainly assesses deviation from zero change. In this test, we can resample the observations by randomly flipping the sign of the log Ji or (i = 1, 2,..., n) under the null hypothesis. For each permutation, voxelwise t tests are computed. We then compute the percentage of voxels inside the chosen ROI with T statistics exceeding a certain threshold. The multiple-comparisons-corrected P value can be determined by counting the number of permutations whose above-defined percentage exceeds that of the un-permuted observed data. This is comparable to set-level inference in the SPM package (Friston et al., 1995). For example, we say that sequence A outperforms B on the whole brain if this corrected P value is smaller than 0.05 (that is, less than 5% of all permutations have the above-defined percentage greater than that of the original data). In this paper, the threshold for T statistics at the voxel level is based on the T table critical value at α = 0.05, with the corresponding degrees of freedom. 10,000 permutations were used to determine the final corrected P value.
However, as discussed earlier, conducting the percentage permutation test on the whole brain for the log Jacobian is bound to yield a negative mean log Jacobian value. This is because the region-wise mean of n log Jacobian maps, each of which with negative region-wise mean is again negative.
| (11) |
Because we actually expect the mean log Jacobian to be negative, it is not ideal to test that the mean log Jacobian is zero (versus negative)—in fact, just doing the typical t test and generating the permutation distribution from the various permutations to assess the significance will almost always (if not always) end up being significant.
Instead, it is better to use a second permutation test, referred to as the extreme statistics permutation test, to conduct corrections for multiple comparisons on the voxelwise mean log Jacobian. In this permutation test, we still resample the distribution as described previously. However, instead of ranking the percentages for all permutations, we collect and rank the maximal and minimal re-sampled T statistics inside the ICBM space for all permutations (as described in Nichols and Holmes, 2001). At a 0.05 α level with 10,000 permutations, the (1 − 0.05) * 10,000th most extreme statistics are then used to threshold the voxelwise T map, that is, T statistics exceeding this threshold are considered significant. In this paper, we only conduct the extreme statistics permutation tests on the mean log Jacobian, while the percentage permutation test is used for assessing both mean and deviation.
Results
The deviation of the logged Jacobian maps from zero will be discussed first followed by statistical testing on the mean absolute change.
Voxelwise deviation
Sequence effect
To determine the order of performance for the different MRI sequences studied in this paper, we divided our maps of change into four groups. These were determined by the coil type and the presence of N3 correction. Within each group, we compared the performance of MP-RAGE, SPGR, IR-SPGR and Synthetic T1 images using permutation tests. The results are shown in Tables 2–5. To visualize the repeatability and deviation of different scan types, Figs. 3 and 4 show the variance maps defined in Eq. (3) and the deviation maps in Eq. (6).
Table 2.
Performance ranking of four sequences acquired using a birdcage coil with N3 correction
| Permutation test (P values) Birdcage with N3 correction
| ||||
|---|---|---|---|---|
| SPGR | MP-RAGE | IR-SPGR | SYN | |
| SPGR | 0.001 (SPGR) | <0.0001 (SPGR) | <0.0001 (SPGR) | |
| MP-RAGE | 0.264 (MP-RAGE) | 0.0081 (MP-RAGE) | ||
| 0.443 (IR-SPGR) | ||||
| IR-SPGR | 0.016 (IR-SPGR) | |||
| SYN | ||||
For all tables, sequence types are listed in the order of performance; the P values are the significance levels in favor of the alternative hypothesis that the sequence in parenthesis has a lower deviation (for example, IR-SPGR has a lower deviation than Synthetic T1 with a significant P value of 0.016). In other words, when the P value is significant, the sequence in parentheses is the one that performs best. Both P values are reported if two pulse sequences are statistically indistinguishable: so, for example, two sequences appear in the MP-RAGE vs. IR-SPGR box. This is because we tested the hypotheses both ways (that is, H1 = MP-RAGE shows less variance than IR-SPGR; and then again, H1 = IR-SPGR shows less variance than MP-RAGE). Note that SPGR in the tables actually refers to SPGR for GE acquisitions and FLASH for Siemens acquisitions.
Table 5.
Performance ranking of four sequences acquired using Phased Array without N3 correction
| Permutation test (P values); Phased Array without N3 correction
| ||||
|---|---|---|---|---|
| SPGR | MP-RAGE | SYN | IR-SPGR | |
| SPGR | 0.050 (SPGR) | 0.038 (SPGR) | <0.0001 (SPGR) | |
| MP-RAGE | 0.132 (MP-RAGE) | 0.014 (MPRRAGE) | ||
| 0.422 (SYN) | ||||
| SYN | <0.0001 (SYN) | |||
| IR-SPGR | ||||
Fig. 3.
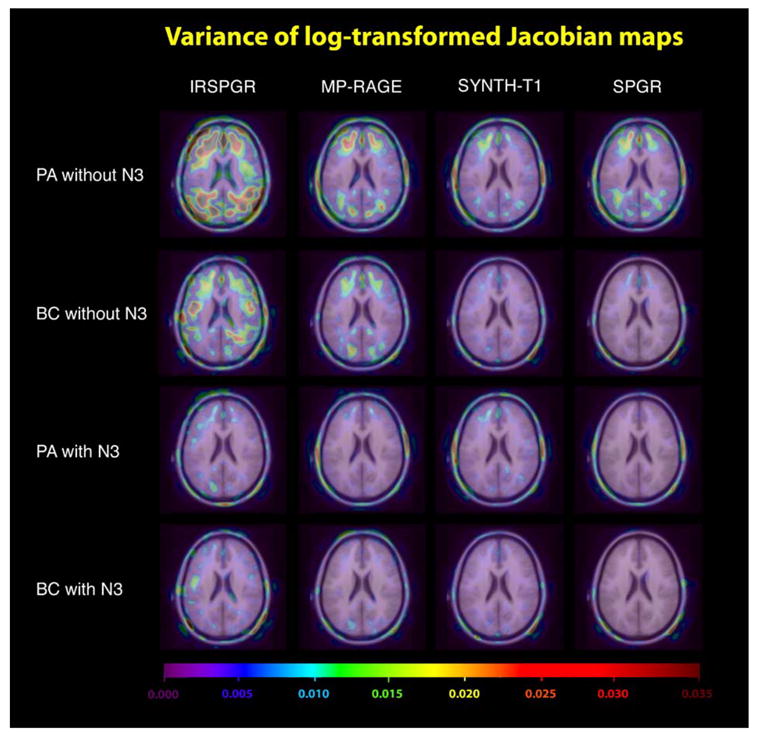
This figure visualizes the repeatability, defined as the voxelwise variance of the log-transformed Jacobian maps, for different sequence types and transmit/receive coils. Note that SPGR in the figures actually refers to SPGR for GE acquisitions and FLASH for Siemens acquisitions, while BC and PA denote bird cage and phased array designs, respectively.
