Abstract
Robertsonian (Rb) translocation chromosomes occur in human and murine cancers and involve the aberrant joining of two acrocentric chromosomes in humans and two telocentric chromosomes in mice. Mechanisms leading to their generation remain elusive, but models for their formation have been proposed. They include breakage of centromeric sequences and their subsequent fusions, centric misdivision, misparing between highly repetitive sequences of p-tel or p-arm repeats, and recombinational joining of centromeres and/or centromeric fusions. Here, we have investigated the role of the oncoprotein c-Myc in the formation of Rb chromosomes in mouse cells harboring exclusively telocentric chromosomes. In mouse plasmacytoma cells with constitutive c-Myc deregulation and in immortalized mouse lymphocytes with conditional c-Myc expression, we show that positional remodeling of centromeres in interphase nuclei coincides with the formation of Rb chromosomes. Furthermore, we demonstrate that c-Myc deregulation in a myc box II-dependent manner is sufficient to induce Rb translocation chromosomes. Because telomeric signals are present at all joined centromeres of Rb chromosomes, we conclude that c-Myc mediates Rb chromosome formation in mouse cells by telomere fusions at centromeric termini of telocentric chromosomes. Our findings are relevant to the understanding of nuclear chromosome remodeling during the initiation of genomic instability and tumorigenesis.
Keywords: Centromere, nuclear organization, Robertsonian chromosome, c-Myc, genomic instability
Introduction
c-Myc deregulation is frequent in human and murine cancers [1–3]. In fact, > 70% of all cancers—including breast, ovarian, prostate, colon, liver, and gastric cancers; neuroblastoma; myeloma; and Burkitt's lymphoma [1,2] (http://www.myccancergene.org/site/cancerDB.asp)—have a known Myc component. Invariably, these cancers display Myc oncoprotein deregulation/overexpression, which is achieved through various mechanisms, such as enhanced mRNA and/or protein stability, myc gene amplification, or myc activation through chromosomal translocation. Because so many cancers show a direct association with Myc, current research by many groups is dedicated to elucidating the role(s) these oncoproteins play in tumor initiation and promotion [3–5] and to exploring them as potential therapeutic targets [6].
c-Myc is a potent inducer of genomic instability (for reviews, see Kuttler and Mai [5] and Mai and Mushinski [7]). Recently, it has become apparent that c-Myc not only acts on the level of genes [8] and chromatin [9,10] but also actively contributes to the remodeling of chromosome and telomere positions in interphase nuclei [11] (for reviews, see Mai and Garini [12,13]). Remodeling of chromosome positions leads to temporary alterations in the spatial organization of chromosomes in the nucleus. As we have shown recently, nuclear reorganization of telomeres may result in the formation of telomeric aggregates and may lead to the formation of dicentric chromosomes [11]. In this study, transient c-Myc deregulation mediated the formation of telomeric aggregates in interphase nuclei. Telomeric aggregates are clusters of telomeres that cannot be further resolved at a resolution of 200 nm [14]. As assessed by metaphase chromosome preparations, such telomeric aggregates represent, in part, telomeric fusions that generate dicentric chromosomes and initiate breakage-bridge-fusion (BBF) cycles. Louis et al. [11] also demonstrated that chromosomes alter their positions when c-Myc is experimentally deregulated. Together, these two events of c-Myc-dependent nuclear remodeling profoundly alter the genomic stability of the cell, directly resulting in BBF cycles and karyotypic abnormalities such as terminal deletions and unbalanced translocations. Consequently, more complex chromosomal changes evolve [11–13].
In this present study, we have investigated whether c-Myc plays an initiating role in the remodeling of centromeric positions in interphase nuclei, using mouse cells. The karyotype of standard laboratory mouse strains consists of 40 telocentric chromosomes. However, some wild-type and laboratory mouse strains contain centromerically fused chromosomes, also known as Robertsonian (Rb) chromosomes [15–18].
The three-dimensional (3D) nuclear organization of mouse centromeres in primary, immortalized, and tumor cells has been recently determined using mouse lymphocytes with telocentric chromosomes [19]. This study showed that nuclear centromeric positions typical of primary mouse lymphocytes are significantly altered in immortalized and malignant mouse lymphocytes. The malignant lymphocytes examined in this study were mouse plasmacytoma cells (i.e., cells with c-myc activation through chromosomal translocation). In this present study, we therefore focused on the potential impact of c-Myc on alterations in 3D nuclear distributions of mouse centromeres. We reasoned that nuclear centromere positions might have been altered due to cellular transformation and/or nuclear remodeling as a result of c-Myc oncogene activation. To distinguish between these possibilities, we investigated, using a model of conditional and constitutive c-Myc oncoprotein deregulation, whether c-Myc deregulation was sufficient to mediate changes in 3D nuclear centromere positions. Moreover, we determined whether a potential c-Myc-dependent remodeling of the overall centromere organization permitted the generation of Rb chromosomes. To address the specificity of c-Myc-dependent effects on chromosome organization, we examined Δ106-Myc, a myc box II deletion mutant Myc protein [20]. The latter does not confer tumorigenic potential when overexpressed in mouse proB lymphocytes (Ba/F3) [21] and is unable to mediate the formation of telomeric aggregates and dicentric chromosomes in these cells [22]. The present study shows that c-Myc, in a myc box II-dependent manner, mediates the formation of Rb chromosomes during the remodeling of centromere positions and by telomere-telomere fusions at the telocentric termini of mouse chromosomes.
Materials and Methods
Cells and Cell Culture Conditions
All cells used are listed in Table 1. Primary splenic lymphocytes and primary plasmacytoma cells (PCT1G1) were directly isolated from mice without any in vitro cultivation from T38HxBalb/c mice (Central Animal Care protocol no. 02-039/1/2/3). PreB lymphocytes [11,23] were grown in RPMI 1640 with 10% fetal bovine serum (FBS), 1% l-glutamine, 1% sodium pyruvate, 1% penicillin-streptomycin, and 0.1% β-mercaptoethanol (Invitrogen/Gibco, Burlington, ON, Canada) at 37°C in a humidified atmosphere and 5% CO2. Cells were maintained at a density of around 105 to 106 cells/ml. The plasmacytoma cell line MOPC460D (a gift from Dr. J. F. Mushinski, National Institutes of Health) was grown in RPMI 1640 with 10% FBS, 1% l-glutamine, 1% sodium pyruvate, 1% penicillin-streptomycin (all the above reagents are from Invitrogen/Gibco), and 100 µl of interleukin-6 hybridoma supernatant per 10-ml plate at 37°C in a humidified atmosphere and 5% CO2. Ba/F3 cells [20,21] with Δ106-MycER were grown in RPMI 1640 (Invitrogen, Burlington ON, Canada), containing 10% FBS (Gibco, Burlington ON, Canada), 1% WEHI supernatant (interleukin-3), and 0.021% plasmocin (Cayla, Toulouse, France). Cells were grown and maintained at a density of approximately 105 to 106 cells/ml.