Fig. 4.
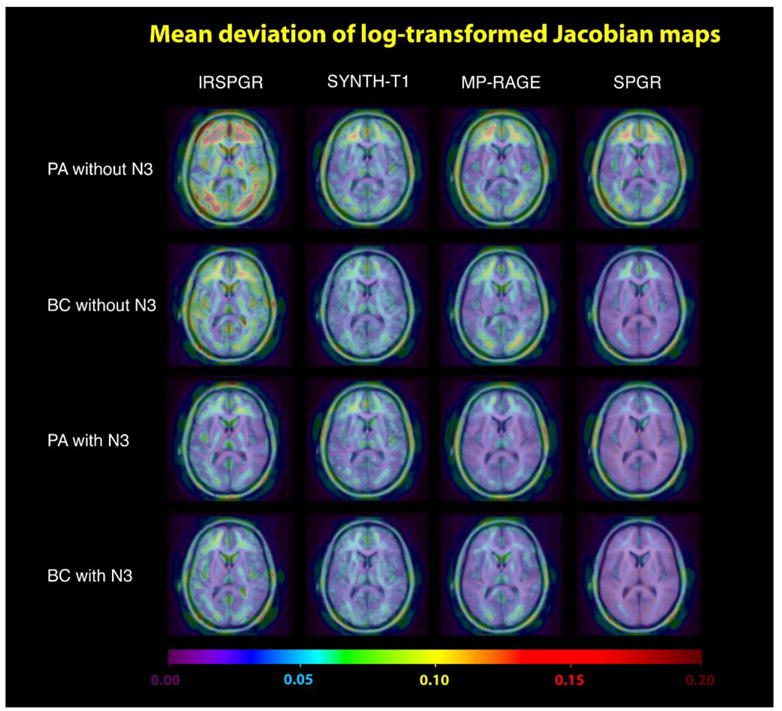
This figure visualizes the performance, measured by the voxelwise absolute mean deviation from zero of the log-transformed Jacobian maps, for different sequence types. Based on the assumption of no brain structural difference, a sequence is defined to have better performance if it has a lower absolute mean deviation. Visually, SPGR acquired using a birdcage coil with N3 correction performs the best overall. This is confirmed using permutation tests as shown in Tables 2–5.
SPGR exhibited the lowest deviation regardless of the coil type or the presence of N3 correction. With N3 correction, the orders of performance were similar for both birdcage and Phased Array. In these two cases, the image type with the greatest deviation was found to be Synthetic T1, while there was no statistical difference between MP-RAGE and IR-SPGR. However, without N3 correction, statistical significance was detected supporting that IR-SPGR has the greatest deviation when Phased Array coils were used (outperformed by both MP-RAGE and Synthetic T1, while MP-RAGE and Synthetic T1 are statistically indistinguishable). With birdcage coils without N3 correction, MP-RAGE was statistically indistinguishable from both Synthetic T1 and IR-SPGR (although Synthetic T1 scans outperformed IR-SPGR in their head-to-head statistical test). Comparing the results across N3 correction, it was noted that Synthetic T1, although it fared well against both MP-RAGE and IR-SPGR without N3 correction, had the greatest deviation once N3 correction is applied. The results suggested that N3 correction had a huge impact on the performance of scans except for Synthetic T1, undoubtedly due to the fact that Synthetic T1 is a calculated T1 image. B1 non-uniformities are represented equally in the spin density and the T1-weighted image volumes used to construct the Synthetic T1 image volume, so the calculated images are thus less sensitive to underlying intensity inhomogeneity in the first place.
Synthetic T1 differences
Inherent differences in the Synthetic T1 imaging technique are also noticeable in Fig. 3 (repeatability map). As shown, Synthetic T1 has a low variance for PAwithout N3 correction (visually better than both SPGR and MP-RAGE), although this high repeatability does not translate to higher performance/lower deviation. The permutation test (Table 5) and the deviation map confirm this. Moreover, N3 both visually and significantly improved the repeatability for SPGR, IR-SPGR and MP-RAGE, but not for Synthetic T1 images. This again is consistent with the fact that the Synthetic T1 imaging technique is fundamentally different from other scan types, that is, it is a quantitative image of the relaxometric parameter T1 rather than a T1-weighted image (Table 6).
Table 6.
Performance ranking of SPGR across coil type and N3 correction
| Permutation test (P values); SPGR across coil type (BC/PA) and N3 correction (+N3/−N3)
| ||||
|---|---|---|---|---|
| BC+N3 | PA+N3 | BC−N3 | PA−N3 | |
| BC+N3 | 0.088 (BC+N3) | <0.0001 (BC+N3) | <0.0001 (BC+N3) | |
| 0.828 (PA+N3) | ||||
| PA+N3 | 0.107 (PA+N3) | <0.0001 (PA+N3) | ||
| 0.066 (BC−N3) | ||||
| BC−N3 | 0.005 (BC−N3) | |||
| PA−N3 | ||||
In this table, +N3 and −N3 denote with and without N3 correction respectively.
N3/coil type effects
To further establish the impact of N3 correction with different transmit/receive coil types on the performance of scans, we used the sequence with the lowest deviation, i.e., SPGR, and compared its performance using 4 different combinations (i.e., with/without N3 correction; BC/PA coil type). The results are shown in Table 4. To summarize, N3 correction visually improves the repeatability for both coil types, as shown in Fig. 3. There was also a statistically significant reduction in deviation for both the BC and PA coil types. Without N3 correction, BC yields a lower deviation than Phased Array (P = 0.005), while with N3, BC only outperforms PA at trend level (P = 0.088). In more detailed comparison, we noticed that BC without N3 correction is statistically indistinguishable from PA with N3 correction (PA outperforms BC: P = 0.107, BC outperforms PA: P = 0.066). N3 correction therefore statistically erased the difference in coil types for SPGR, supporting the hypothesis that, for TBM, the most important differences among coil types are intensity inhomogeneities correctable using N3.
Table 4.
Performance ranking of four sequences acquired using a birdcage without N3 correction
| Permutation test (P values); Birdcage without N3 correction
| ||||
|---|---|---|---|---|
| SPGR | SYN | MP-RAGE | IR-SPGR | |
| SPGR | <0.0001 (SPGR) | 0.0009 (SPGR) | <0.0001 (SPGR) | |
| SYN | 0.158 (SYN) | <0.0001 (SYN) | ||
| 0.117 (MP-RAGE) | ||||
| MP-RAGE | 0.104 (MP-RAGE) | |||
| 0.532 (IR-SPGR) | ||||
| IR-SPGR | ||||
B0 correction
Since only 6 subjects included in this study (Table 1) had both B0-corrected (i.e., MEDIC) and non-B0-corrected data, we combined BC and PA (i.e., 12 pairs of serial scans) to study the B0 correction effect. Images of these 12 pairs with GW and N3 correction were analyzed (Fig. 5), and the permutation results were inconclusive at the 0.05 level (B0 correction outperforms no correction: P = 0.641; no B0 correction outperforms B0 correction: P = 0.063).
Fig. 5.
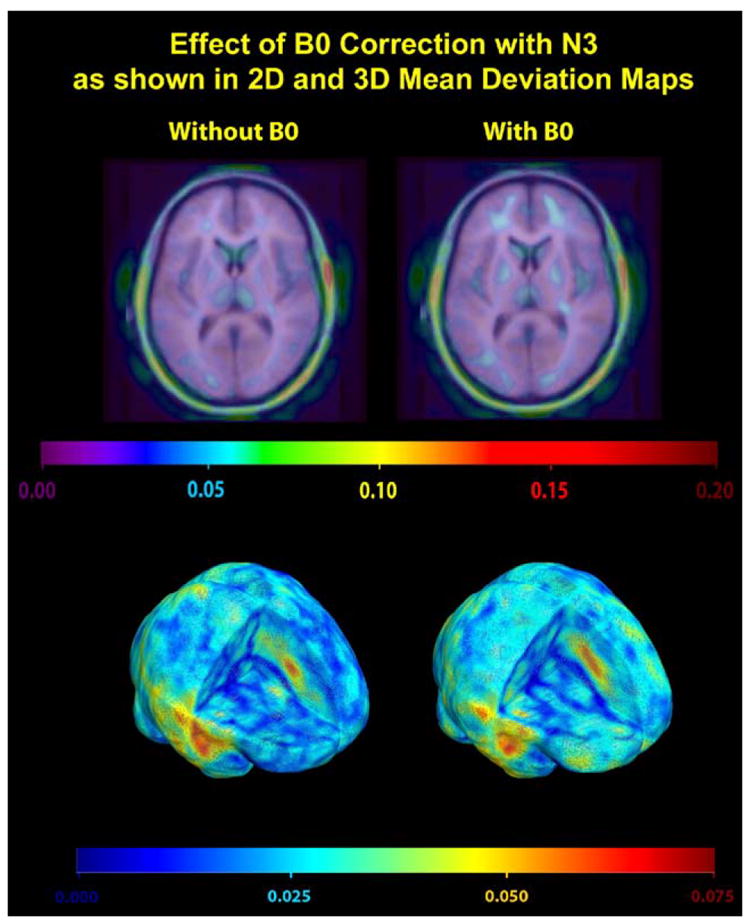
This figure visualizes the comparison of B0-corrected images (MEDIC) vs. GW and B1 intensity-corrected MP-RAGE images with N3 correction (N = 12 scans). A permutation test established no statistically significant differences between the two image types, although an effect may be detectable in a larger sample of scans. As in Fig. 4, the performance, coded here in color, is defined as the voxelwise absolute mean deviation from zero of the log-transformed Jacobian maps.
Voxelwise mean log Jacobian maps
Both the percentage and the extreme statistics permutation tests were conducted on all four sequence types acquired using BC with and without N3. These results are summarized in Table 7. In the case of a percentage permutation test, two P values (p1 and p2) are reported that test the positivity and the negativity of the mean log Jacobian respectively. Based on the percentage permutation test, all sequences with N3 correction were confirmed to have a statistically significant negative mean as suggested in our discussions on region-wise mean, while only SPGR and Synth-T1 had a significantly negative mean when no N3 correction is applied. We hypothesized that this significance without N3 correction was probably due to the overall lower deviation of SPGR and Synth-T1 acquired using BC, consistent with the findings in Table 4. Moreover, on closer inspection, all MRI sequences have p2 values lower than p1, suggesting the negativity of the region-wise mean, with a further decrease in p2 values after N3 correction (except for Synth-T1), suggesting a greater effect size. These findings again support the hypothesis that N3 correction greatly reduces the deviation and increases power in statistical tests using TBM.
Table 7.