Table 1.
List of Cells Used in This Study.
| Cells Studied | Characteristics | Karyotype |
| Primary mouse lymphocytes | Diploid | 40,XX in all cells |
| Immortalized preB cells carrying MycER | Diploid; nontumorigenic in the absence of MycER activation [11,23] | 40,XX in all cells |
| Plasmacytoma cell line MOPC460D | Near-tetraploid; from female BALB/c mouse; tumorigenic [11] | 75–82,XXXX,T(12;15) |
| Primary plasmacytoma cell line PCT1G1 | Near tetraploid; v-abl/myc-induced; tumorigenic (unpublished data) | 81,XXXX,T(X;11); TsT(X;11) |
| Immortalized proB cells (Ba/F3) | Near-tetraploid; nontumorigenic; from male BALB/c mouse [20,21] | 69–80,XXYY |
| Carrying MycER | Near-tetraploid; tumorigenic only in the presence of MycER activation [20,21] | 69–80,XXYY |
| Carrying myc box II deletion mutant Δ106-MycER | Near-tetraploid; nontumorigenic in the presence or in the absence of Δ106-MycER activation [20,21] | 69–80,XXYY |
More details on these cells can be found in Materials and Methods and in the accompanying references.
Conditional c-Myc Expression
MycER in preB cells and MycER or Δ106-MycER in Ba/F3 cells were activated by 4-hydroxytamoxifen (4HT; Sigma-Aldrich, Oakville, ON, Canada) at a final concentration of 100 nM in 105 cells/ml [11]. Cells were split 24 hours prior to induction.
Induction of c-Myc was confirmed by fluorescence immunohistochemistry, as described previously [24]. A cytospin was made for each time point and for positive and negative controls (c-Myc-overexpressing mouse plasmacytoma cells and non-Myc-activated resting B lymphocytes, respectively). Anti-c-Myc (N262) primary antibody was used at a dilution of 1:100 (Santa Cruz Biotechnology, Santa Cruz, CA) and visualized by goat anti-rabbit IgG fluorescein isothiocyanate (FITC) secondary antibody at a dilution of 1:100 (Sigma-Aldrich). Imaging and analysis were performed as described in Fest et al. [21].
Cell Fixation and Chromosome Preparations
Cells were directly harvested from mice or from cell culture. They were spun down at 200g for 10 minutes and resuspended in 5 ml of 0.075 M KCl for 10 minutes at room temperature for subsequent 3D fixation of nuclei or for 30 minutes at room temperature for chromosome preparation. For 3D fixation, a hypotonic solution was overlaid with 1 ml of freshly prepared fixative (methanol/acetic acid, 3:1), inverted carefully for a couple of times, and centrifuged at 200g for 10 minutes at room temperature. Thereafter, pellets were resuspended with 5 ml of fixative and washed for two more times as above. This fixation method yielded nuclei ready for 3D image acquisition, and results were identical to those with 3.7% formaldehyde fixation, as also stated elsewhere [25]. The ellipsoid nature of lymphocytes was confirmed by confocal microscopy [22].
For chromosome fixation, the drop fixation method [26] was used. For all assays involving metaphase chromosomes, a minimum of twenty metaphases was scored.
Peptide Nuclei Acid Fluorescence In Situ Hybridization
(PNA-FISH) with Centromeres and Telomeres PNA-FISH was performed on both 3D interphase and two-dimensional (2D) metaphase samples derived from the above cells. A PNA human centromeric probe (Applied Biosystems, Foster City, CA) was custom-made to the sequences listed below.
The PNA centromere probe sequences used in this study are as follows:
Sequence 1: (N-terminus)Flu-OEE-ATTCGTTGGAAACGGGA-EE(C-terminus)
Sequence 2: (N-terminus)Flu-OEE-CACAAAGAAGTTTCTGAG-EE(C-terminus)
Sequence 3: (N-terminus)Flu-OEE-CAGACAGAAGCATTCTCA-EE(C-terminus)
Sequence 4: (N-terminus)Flu-OEE-TGCATTCAACTCACAGAG-EE(C-terminus).
This probe cocktail hybridized to all mouse centromeres (Figure 3).
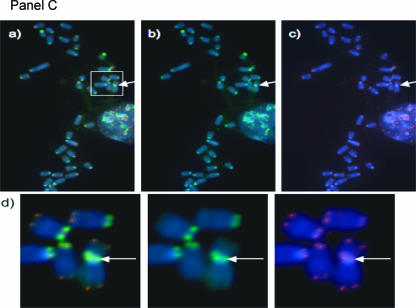
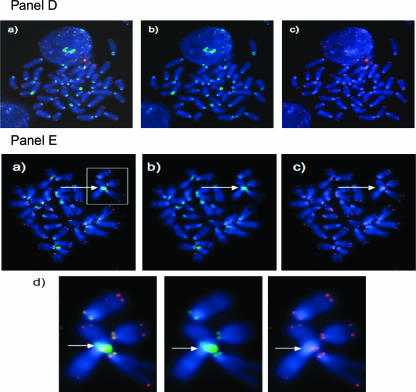
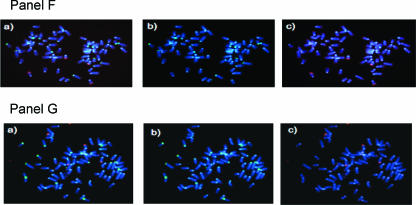
Figure 3.