Significance levels when testing the positivity or negativity of the region-wise mean using permutation tests for different sequence types
| P1 | P2 | Maximum T | Minimum T | T threshold | |
|---|---|---|---|---|---|
| SPGR BC+N3 | 0.394 | 0.008 | 4.42 | −6.45 | ±7.35 |
| SPGR BC−N3 | 0.471 | 0.015 | 5.55 | −8.33 | ±7.3 |
| MP-RAGE BC+N3 | 0.362 | 0.046 | 9.90 | −7.30 | ±7.03 |
| MP-RAGE BC−N3 | 0.976 | 0.301 | 4.93 | −6.49 | ±6.90 |
| IR-SPGR BC+N3 | 0.173 | 0.038 | 7.49 | −9.99 | ±10.58 |
| IR-SPGR BC−N3 | 0.992 | 0.303 | 5.54 | −8.96 | ±10.8 |
| SYN BC+N3 | 0.827 | 0.039 | 5.40 | −6.02 | ±7.29 |
| SYN BC−N3 | 0.772 | 0.021 | 5.28 | −6.22 | ±7.28 |
P1 (P2) is the significance level of the alternative hypothesis that the region-wise mean is positive (negative), while Max (Min) T is the maximal (minimal) voxelwise T statistic inside the ROI defined by a mask of the average brain template (ICBM-53) in ICBM space. The T threshold is the corrected cut-off T value obtained using 10,000 permutations on the maximum/minimum statistic (at the P = 0.05 level). Voxels with T values more extreme than the corrected cut-off T threshold are considered active (since we should detect no change within a 2-week period, these voxels are considered false positives under the null hypothesis).
Effects of N3
The extreme statistics permutation test yielded more interesting results (Table 7). We noticed that some voxels in the maps for SPGR without N3 correction and MP-RAGE with N3 correction were found to have T statistics more extreme than their respective multiple-comparisons-corrected T threshold (although we should assume that these voxels are false positives). Thus, N3 correction does not seem to change the negative region-wise mean. However, it might alter the regional statistical properties of some voxels. This suggests that N3 correction should be applied with caution, especially when interpreting group mean log Jacobian map encoding regional tissue loss or expansion, as the application of N3 might influence the statistical properties/conclusions at some voxels. Therefore, it is recommended that mean log Jacobian maps be constructed both with and without N3 correction followed by multiple comparisons correction. One should then carefully examine regions identified as undergoing tissue shrinkage or expansion based on maps obtained by either using or not using N3. A region should not be declared to have statistically significant local tissue change when the two results are not consistent.
Discussion
In this paper, we examined the robustness of different MRI scan types for mapping brain changes using tensor-based morphometry. We found that SPGR acquired using the birdcage design with N3 correction was the most stable sequence with least deviation. While in theory a phased array design increases the signal to noise ratio relative to a birdcage design, the latter yielded a lower deviation in our comparison test. This is probably because regularizers are always applied to deformation fields in TBM analyses. Thus, the noise level of the images, so long as it is within a certain acceptable range, plays a less crucial role in determining the performance of a particular scan type. On the other hand, phased array receive coils are more prone to image intensity inhomogeneities. In theory, the B1 correction technique should largely remove this, but our statistical tests did not support the assumption that a B1-corrected phased array design clearly outperforms a birdcage design (notice that, in this paper, no statistical testing was directly performed on the effect of B1 correction as the B1 correction was built-in for all phased array images). In fact, without N3 correction, B1-corrected phased array images had greater deviation than those acquired using a birdcage transmit/receive coil. This difference became non-significant after applying N3. It is therefore safe to assume that B1 correction does not entirely remove the RF inhomogeneity as N3 correction further improves phased array images. N3 also removes most of the differences between coil types in terms of RF homogeneity, supported by the disappearance of performance differences after the application of N3. For TBM, the most relevant difference between coil types is RF homogeneity (rather than signal to noise ratio), and N3 is an effective correction in removing this artifact.
Synthetic T1 imaging gave inherently different results relative to other non-calculated pulse sequences. Synthetic T1 images showed good repeatability even without N3, but not much improvement was seen after applying N3. Moreover, repeatability was visually more comparable between Synthetic T1 images acquired using different coil types than other sequence types. The high repeatability of Synthetic T1 does not translate into a better performance, which is harder to interpret. One possible explanation is that there is a difference in the baseline mean log J field for this scan type compared to other non-calculated scan types. We also studied the effect of B0 correction for spatial distortion, and the results could not confirm a statistical improvement, although a larger sample of scans might be required to detect a subtle effect.
Although we concluded that high SNR matters less than good intensity homogeneity for tensor-based morphometry, the lower SNR for the MP-RAGE scans may have handicapped that sequence relative to SPGR in these analyses. We ultimately changed the acquisition parameters to boost SNR on the 1.5 T birdcage coils to correct this, and the data used for the analyses here may have under-represented the optimal performance of MP-RAGE relative to SPGR in terms of SNR. Because the poor SNR in the birdcage scans was easily correctable, it was not considered a fundamental feature/flaw of the MP-RAGE sequence.
This paper only reports the TBM analysis of longitudinal data acquired with the purpose of determining the optimal MRI pulse sequence for the Alzheimer’s Disease Neuroimaging Initiative. In addition to the longitudinal data, cross-sectional studies comparing controls to subjects with Alzheimer’s Disease were acquired. Furthermore, all the MRI data were also analyzed by the following methods that rely on, and exploit, different aspects of image quality: atlas-based measurements of hippocampal volume (Haller et al., 1997; Hsu et al., 2002), the boundary shift integral (Fox and Freeborough, 1997; Fox et al., 2000), voxel-based morphometry using Statistical Parametric Mapping (VBM; Ashburner and Friston, 2000), cortical thickness measures (Fischl and Dale, 2000) and tensor-based morphometry (TBM; Studholme et al., 2001; Leow et al., 2005a,b). The results of these studies, and the data and rationale for selecting MRI pulse sequences for the Alzheimer’s Disease Neuroimaging Initiative, will be reported elsewhere. Fundamentally, one of the inevitable and predicted problems of evaluating reproducibility of MRI scanning at short intervals is that we were able to assess the sequences in terms of insensitivity to noise (in that controls scanned 2 weeks apart should show little change) but we were unable to assess the sequences’ sensitivity to real change (e.g., the ability to pick up change in Alzheimer’s disease over a year). The final decision on specific imaging protocols therefore involved both quantitative and qualitative assessments of sequence performances and was not therefore based solely on evaluations of change over short intervals as detected using TBM.
In this study, after some discussion, we did not randomize the order of the pulse sequences because this would have been difficult logistically. This leaves open the possibility that some systematic bias might have been introduced (e.g., greater motion in later sequences). In mitigation, the subjects were normal controls, and so the problems of excessive motion or loss of concentration are not as marked as in AD patients. The imaging protocol was reviewed and approved by a panel of experienced MR professionals and was designed to reduce human errors related to data acquisition in the preparatory phase. It was decided that randomizing the scan order would increase the complexity of prescribing the acquisition parameters and increase the likelihood of technologist errors in collecting the data.
It is also not known which biological sources of variation contributed most to the residual deformations observed in this serial MRI data. Regardless of the imaging protocol, the ultimate geometrical stability of the brain scanned over time is limited by biological changes in brain tissue hydration (which also may impact T1 or T2 contrast), mechanical effects such as ventricular deformation (which may be minimized by consistent placement of the subject in the scanner), as well as other short-term physiological changes. Oatridge et al. (2001) have reported a change in CSF volume later in the menstrual cycle in women, and other studies have noted minor brain volume variations during normal pregnancy or with jet-lag or alcohol intake (Oatridge et al., 1998, 1999). Better understanding of these relatively short-term reversible biological effects may lead to improved statistical methods to model and adjust for them in studies of serial anatomical change.
In this paper, we applied logarithmic transformation to all Jacobian maps before conducting statistical analysis. Log transformation of a Jacobian determinant field has become standard practice in most TBM papers. The Jacobian determinant of a diffeomorphic (smooth) map is bounded below by zero but unbounded above, so, at any voxel, its null distribution would be a better fit to a symmetric normal distribution if the Jacobians are logged.
Another observation supporting the use of logarithmic transformation comes from registering two images where no difference other than noise is present. We expect the chosen (unbiased) statistic to pick up no statistically significant change between these two images. In a classical statistical setting, one would hope that statistic used to estimate change might follow a Gaussian distribution with zero mean. This again suggests that some symmetrizing should be applied to Jacobian maps, leading to the use of logarithmic transform. Unfortunately, the resulting distribution does not have zero mean: as we showed in Leow et al. (2005a,b), log-transformed Jacobian maps always lead to biased estimates (i.e., have negative means under null conditions), and this problem occurs even if log Jacobian maps are analyzed at the voxel-by-voxel level.
We also showed, using Kullback–Liebler distances on material density functions, that the integral of the logged Jacobian map of any volume-preserving transformation (not just inverse-consistent mappings) with respect to an image domain is always negative inside this domain. Moreover, inverse-consistent mappings constructed by symmetrizing regularizers (in the form of differential operators) integrate to a less negative number than their inconsistent counterparts applied to the same data.
This negative mean bias is not introduced by integrating the logged Jacobian field over a region as the same bias even occurs when considering log-transformed Jacobian maps at the voxelwise level (averaging across subjects at a single voxel). By utilizing multiple copies of the artificial Jacobian map presented in Leow et al. (2005a,b) (squeezing half of the domain of interest to an arbitrarily small size while preserving the overall volume/size of the domain), we can now easily construct a collection of Jacobian maps defined in a region stable in volume/size, whose (voxelwise) mean log Jacobian approaches minus infinity at every voxel. This would incorrectly reject the null hypothesis that the overall volume/size of this domain is unchanged and shows that log Jacobian maps are biased in the sense that they are not zero mean across subjects at each voxel.
One way of alleviating this bias is to use inverse-consistent mappings and to integrate the log Jacobian over a spatial domain, as is done in this paper. We acknowledge that this integral produces a summary quantity whose geometric meaning is harder to grasp, and one could argue that it is preferable to integrate the volume change over a region first before performing the log operation (which would yield the log of deformed volume). Here, we preferred to integrate the log Jacobian, as we did not wish to apply one approach voxelwise, and a different approach when considering statistics across a region. As such, the integral of the logged Jacobian is a regional, if somewhat abstract, summary of the fluctuations in the log Jacobian measure over a region. A somewhat related approach is taken in Pennec et al. (2005), where the deformation tensor of a mapping is logged and integrated over the image domain, and this integral is used as a penalty function (cost functional) to regularize the deformation.
We would also like to comment on our choice of cost function (mutual information) for nonlinear registration. As would be expected for any registration algorithm driven by mutual information, there is a gain in the mutual information of the registered images, for each sequence type, after nonlinear registration. The algorithm works by gradient descent on the deformation parameters to make the mutual information increase from its initial value, subject to other constraints on the smoothness and symmetry of the deformation. Note that the mutual information is not necessarily monotonically increasing with better registration as the registration quality is quantified by the sum of two terms, the mutual information and the energy of the applied deformation field, which describes how much image distortion is required to attain the measured level of signal correspondence. Depending on the application, both geometric and intensity similarity may be important factors in considering image reproducibility, so it is possible that there will be no clear best sequence: some imaging sequences may provide better geometric similarity over time and others may tend to be more similar in terms of intensity. Further work is needed to help assess intensity similarity. For example, an information-theoretic metric might be used to estimate the degree to which the image at time 1 predicts the image at time 2 or how many bits of information are needed to represent the residual information. The final value for the mutual information of two registered images may be difficult to compare objectively across imaging sequences as it depends on the deformation energy allowed in the registration process.
Finally, even though the TBM analysis of longitudinal data from normal subjects in this study indicated that SPGR gave the most robust results (when N3 correction was not used), this should not be interpreted to mean that SPGR is the ‘‘best sequence’’ for longitudinal studies. Selection of MRI pulse sequences is extremely dependent on the needs of the study including the specific hypotheses, patients to be studied, equipment available, analysis techniques and other factors. Therefore, the major message of this report is that TBM is a useful quantitative tool when comparing different methods for studying longitudinal change of the brain. Our results provide statistical information on the baseline repeatability, reproducibility and variability of changes detected in different scan types at an interval short enough to be insensitive to any disease- or age-related structural brain change. The order of performance of different scan types was determined, providing researchers with relevant baseline information when deciding on a particular sequence/scanner or correction type. Our results will be used as a reference for our future serial scan studies of disease in individuals and groups, reducing the possibility of detecting false positive signals.
Table 3.