Centromere-telomere FISH performed on metaphases of mouse lymphocytes. Representative images are shown: Panel (A) primary lymphocyte metaphase from a T38HxBalb/c mouse; Panel (B) metaphase of MOPC460D; Panel (C) partial metaphase of the primary mouse plasmacytoma PCT1G1; Panel (D) preB cell metaphase without MycER activation; Panel (E) preB metaphase 30 hours after MycER activation; Panel (F) Ba/F3 cells without Δ106-MycER activation; Panel (G) Ba/F3 cells 30 hours after Δ106-MycER activation. In each panel, (a) represents dual-color FISH hybridization signals of telomeres (red) and centromeres (green); (b) shows only centromeric signals; and (c) shows only telomeric signals. Rb chromosomes shown in Panel (B), Panel (C), and Panel (E) are highlighted with white boxes and arrows, and then enlarged (d) to observe CTC signals. Note small telomeric signals at Rb fusion in Panel (B, d). The overall telomeric length in MOPC460D is reduced in comparison to the telomeric length of primary B cells and preB cells (data not shown; Figure 1, Panel C).
A PNA telomeric probe was purchased from DAKO (Glostrup, Denmark). 3D fixed interphase nuclei were fixed onto slides using 3.7% formaldehyde/phosphate-buffered saline. The PNA human centromeric probe was denatured at 80°C for 5 minutes and then added to slides in conjunction with the PNA telomeric probe. The slides were denatured at 80°C for 3 minutes, subsequently hybridized for 2 hours at 30°C using the Hybrite system (Vysis; Abbott Diagnostics, Des Plains, IL), and then washed in 70% formamide/2 x SSC. DAPI (0.1 µg/ml) was applied, and, finally, one drop of Vectashield (Vector Laboratories, Burlington, ON, Canada) was added. All slides were imaged right away to avoid changes in imaging conditions and were handled as described in 3D Image Acquisition section.
3D Image Acquisition
Image acquisition was performed on 30 interphase nuclei per cell line using an Axioplan 2 microscope (Carl Zeiss, Inc. Canada) and an AxioCam HR charge-coupled device (Carl Zeiss, Inc. Canada). A 63x/1.4 oil objective lens (Carl Zeiss, Inc. Canada) was used at acquisition times of 300 milliseconds for FITC (centromere), 200 milliseconds for Cy3 (telomere), and 20 to 50 milliseconds for DAPI (nuclei). Eighty to 90 z-stacks were acquired at a sampling distance of xy: 107 nm and z:200 nm for each slice of the stack. Axiovision 3.1 software (Carl Zeiss, Inc. Canada) and constrained iterative algorithm [27] were used for deconvolution.
Scoring of Centromere-Telomere Signals in 3D Nuclei
Centromere-telomere hybridization signals were scored as follows: In nuclei with declustered centromeres, telomere signals that flanked centromeres from one or two sides were counted as normal. In contrast, when telomere signals were flanked by centromere signals on two sides, such signals were scored as aberrant. Telomere-centromere-telomere (TCT) signals thus represent a normal nuclear organization, whereas centromere-telomere-centromere (CTC) signals represent an aberrant nuclear organization. Nuclear domains with clustered centromeres were not included in this analysis. Note that telomeric signals at a distance of ≤ 200 nm will be detected as one signal [14].
Spectral Karyotyping (SKY) Analysis
SKY was performed using the ASI (Applied Spectral Imaging, Vista, CA) kit for mice in accordance with the supplier's hybridization protocols.We used the Spectra Cube (ASI) on an Axioplan 2 microscope (Carl Zeiss, Inc. Canada) with a 63x/1.4 oil objective and the Case Data Manager 4.0 software (ASI) for PC to perform analyses. A minimum of 20 metaphases was examined for preB (induced and noninduced), MOPC460D, T38HxBalb/c, and Δ106 (induced and non-induced). Metaphases were then analyzed for Rb fusions, and matching control time points were statistically compared using Fisher's exact test. P < .05 was considered significant.
Results
Myc and myc Box II-Dependent Nuclear Remodeling of Centromere Positions
The 3D nuclear distribution frequencies of centromeres are significantly changed during immortalization and malignant transformation: Centromeres assume higher distribution frequencies toward central nuclear positions in mouse tumor cells than in immortalized and normal mouse lymphocytes [19]. To determine the consequences of altered centromeric organization for the structural organization of chromosomes, we performed a detailed analysis of the nuclear organization of centromeres and telomeres in primary, immortalized, and malignant mouse lymphocytes (Table 1). These cell lines were chosen to enable the analysis of centromeric positions and their remodeling within the same cell lineage, as cell type-specific variations in centromere positions do not lend themselves to a direct comparison of their respective centromeric distribution frequencies.
Figure 1 highlights representative images of primary mouse lymphocytes, immortalized mouse preB and proB lymphocytes, and tumor cells (mouse plasmacytoma cells) in the absence or in the presence of constitutive or conditional wild-type or myc box II-deletion mutant (Δ106) Myc expression (Table 1). After dual-color hybridization with centromeres (green) and telomeres (red), the nuclei of primary lymphocytes and immortalized preB cells show a predominantly peripheral organization of centromeres; their telomeres are found throughout nuclear space (Figure 1, A and B). In contrast to this nuclear organization, the nuclei of MOPC460D tumor cells with constitutive deregulation of c-Myc protein due to T(12;15) exhibit a more central nuclear distribution of centromeres (Figure 1C). Their telomeres (red) are shorter, and some are found in close association with centromeres (Figure 1C, yellow arrows; data not shown).
Figure 1.
Nuclear centromere and telomere distribution patterns. Representative images of dual-color hybridization of mouse lymphocytes with a centromere probe (green) and a telomere probe (red) on interphase nuclei (blue). Panel (A–G) Different lymphocyte cell nuclei: Panel (A) primary lymphocyte nucleus of T38HxBalb/c; Panel (B) nucleus of preB cell; Panel (C) nucleus of MOPC460D; Panel (D) preB cell nucleus without MycER activation 30 hours after mock treatment with ethanol; Panel (E) preB cell nucleus 30 hours after MycER activation; Panel (F) Ba/F3 cell nucleus without Δ106-MycER activation 30 hours after mock treatment with ethanol; Panel (G) Ba/F3 cell nucleus 30 hours after Δ106-MycER activation. Each panel illustrates a nucleus in 2D Panel (A–G, a) and in 3D Panel (A–G, b–d). Among 3D panels, (b) represents dual-color hybridization, with centromeres in green and telomeres in red; (c) illustrates centromeric signals only; and (d) shows telomeric hybridization signals. Panels (C and E, e) Enlarged 3D views of CTC hybridization signals (green-red-green) that are also highlighted by yellow arrows in Panel (C) and Panel (E), respectively. In addition, telomere-centromere hybridization signals are highlighted in small cartoons on the right side of each panel. Scale bars are given in 3D panels and represent sizes in nanometers.