Performance ranking of four sequences acquired using Phased Array coil with N3 correction
| Permutation test (P values); Phased Array with N3 correction
| ||||
|---|---|---|---|---|
| SPGR | MP-RAGE | IR-SPGR | SYN | |
| SPGR | 0.0003 (SPGR) | 0.0027 (SPGR) | <0.0001 (SPGR) | |
| MP-RAGE | 0.544 (MP-RAGE) | 0.0004 (MP-RAGE) | ||
| 0.263 (IR-SPGR) | ||||
| IR-SPGR | 0.003 (IR-SPGR) | |||
| SYN | ||||
Acknowledgments
This project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI; Principal Investigator: Michael Weiner; NIH grant number U01 AG024904). ADNI is funded by the National Institute of Aging, the National Institute of Biomedical Imaging and Bioengineering (NIBIB) and the Foundation for the National Institutes of Health, through generous contributions from the following companies and organizations: Pfizer Inc., Wyeth Research, Bristol-Myers Squibb, Eli Lilly and Company, Glaxo-SmithKline, Merck and Co. Inc., AstraZeneca AB, Novartis Pharmaceuticals Corporation, the Alzheimer’s Association, Eisai Global Clinical Development, Elan Corporation plc, Forest Laboratories and the Institute for the Study of Aging (ISOA), with participation from the U.S. Food and Drug Administration. The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. Algorithm development for this study was also funded by the NIA, NIBIB, the National Library of Medicine and the National Center for Research Resources (AG016570, EB01651, LM05639, RR019771 to PT). Author contributions were as follows: AL, AK and PT performed the image analyses and CJ and MW designed the overall evaluation of serial MRI reproducibility as part of the preparatory imaging phase of ADNI. AT, AD, MB, PB, JG, CW, JW, BB, NF, DH, JK, NS, CS and GA assisted with the image acquisition, design of the study, quality control, pre-processing, analysis and databasing, and AF recruited subjects at UCSD. We also acknowledge the help of Heidi A. Ward, Ph.D. (GE Healthcare) in investigating IR-SPGR image quality.
References
- Ashburner J, Friston KJ. Voxel-based morphometry—The methods. NeuroImage. 2000 Jun;11(6):805–821. doi: 10.1006/nimg.2000.0582. Pt 1. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Hutton C, Frackowiak R, Johnsrude I, Price C, Friston K. Identifying global anatomical differences: deformation-based morphometry. Hum Brain Mapp. 1998;6(5–6):348–357. doi: 10.1002/(SICI)1097-0193(1998)6:5/6<348::AID-HBM4>3.0.CO;2-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Csernansky J, Davatzikos C, Fox NC, Frisoni G, Thompson PM. Computer-assisted imaging to assess brain structure in healthy and diseased brains. Lancet Neurol. 2003 February;2(2):79–88. doi: 10.1016/s1474-4422(03)00304-1. [DOI] [PubMed] [Google Scholar]
- Bullmore ET, Suckling J, Overmeyer S, Rabe-Hesketh S, Taylor E, Brammer MJ. Global, voxel, and cluster tests, by theory and permutation, for a difference between two groups of structural MR images of the brain. IEEE Trans Med Imag. 1999;18:32–42. doi: 10.1109/42.750253. [DOI] [PubMed] [Google Scholar]
- Cachier P, Rey D. Symmetrization of the non-rigid registration problem using inversion-invariant energies: application to multiple sclerosis. Proc MICCAI. 2000:472–481. [Google Scholar]
- Cao J, Worsley KJ. The geometry of the Hotelling’s T-squared random field with applications to the detection of shape changes. Ann Stat. 1999;27:925–942. [Google Scholar]
- Chung MK, Worsley KJ, Paus T, Cherif C, Collins DL, Giedd JN, Rapoport JL, Evans AC. A unified statistical approach to deformation-based morphometry. NeuroImage. 2001 Sep;14(3):595–606. doi: 10.1006/nimg.2001.0862. [DOI] [PubMed] [Google Scholar]
- Collins DL, Neelin P, Peters TM, Evans AC. Automatic 3D intersubject registration of MR volumetric data into standardized Talairach space. J Comput Assist Tomogr. 1994 March;18(2):192–205. [PubMed] [Google Scholar]
- Crum WR, Scahill RI, Fox NC. Automated hippocampal segmentation by regional fluid registration of serial MRI: validation and application in Alzheimer’s disease. NeuroImage. 2001;13:847–855. doi: 10.1006/nimg.2001.0744. [DOI] [PubMed] [Google Scholar]
- Deoni SC, Rutt BK, Peters TM. Rapid combined T1 and T2 mapping using gradient recalled acquisition in the steady state. Magn Reson Med. 2003 Mar;49(3):515–526. doi: 10.1002/mrm.10407. [DOI] [PubMed] [Google Scholar]
- Fillard P, Pennec X, Thompson PM, Ayache N. Extrapolation of Sparse Tensor Fields: Application to the Modeling of Brain Variability, Information Processing in Medical Imaging (IPMI) 2005. Glenwood Springs; Colorado: 2005. July 11–15, [DOI] [PubMed] [Google Scholar]
- Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A 97. 2000 Sep 26;(20):11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat DH, van der Kouwe AJ, Makris N, Segonne F, Quinn BT, Dale AM. Sequence-independent segmentation of magnetic resonance images. NeuroImage. 2004;23:S69–S84. doi: 10.1016/j.neuroimage.2004.07.016. Suppl 1. [DOI] [PubMed] [Google Scholar]
- Fox NC, Freeborough PA. Brain atrophy progression measured from registered serial MRI: validation and application to Alzheimer’s disease. J Magn Reson Imaging. 1997;7:1069–1075. doi: 10.1002/jmri.1880070620. [DOI] [PubMed] [Google Scholar]
- Fox NC, Freeborough PA, Mekkaoui KF, Stevens JM, Rossor MN. Cerebral and cerebellar atrophy on serial magnetic resonance imaging in an initially symptom free subject at risk of familial prion disease. BMJ. 1997 Oct. 4;315(7112):856–857. doi: 10.1136/bmj.315.7112.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox NC, Scahill RI, Crum WR, Rossor MN. Correlation between rates of brain atrophy and cognitive decline in AD. Neurology. 1999;52:1687–1689. doi: 10.1212/wnl.52.8.1687. [DOI] [PubMed] [Google Scholar]
- Fox NC, Cousens S, Scahill R, Harvey RJ, Rossor MN. Using serial registered brain magnetic resonance imaging to measure disease progression in Alzheimer disease: power calculations and estimates of sample size to detect treatment effects. Arch Neurol. 2000;57:339–344. doi: 10.1001/archneur.57.3.339. [DOI] [PubMed] [Google Scholar]
- Fox NC, Crum WR, Scahill RI, Stevens JM, Janssen JC, Rossor MN. Imaging of onset and progression of Alzheimer’s disease with voxel-compression mapping of serial magnetic resonance images. Lancet. 2001;358:201–205. doi: 10.1016/S0140-6736(01)05408-3. [DOI] [PubMed] [Google Scholar]
- Freeborough PA, Fox NC. The boundary shift integral: an accurate and robust measure of cerebral volume changes from registered repeat MRI. IEEE Trans Med Imag. 1997;16(5):623–629. doi: 10.1109/42.640753. [DOI] [PubMed] [Google Scholar]
- Freeborough PA, Woods RP, Fox NC. Accurate registration of serial 3D MR brain images and its application to visualizing change in neurodegenerative disorders. J Comput Assist Tomogr. 1996;20(6):1012–1022. doi: 10.1097/00004728-199611000-00030. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Holmes AP, Worsley KJ, Poline JP, Frith CD, Frackowiak RSJ. Statistical parametric maps in functional imaging: a general linear approach. Hum Brain Mapp. 1995;2:189–210. [Google Scholar]
- Gaser C, Volz HP, Kiebel S, Riehemann S, Sauer H. Detecting structural changes in whole brain based on nonlinear deformations—Application to schizophrenia research. NeuroImage. 1999 Aug;10(2):107–113. doi: 10.1006/nimg.1999.0458. [DOI] [PubMed] [Google Scholar]
- Ge Y, Grossman RJ, Udupa JK, Wei L, Mannon LJ, Polansky M, Kolson DL. Longitudinal quantitative analysis of brain atrophy in relapsing–remitting and secondary-progressive multiple sclerosis. International Soc of Magnetic Resonance in Medicine. 1999 doi: 10.1148/radiology.214.3.r00mr30665. [DOI] [PubMed] [Google Scholar]
- Hajnal JV, Saeed N, Oatridge A, Williams EJ, Young IR, Bydder GM. Detection of subtle brain changes using subvoxel registration and subtraction of serial MR images. J Comput Assist Tomogr. 1995a;19(5):677–691. doi: 10.1097/00004728-199509000-00001. [DOI] [PubMed] [Google Scholar]
- Hajnal JV, Saeed N, Soar EJ, Oatridge A, Young IR, Bydder GM. A registration and interpolation procedure for subvoxel matching of serially acquired MR images. J Comput Assist Tomogr. 1995b;19(2):289–296. doi: 10.1097/00004728-199503000-00022. [DOI] [PubMed] [Google Scholar]
- Hajnal JV, Hill DLG, Hawkes DJ, editors. Medical Image Registration. CRC Press; New York: 2001. [Google Scholar]
- Haller JW, Banerjee A, Christensen GE, et al. Three-dimensional hippocampal MR morphometry with high-dimensional transformation of a neuroanatomic atlas. Radiology. 1997;202:504–510. doi: 10.1148/radiology.202.2.9015081. [DOI] [PubMed] [Google Scholar]
- Hsu YY, Schuff N, Du AT, et al. Comparison of automated and manual MRI volumetry of hippocampus in normal aging and dementia. J Magn Reson Imaging. 2002;16:305–310. doi: 10.1002/jmri.10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke AL, Zubicaray Gd, Rose SE, Griffin M, Chalk JB, Galloway GJ. 4D deformation modeling of cortical disease progression in Alzheimer’s dementia. Magn Reson Med. 2001 Oct;46(4):661–666. doi: 10.1002/mrm.1243. [DOI] [PubMed] [Google Scholar]
- Jezzard P, Balaban RS. Correction for geometric distortion in echo planar images from B0 field variations. Magn Reson Med. 1995;34:65–73. doi: 10.1002/mrm.1910340111. [DOI] [PubMed] [Google Scholar]
- Jovicich J, Czanner S, Greve D, Haley E, van der Kouwe A, Gollub R, Kennedy D, Schmitt F, Brown G, MacFall J, Fischl B, Dale AM. Reliability in multi-site structural MRI studies: effects of gradient non-linearity correction on phantom and human data. NeuroImage (electronic publication ahead of print) doi: 10.1016/j.neuroimage.2005.09.046. in press. doi: 10.1016/j.neuroimage.2005.09.046. [DOI] [PubMed] [Google Scholar]
- Lemieux L, Wieshmann UC, Moran NF, Fish DR, Shorvon SD. The detection and significance of subtle changes in mixed-signal brain lesions by serial MRI scan matching and spatial normalization. Med Image Anal. 1998;2(3):227–242. doi: 10.1016/s1361-8415(98)80021-2. [DOI] [PubMed] [Google Scholar]
- Leow AD, Huang SC, Geng A, Becker JT, Davis SW, Toga AW, Thompson PM. Inverse consistent mapping in 3D deformable image registration: its construction and statistical properties, Information Processing in Medical Imaging (IPMI) 2005. Glenwood Springs; Colorado: 2005a. July 11–15, 2005, pp. 493–503. LNCS 2565. [DOI] [PubMed] [Google Scholar]
- Leow AD, Yu CL, Lee SJ, Huang SC, Nicolson R, Hayashi KM, Protas H, Toga AW, Thompson PM. Brain structural mapping using a novel hybrid implicit/explicit framework based on the level-set method. NeuroImage. 2005b Feb 1;24(3):910–927. doi: 10.1016/j.neuroimage.2004.09.022. [DOI] [PubMed] [Google Scholar]