Each telocentric mouse chromosome is expected to have four telomeres because there are four chromatid ends (i.e., two at each end of the long arm and two at the short centromeric end of a chromosome). Therefore, the expected hybridization signals of telomeres and centromeres in mouse interphase nuclei would be found in the following sequence: Telomeres (red) will be observed both adjacent to a centromere (green) and distant from a centromere (Figure 1A, cartoon). This is due to the presence of telomeres at the short and long ends of mouse chromosomes, respectively. Nuclear telomere (red)-centromere (green)-telomere (red) (TCT) signals may also touch each other if a chromosome is bent.
Our data show that primary mouse lymphocytes display expected TCT signals (Figure 1A). Similarly, immortalized preB cells exhibit TCTsignals (Figure 1B). However, the nuclei of MOPC460D plasmacytoma cells display a different organization; TCT signals are frequently altered into centromere (green)-telomere (red)-centromere (green) (CTC) signals, suggesting a centromere-centromere association, with telomeric signals bridging the centromeres (Figure 1C, yellow arrows, cartoon, and enlarged CTC images in e) (P < .0001).
To examine the 3D distribution patterns of centromeres and telomeres in conditionally Myc-expressing cells, we studied preB cells stably transfected with MycER [11,23] (Table 1). These cells allow for the conditional expression of c-Myc (Figure W1, A and B). Moreover, in the absence of c-Myc activation, these cells are diploid (Table 1). Figure 1, D and E, illustrates the data obtained for preB cells in the absence of MycER activation (D) and in its presence (E). The nuclear organization of CTC is apparent only after MycER activation (Figure 1E, yellow arrow, cartoon, and zoomed image e) (P = .02), suggesting an Myc-dependent nuclear remodeling of centromeres. To verify the Myc dependency of this process, a myc box II deletion mutant Δ106-MycER was tested under identical conditions (Figure 1, F and G; Figure W1, C and D). This deletion mutant, when overexpressed, is unable to initiate in vivo tumorigenesis of spontaneously immortalized proB lymphocytes (Ba/F3) [21]. Δ106-MycER did not induce the nuclear remodeling of centromeres, and no CTC signal was found (Figure 1, F and G, cartoon). For the cells studied here, we conclude that the formation of CTC appears to be dependent on the presence of Myc and on the presence of myc box II.
c-Myc and myc Box II-Dependent Formation of Rb Chromosomes
The analysis of TCT versus CTC signals in 3D images is complex, and not all potential CTC signals will be found or will be correctly assessed due to the clustering of centromeres at the nuclear periphery (see also Sarkar et al. [19], Solovei et al. [28], and Weierich et al. [29]). We therefore decided to tackle the question of centromere remodeling by molecular cytogenetics. Using this approach, we investigated whether the altered nuclear organization of centromeres impacts on the structural organization of chromosomes, particularly on the formation of Rb chromosomes. In mouse Rb chromosomes, telocentric chromosomes become biarmed due to the fusion of centromeres of the two individual telocentric chromosomes.
To address the question of Rb chromosome formation in our cell models, we performed SKY of primary mouse lymphocytes, mouse plasmacytoma cells (MOPC460D), mouse diploid immortalized preB cells with and without MycER activation, and near-tetraploid mouse BaF/3 cells (immortalized proB cells) in the presence or in the absence of wild-type MycER or Δ106-MycER activation (Table 1 and Figure 2; Figure W1). Twenty metaphases were examined for each cell type. In contrast to primary lymphocytes of T38HxBalb/c mice that did not exhibit Rb chromosomes (Figure 2A), MOPC460D tumor cells showed a significant number of Rb chromosomes per metaphase (P < .0001): Fifteen of 20 MOPC460D metaphases showed one or more Rb chromosomes; 26 Rb chromosomes were observed in 15 metaphases (Figure 2B, white arrows).
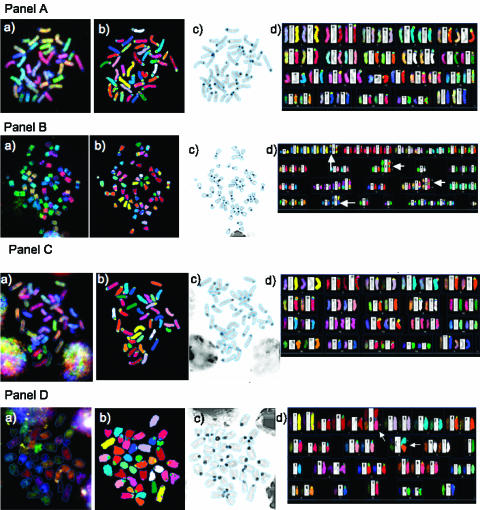
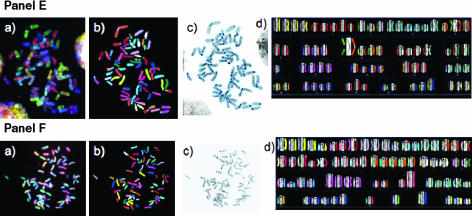
Figure 2.
SKY of metaphases derived from the cells of this study. Representative images from SKY analyses are shown: Panel (A) primary lymphocytes from T38HxBalb/c mice; Panel (B) MOPC460D; Panels (C and D) preB cells in the absence Panel (C) or in the presence Panel (D) of MycER activation; Panels (E and F) Ba/F3 Δ106-MycER without Panel (E) or with Panel (F) Δ106-MycER activation. White arrows point to Rb fusion chromosomes. The Rb (8;8) in Ba/F3 cells is constitutional and has been noted previously [21]. Each panel shows a representative image for each cell type; however, layout is the same for all: (a) the raw image of a metaphase; (b) the classified image of the metaphase; (c) the inverted DAPI-banded image of the metaphase; and (d) the karyotype table of the metaphase.
It is noteworthy that MOPC460D cells displayed a nonrandom involvement of specific chromosomes in the formation of Rb chromosomes, such as chromosomes 1, 3, 8, 14, 15, 17, and X (Table 2). Out of these, chromosome 15 was most frequent (found 24 times in Rb fusions in the 15 Rb chromosome-carrying metaphases), followed by chromosomes 1 and 14 (found 21 times in 15 Rb chromosome-carrying metaphases) (Table 2).
Table 2.
Summary of Chromosomes Participating in the Formation of Rb Chromosomes in MOPC460D Cells in a Nonrandom Manner.
| Chromosome Number | Times Involved in Rb Fusions | P |
| 1 | 21 | < .0001 |
| 3 | 11 | .001 |
| 8 | 18 | < .0001 |
| 14 | 21 | < .0001 |
| 15 | 24 | < .0001 |
| 17 | 14 | .0001 |
| X | 11 | .001 |
The numbers given are derived from the analysis of 20 metaphases. Fifteen of these 20 metaphases carried one or more Rb chromosomes. The involvement of each chromosome in the formation of Rb chromosomes is given, and the respective significance is indicated. For further details, see text and Materials and Methods section.
To analyze the impact ofMyc on the formation of Rb chromosomes, we used preB cells and studied them in the absence and in the presence of MycER activation (Figure 2, Panels C and D, respectively). Although non-MycER-activated preB cells did not exhibit Rb chromosomes (Figure 2, Panel C), MycER-activated preB cells showed Myc-dependent formation of Rb chromosomes. Thirty-three percent of metaphases (6/20) showed formation of Rb chromosomes within 30 hours (Figure 2, Panel D, arrows; P = .02). During this observation period, we did not note any specific chromosome combinations that were involved in the formation of Rb chromosomes.
The Myc dependency of this structural chromosomal change was confirmed with the myc box II deletion mutant Δ106. The conditional expression of Δ106-MycER did not lead to the formation of Rb chromosomes in Ba/F3 cells (Figure 2, Panels E and F, respectively), whereas the induction of wild-type MycER in Ba/F3 cells did. Chromosomes X and 5 were most frequently involved in the formation of Rb chromosomes subsequent to wild-type MycER activation of Ba/F3 cells (P = .0001 and P = .02, respectively, 30 hours after Myc activation) (data not shown).
The above data suggest that: 1) nuclear centromere organization impacts on chromosomal order permitting the formation of Rb chromosomes; and 2) Myc deregulation leads to the formation of Rb chromosomes in a myc box II-dependent manner.
Rb Chromosomes Form By Centromere-Telomere-Fusion in an Myc-Dependent Manner
Using dual-color FISH with centromeres and telomeres on metaphase chromosomes, we next examined whether the Rb chromosomes seen displayed centromere-telomere fusions. Using the cell lines listed in Table 1, we analyzed 20 metaphases per cell type and determined the presence of telomeric signals at the fusion points of centromeres in Rb fusion chromosomes (Figure 3). We noted the presence of telomeric hybridization signals on Rb chromosomes formed after MycER activation in preB cells (Figure 3, Panel E, arrows and zoomed images), MOPC460D cells (Figure 3, Panel B, arrows and zoomed images), and PCT1G1 cells (a primary mouse plasmacytoma cell line; Figure 3, Panel C, (arrows and zoomed images)). In contrast, no Rb chromosomes were seen in primary lymphocytes (Figure 3, Panel A), non-MycER-activated preB cells (Figure 3, Panel D), or Δ106-MycER-activated and control Ba/F3 cells (Figure 3, Panels G and F, respectively). We conclude that constitutive or conditional wild-type Myc deregulation, but not deregulated Δ106-Myc protein expression, led to the formation of Rb chromosomes that carry telomeric signals at their fusion points.
The above data further suggest that critically shortened telomeres are unlikely to cause the formation of Rb translocation chromosomes in this experimental setting. Instead, it is likely that c-Myc-dependent uncapping of telomeric sequences plays an initiating role in this event. This conclusion is very likely due to the short time period required to permit such fusions: 30 hours after a single c-Myc deregulation were sufficient to allow for the formation of Rb translocation chromosomes that carry telomeric signals in fused centromeres. The latter was observed for both preB and Ba/F3 cells.
Discussion
Rb Chromosomes in Different Species and in Cancer
Rb chromosomes represent structural genetic changes that occur in many species, including plants [30], cattle [31], some strains of mice [16,32], fish [33], and humans [34]. In humans, such Rb chromosomes are among the most common structural aberrations in aborted fetuses and newborns [35–37]. Moreover, Rb chromosomes in humans have been found as acquired or constitutional genetic lesions in hematologic cancers [34,38] and in solid tumors [39,40]. In addition, they have been reported at the onset of acute myelogenous leukemia [41].
Does It Matter to Have De Novo Rb Chromosomes in a Cell?
One could assume that Rb chromosomes merely remodel nuclear organization, thereby placing two chromosomes into a “forced” unit and into a new nuclear position or environment, without any further impact on the cell. Several lines of evidence suggest, however, that this new fused entity can be different and that the remodeling of two chromosomes into one Rb chromosome may possibly have wide-ranging effects. For example, it has been described that the formation of an Rb chromosome suppresses somatic recombination [42]. Another study linked Rb chromosomes to altered nuclear architecture and subfertility in mice [43]. In tumor induction studies with Rb-carrying or non-Rb-carrying BALB/c mice and congenics, it has been shown that the type of c-myc-activating chromosomal translocations in mouse plasmacytoma cells is altered in Rb6.15-carrying mice compared to BALB/c mice with telocentric chromosomes [44]. In humans, Rb translocations occur between acrocentric chromosomes of the D and G groups 13–15 and 21–22. These Rb translocations most frequently involve chromosomes 13 and 14. Carriers of such chromosomes are at risk for chromosomal nondisjunction leading to offspring with trisomy or uniparental disomy (UPD) following pregnancy rescue. Although UPD for chromosomes 13, 21, and 22 does not show apparent phenotypes, UPD for chromosomes 14 and 15 results in abnormal phenotypes [37]. Altogether, the above data suggest that a nuclear reorganization of two single chromosomes into one Rb chromosome may have a broad impact on the overall physiological state of Rb-carrying cells, on the function of the organism, and on oncogenesis.