- Miller MI, Trouve A, Younes L. On the metrics and Euler–Lagrange equations of computational anatomy. Annu Rev Biomed Eng. 2002;4:375–405. doi: 10.1146/annurev.bioeng.4.092101.125733. [DOI] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L. The Alzheimer’s Neuroimaging Initiative; Proceedings of the ADPD Meeting; Sorrento. 2005. submitted for publication (a) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L. The Alzheimer’s Disease Neuroimaging Initiative. Neuroradiological Clinics of North America. doi: 10.1016/j.nic.2005.09.008. submitted for publication (b) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Petersen RC, Jack CR, Jagust W, Trojanowski JQ, Toga AW, Beckett L. Ways towards an early diagnosis in Alzheimer’s disease: the Alzheimer’s Disease Neuroimaging Initiative, Alzheimer’s and Dementia. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayana PA, Brey WW, Kulkarni MV, Sievenpiper CL. Compensation for surface coil sensitivity variation in magnetic resonance imaging. Magn Reson Imaging. 1988 May–Jun;6(3):271–274. doi: 10.1016/0730-725x(88)90401-8. [DOI] [PubMed] [Google Scholar]
- Nichols TE, Holmes AP. Nonparametric analysis of PET functional neuroimaging experiments. Hum Brain Mapp. 2001;15:1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatridge A, Saeed N, Hajnal JV, Puri BK, Mitchell L, Holdcroft A, Bydder GM. Quantification of brain changes seen on serially registered MRI in normal pregnancy and pre-eclampsia. Proc Int Soc Magn Reson Med. 1998;6:1381. [Google Scholar]
- Oatridge A, Saeed N, Hajnal JV, Puri BK, Holdcroft A, Fusi L, Bydder GM. Quantification of brain and ventricular size in pregnancy and pre-eclampsia. Proc Int Soc Magn Reson Med. 1999;7:373. [Google Scholar]
- Oatridge A, Hajnal JV, Bydder GM. Registration and subtraction of serial magnetic resonance images of the brain: image interpretation and clinical applications. In: Hajnal JV, Hawkes DJ, Hill DLG, editors. Medical Image Registration. CRC Press; Florida: 2001. [Google Scholar]
- O’Brien JT, Paling S, Barber R, Williams ED, Ballard C, McKeith IG, Gholkar A, Crum WR, Rossor MN, Fox NC. Progressive brain atrophy on serial MRI in dementia with Lewy bodies, AD, and vascular dementia. Neurology. 2001;56(10):1386–1388. doi: 10.1212/wnl.56.10.1386. [DOI] [PubMed] [Google Scholar]
- Pennec X, Stefanescu R, Arsigny V, Fillard P, Ayache N. Riemannian elasticity: a statistical regularization framework for nonlinear registration. MICCAI. 2005:943–950. doi: 10.1007/11566489_116. LNCS 3750. [DOI] [PubMed] [Google Scholar]
- Rey D, Subsol G, Delingette H, Ayache N. Automatic detection and segmentation of evolving processes in 3D medical images: application to multiple sclerosis. Med Image Anal. 2002 Jun;6(2):163–179. doi: 10.1016/s1361-8415(02)00056-7. [DOI] [PubMed] [Google Scholar]
- Schmid MR, Pfirrmann CW, Koch P, Zanetti M, Kuehn B, Hodler J. Imaging of patellar cartilage with a 2D multiple-echo data image combination sequence. AJR Am J Roentgenol. 2005 Jun;184(6):1744–1748. doi: 10.2214/ajr.184.6.01841744. [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, Evans AC. A non-parametric method for automatic correction of intensity non-uniformity in MRI data. IEEE Trans Med Imag. 1998;17:87–97. doi: 10.1109/42.668698. [DOI] [PubMed] [Google Scholar]
- Smith SM, De Stefano N, Jenkinson M, Matthews PM. Measurement of brain change over time, FMRIB Technical Report TR00SMS1. 2002 http://www.fmrib.ox.ac.uk/analysis/research/siena/siena/siena.html.
- Studholme C, Cardenas V, Schuff N, Rosen H, Miller B, Weiner MW. Detecting spatially consistent structural differences in Alzheimer’s and frontotemporal dementia using deformation morphometry. MICCAI. 2001:41–48. [Google Scholar]
- Thompson PM, Toga AW. A surface-based technique for warping 3-dimensional images of the brain. IEEE Trans Med Imag. 1996a Aug;15(4):1–16. doi: 10.1109/42.511745. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Toga AW. Visualization and mapping of anatomic abnormalities using a probabilistic brain atlas based on random fluid transformations. In: Höhne K-H, Kikinis R, editors. Lecture Notes in Computer Science (LNCS) Vol. 1131. Springer-Verlag; 1996b. pp. 383–392. [Google Scholar]
- Thompson PM, Toga AW. A framework for computational anatomy (Invited Paper) Comput Vis Sci. 2002;5:1–12. [Google Scholar]
- Thompson PM, MacDonald D, Mega MS, Holmes CJ, Evans AC, Toga AW. Detection and mapping of abnormal brain structure with a probabilistic atlas of cortical surfaces. J Comput Assist Tomogr. 1997 Jul–Aug;21(4):567–581. doi: 10.1097/00004728-199707000-00008. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Giedd JN, Woods RP, MacDonald D, Evans AC, Toga AW. Growth patterns in the developing brain detected by using continuum-mechanical tensor maps. Nature. 2000;404(6774):190–193. doi: 10.1038/35004593. [DOI] [PubMed] [Google Scholar]
- Woods RP. Characterizing volume and surface deformations in an atlas framework: theory, applications, and implementation. NeuroImage. 2003 Mar;18(3):769–788. doi: 10.1016/s1053-8119(03)00019-3. [DOI] [PubMed] [Google Scholar]
- Worsley KJ. Local maxima and the expected Euler characteristic of excursion sets of Chi-squared, F and t fields. Adv Appl Probab. 1994;26:13–42. [Google Scholar]
- Worsley KJ, Andermann M, Koulis T, MacDonald D, Evans AC. Detecting changes in nonisotropic images. Hum Brain Mapp. 1999;8(2–3):98–101. doi: 10.1002/(SICI)1097-0193(1999)8:2/3<98::AID-HBM5>3.0.CO;2-F. [DOI] [PMC free article] [PubMed] [Google Scholar]