Randomness and Nonrandomness of Rb Chromosomes Formed
Our present data show that mouse plasmacytoma cells (MOPC460D) with constitutive c-Myc deregulation and in long-term culture develop significant numbers of Rb chromosomes. Specific Rb chromosomes are found more frequently than others. For example, chromosomes 1, 3, 8, 14, 15, 17, and X are involved in Rb translocations almost all the time, although with different individual frequencies (Table 2). It is possible that the nonrandom composition of Rb fusions was selected during long-term culture, in combination with constitutively elevated levels of c-Myc protein. However, the above data also support current concepts suggesting that chromosomal proximity in interphase nuclei is required for intrachromosomal rearrangements (for review, see Meaburn et al. [45]). One could therefore assume that those chromosomes that were involved in Rb fusions more often than others resided in centromere clusters or in close nuclear proximity to each other in interphase nuclei. In fact, a previous study undertaken by our group suggested that specific chromosomes involved in unbalanced translocations were found in close nuclear proximity, showing considerable overlap as a result of c-Myc deregulation [11]. Moreover, studies into the 3D organization of centromeres suggest that their distribution is altered in tumor cells such as MOPC460D, where centromeres are no longer found with high frequency toward the nuclear periphery (as is commonly observed for primary lymphocytes) but are instead located in a more central nuclear space [19]. If we also consider that mouse chromosomes have a > 99% sequence homology in their telocentric regions [46], fusion events become very possible. They have indeed been reported in detail for Mus musculus speciation [15–18]. Thus, we propose that chromosomal positions and telocentric sequences contribute to both the specificity and the randomness of Rb chromosome formation in mouse cells. In accordance with this notion, we have observed that a single MycER activation in preB cells led to the formation of Rb chromosomes but did not result in the generation of nonrandom combinations of Rb chromosomes. Of note, PCT1G1, a primary plasmacytoma cell line that displayed Rb chromosomes (Table 1 and Figure 3), also did not show nonrandom constitution of Rb chromosomes, whereas a single wild-type MycER activation in Ba/F3 cells led to preferential formation of certain Rb chromosome combinations. From these findings, we conclude that c-Myc deregulation permits the formation of random and nonrandom Rb chromosomes within the given context of nuclear chromosomal and centromeric positions and of telocentric sequences.
Mechanisms of Rb Chromosome Formation in the Context of c-Myc-Dependent Oncogenic Nuclear Remodeling
Previous studies have suggested that Rb chromosomes form after recombination [46,47], centric misdivision and rejoining [30], or fusion [48]. Our data support the concept of fusion but add a new dimension: Rb fusions are initiated by c-Myc oncogene deregulation and depend on myc box II. After c-Myc deregulation, Rb chromosomes are generated when centromeric telomeres of mouse telocentric chromosomes fuse. This telomeric fusion is a direct consequence of the recently described Myc-dependent formation of telomeric aggregates [11] and of the nuclear remodeling of centromeres (this study and Sarkar et al. [19]). In this context, c-Myc-induced telomeric aggregates will lead to end-to-end fusions of telomeres on both ends of the chromosomes. Telocentric telomere fusions will generate Rb chromosomes, whereas telomeric fusions of the long arms of two chromosomes will create dicentric chromosomes. The latter usually initiates BBF cycles, which we have described recently [11].
The novel finding of this study, thus, is a direct link between Rb chromosome formation and c-Myc deregulation that occurs in a myc box II-dependent manner and through telomeric fusions at telocentric ends of mouse chromosomes. Whether previously observed mouse and rat Rb fusions involve myc box II and telomeric fusions is currently unknown. For example, Rb chromosomes were observed in Rat1A fibroblasts [49,50] and in transgenic MMTV-myc/p53 mice [51]. One of the primary tumors that formed in these mice, 67a5, contained RbX.15 and Rb11.15 [51]. Furthermore, Rb chromosomes were common in a model of mouse skin tumorigenesis (unpublished data).
Theoretically, the c-Myc-dependent telomere-mediated centromeric fusion process that creates mouse Rb chromosomes does not require additional mechanisms. However, it is likely that c-Myc's ability to induce DNA breaks [52–54] may contribute to a second molecular pathway of Rb chromosome formation. Whether the latter mechanism would act alone or in concert with the former is currently unknown.
The requirement of myc box II for mouse Rb chromosome formation confirms that the process of Rb chromosome formation is Myc-dependent and involves telomeric fusions [22]. The present study opens new avenues into investigations about myc box II-related Myc-cooperating proteins that may play a role in this Myc-induced nuclear remodeling of centromeres.
Finally, this study highlights the fact that c-Myc-dependent telomeric fusions at telocentric mouse chromosomes do not require critically short telomeres. This conclusion is based on the following: 1) compared to human telomeres, mouse telomeres are long (in the range of 20–60 kb, depending on the mouse strain) [55]; and 2) telomeric fusions at telocentric chromosomes occurred within 30 hours of experimentally induced c-Myc deregulation. This fact precludes the idea that, in this experimental context, after multiple divisions and/or mouse generations, telomeres reached a critically short state that predisposed them to fusions. Instead, these findings suggest a direct impact of c-Myc on the capping of telomeres. Future studies will address these potential interactions.
Supplementary Material
Acknowledgement
We thank FrancisWiener for critical reading of the manuscript.
Abbreviations
- FISH
fluorescence in situ hybridization
- SKY
spectral karyotyping
- Rb
Robertsonian
- 3D
three-dimensional
- 2D
two-dimensional
- BBF
breakage-bridge-fusion
- TCT
telomere-centromere-telomere
- CTC
centromere-telomere-centromere
Footnotes
This article refers to supplementary material, which is designated by “W” (i.e., Figure W1) and is available online at www.bcdecker.com.
We thank our granting agencies for financial support. These include the National Cancer Institute of Canada, CancerCare Manitoba Foundation, and the Canadian Institutes of Health Research Strategic Training Program “Innovative Technologies in Multidisciplinary Health Research Training.”
References
- 1.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. (Review) [DOI] [PubMed] [Google Scholar]
- 2.Popescu NC, Zimonjic DB. Chromosome-mediated alterations of the MYC gene in human cancer. J Cell Mol Med. 2002;6:151–159. doi: 10.1111/j.1582-4934.2002.tb00183.x. (Review) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arvanitis C, Felsher DW. Conditional transgenic models define how MYC initiates and maintains tumorigenesis. Semin Cancer Biol. 2006;16:313–317. doi: 10.1016/j.semcancer.2006.07.012. (Epub 2006 Jul 21) [DOI] [PubMed] [Google Scholar]
- 4.Lutz W, Leon J, Eilers M. Contributions of Myc to tumorigenesis. Biochim Biophys Acta. 2002;1602:61–71. doi: 10.1016/s0304-419x(02)00036-7. (Review) [DOI] [PubMed] [Google Scholar]
- 5.Kuttler F, Mai S. c-Myc, genomic instability and disease. In: Volff J-N, editor. Vol 1: Genome Dynamics. Genome and Disease. Würzburg, Germany: Karger Publishers; 2006. pp. 171–191. [DOI] [PubMed] [Google Scholar]
- 6.Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol. 2006;16:318–330. doi: 10.1016/j.semcancer.2006.07.015. (Review; Epub 2006 Aug 3) [DOI] [PubMed] [Google Scholar]
- 7.Mai S, Mushinski JF. c-Myc-induced genomic instability. J Environ Pathol Toxicol Oncol. 2003;22:179–199. doi: 10.1615/jenvpathtoxoncol.v22.i3.30. (Review) [DOI] [PubMed] [Google Scholar]
- 8.Oster SK, Ho CS, Soucie EL, Penn LZ. The myc oncogene: MarvelouslY Complex. Adv Cancer Res. 2002;84:81–154. doi: 10.1016/s0065-230x(02)84004-0. (Review) [DOI] [PubMed] [Google Scholar]
- 9.Frank SR, Parisi T, Taubert S, Fernandez P, Fuchs M, Chan HM, Livingston DM, Amati B. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 2003;4:575–580. doi: 10.1038/sj.embor.embor861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knoepfler PS, Zhang XY, Cheng PF, Gafken PR, McMahon SB, Eisenman RN. Myc influences global chromatin structure. EMBO J. 2006;25:2723–2734. doi: 10.1038/sj.emboj.7601152. (Epub 2006 May 25) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Louis SF, Vermolen BJ, Garini Y, Young IT, Guffei A, Lichtensztejn Z, Kuttler F, Chuang TC, Moshir S, Mougey V, et al. c-Myc induces chromosomal rearrangements through telomere and chromosome remodeling in the interphase nucleus. Proc Natl Acad Sci USA. 2005;102:9613–9618. doi: 10.1073/pnas.0407512102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mai S, Garini Y. Oncogenic remodeling of the threedimensional organization of the interphase nucleus: c-Myc induces telomeric aggregates whose formation precedes chromosomal rearrangements. Cell Cycle. 2005;4:1327–1331. doi: 10.4161/cc.4.10.2082. (Epub 2005 Oct 5) [DOI] [PubMed] [Google Scholar]
- 13.Mai S, Garini Y. The significance of telomeric aggregates in the interphase nuclei of tumor cells. J Cell Biochem. 2006;97:904–915. doi: 10.1002/jcb.20760. (Review) [DOI] [PubMed] [Google Scholar]
- 14.Chuang TC, Moshir S, Garini Y, Chuang AY, Young IT, Vermolen B, van den Doel R, Mougey V, Perrin M, Braun M, et al. The three-dimensional organization of telomeres in the nucleus of mammalian cells. BMC Biol. 2004;3:2–12. doi: 10.1186/1741-7007-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gropp A, Winking H, Redi C, Capanna E, Britton-Davidian J, Noack G. Robertsonian karyotype variation in wild house mice from Rhaeto-Lombardia. Cytogenet Cell Genet. 1982;34:67–77. doi: 10.1159/000131794. [DOI] [PubMed] [Google Scholar]
- 16.Gazave E, Catalan J, Ramalhinho Mda G, Mathias Mda L, Nunes AC, Dumas D, Britton-Davidian J, Auffray JC. The non-random occurrence of Robertsonian fusion in the house mouse. Genet Res. 2003;81:33–42. doi: 10.1017/s001667230200602x. [DOI] [PubMed] [Google Scholar]
- 17.Garagna S, Marziliano N, Zuccotti M, Searle JB, Capanna E, Redi CA. Pericentromeric organization at the fusion point of mouse Robertsonian translocation chromosomes. Proc Natl Acad Sci USA. 2001;98:171–175. doi: 10.1073/pnas.98.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Capanna E, Castiglia R. Chromosomes and speciation in Mus musculus domesticus. Cytogenet Genome Res. 2004;105:375–384. doi: 10.1159/000078210. [DOI] [PubMed] [Google Scholar]
- 19.Sarkar R, Guffei A, Vermolen BJ, Garini Y, Mai S. Alterations in centromere positions in immortalized and malignant mouse lymphocytes. Cytometry Part A. 2007;71(6):386–392. doi: 10.1002/cyto.a.20395. [DOI] [PubMed] [Google Scholar]
- 20.Fest T, Mougey V, Dalstein V, Hagerty M, Milette D, Silva S, Mai S. c-MYC overexpression in Ba/F3 cells simultaneously elicits genomic instability and apoptosis. Oncogene. 2002;21:2981–2990. doi: 10.1038/sj.onc.1205274. [DOI] [PubMed] [Google Scholar]
- 21.Fest T, Guffei A, Williams G, Silva S, Mai S. Uncoupling of genomic instability and tumorigenesis in a mouse model of Burkitt's lymphoma expressing a conditional box II-deleted Myc protein. Oncogene. 2005;24:2944–2953. doi: 10.1038/sj.onc.1208467. [DOI] [PubMed] [Google Scholar]
- 22.Caporali A, Wark L, Mai S. Telomeric aggregates and end-to-end chromosomal fusions require my box II. Oncogene. 2007;26:1398–1406. doi: 10.1038/sj.onc.1209928. (Epub 2006 Sep 4) [DOI] [PubMed] [Google Scholar]
- 23.Mai S, Hanley-Hyde J, Rainey GJ, Kuschak TI, Paul JT, Littlewood TD, Mischak H, Stevens LM, Henderson DW, Mushinski JF. Chromosomal and extrachromosomal instability of the cyclin D2 gene is induced by Myc overexpression. Neoplasia. 1999;1:241–252. doi: 10.1038/sj.neo.7900030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fukasawa K, Wiener F, Vande Woude GF, Mai S. Genomic instability and apoptosis are frequent in p53 deficient young mice. Oncogene. 1997;11:1295–1302. doi: 10.1038/sj.onc.1201482. [DOI] [PubMed] [Google Scholar]
- 25.Kim SH, McQueen PG, Lichtman MK, Shevach EM, Parada LA, Misteli T. Spatial genome organization during T-cell differentiation. Cytogenet Genome Res. 2004;105:292–301. doi: 10.1159/000078201. [DOI] [PubMed] [Google Scholar]
- 26.Mai S, Wiener F. Murine FISH. In: Beatty B, Mai S, Squire J, editors. FISH: A Practical Approach. Oxford: Oxford University Press; 2002. pp. 55–67. [Google Scholar]
- 27.Schaefer LH, Schuster D, Herz H. Generalized approach for accelerated maximum likelihood based image restoration applied to three-dimensional fluorescence microscopy. J Microsc. 2001;204:99–107. doi: 10.1046/j.1365-2818.2001.00949.x. [DOI] [PubMed] [Google Scholar]
- 28.Solovei I, Schermelleh L, During K, Engelhardt A, Stein S, Cremer C, Cremer T. Differences in centromere positioning of cycling and postmitotic human cell types. Chromosoma. 2004;112:410–423. doi: 10.1007/s00412-004-0287-3. [DOI] [PubMed] [Google Scholar]
- 29.Weierich C, Brero A, Stein S, von Hase J, Cremer C, Cremer T, Solovei I. Three-dimensional arrangements of centromeres and telomeres in nuclei of human and murine lymphocytes. Chromosome Res. 2003;11:485–502. doi: 10.1023/a:1025016828544. [DOI] [PubMed] [Google Scholar]
- 30.Friebe B, Zhang P, Linc G, Gill BS. Robertsonian translocations in wheat arise by centric misdivision of univalents at anaphase I and rejoining of broken centromeres during interkinesis of meiosis II. Cytogenet Genome Res. 2005;109:293–297. doi: 10.1159/000082412. [DOI] [PubMed] [Google Scholar]
- 31.Mastromonaco GF, Coppola G, Crawshaw G, DiBerardino D, King WA. Identification of the homologue of the bovine Rob(1;29) in a captive gaur (Bos gaurus) Chromosome Res. 2004;12:725–731. doi: 10.1023/B:CHRO.0000045800.44911.67. [DOI] [PubMed] [Google Scholar]
- 32.Nachman MW, Searle JB. Why is the house mouse karyotype so variable? Trends Ecol Evol. 1995;10:397–402. doi: 10.1016/s0169-5347(00)89155-7. [DOI] [PubMed] [Google Scholar]
- 33.Gold JR, Gall GA. Chromosome cytology and polymorphism in the California High Sierra golden trout (Salmo aguabonita) Can J Genet Cytol. 1975;17:41–53. doi: 10.1139/g75-005. [DOI] [PubMed] [Google Scholar]
- 34.Welborn J. Acquired Robertsonian translocations are not rare events in acute leukemia and lymphoma. Cancer Genet Cytogenet. 2004;151:14–35. doi: 10.1016/j.cancergencyto.2003.09.019. [DOI] [PubMed] [Google Scholar]
- 35.Jacobs PA. Mutation rates of structural chromosome rearrangements in man. Am J Hum Genet. 1981;33:44–54. [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen J, Wohlert M. Chromosome abnormalities found among 34,910 newborn children: results from a 13-year incidence study in Århus, Denmark. Hum Genet. 1991;87:81–83. doi: 10.1007/BF01213097. [DOI] [PubMed] [Google Scholar]
- 37.Kim SR, Shaffer LG. Robertsonian translocations: mechanisms of formation, aneuploidy, and uniparental disomy and diagnostic considerations. Genet Test. 2002;6:163–168. doi: 10.1089/109065702761403315. [DOI] [PubMed] [Google Scholar]
- 38.Qian J, Xue Y, Sun J, Guo Y, Pan J, Wu Y, Wang W, Yao L. Constitutional Robertsonian translocations in (9;22)-positive chronic myelogeneous leukemia. Cancer Genet Cytogenet. 2002;132:79–80. doi: 10.1016/s0165-4608(01)00522-2. [DOI] [PubMed] [Google Scholar]
- 39.Padilla-Nash HM, Heselmeyer-Haddad K, Wangsa D, Zhang H, Ghadimi BM, Macville M, Augustus M, Schrock E, Hilgenfeld E, Ried T. Jumping translocations are common in solid tumor cell lines and result in recurrent fusions of whole chromosome arms. Genes Chromosomes Cancer. 2001;30:349–363. doi: 10.1002/gcc.1101. [DOI] [PubMed] [Google Scholar]
- 40.Bayani J, Zielenska M, Pandita A, Al-Romaih K, Karaskova J, Harrison K, Bridge JA, Sorensen P, Thorner P, Squire JA. Spectral karyotyping identifies recurrent complex rearrangements of chromosomes 8, 19 and 20 in osteosarcomas. Genes Chromosomes Cancer. 2003;36:7–16. doi: 10.1002/gcc.10132. [DOI] [PubMed] [Google Scholar]
- 41.Shimokawa T, Sakai M, Kojima Y, Takeyama H. Acute myelogeneous leukemia (M5a) that demonstrated chromosomal abnormality of Robertsonian 13;21 translocation at onset. Intern Med. 2004;43:508–511. doi: 10.2169/internalmedicine.43.508. [DOI] [PubMed] [Google Scholar]
- 42.Haigis KM, Dove WF. A Robertsonian translocation suppresses a somatic recombination pathway to loss of heterozygosity. Nat Genet. 2003;33:33–39. doi: 10.1038/ng1055. (Epub 2002 Nov 25) [DOI] [PubMed] [Google Scholar]
- 43.Garagna S, Zuccotti M, Thornhill A, Fernandez-Donoso R, Berrios S, Capanna E, Redi CA. Alteration of nuclear architecture in male germ cells of chromosomally derived subfertile mice. J Cell Sci. 2001;114:4429–4434. doi: 10.1242/jcs.114.24.4429. [DOI] [PubMed] [Google Scholar]
- 44.Silva S, Wiener F, Klein G, Janz S. Location of Myc, Igh, and Igk on Robertsonian fusion chromosomes is inconsequential for Myc translocations and plasmacytoma development in mice, but Rb(6.15)-carrying tumors prefer Igk-Myc inversions over translocations. Genes Chromosomes Cancer. 2005;42:416–426. doi: 10.1002/gcc.20149. [DOI] [PubMed] [Google Scholar]
- 45.Meaburn KJ, Misteli T, Soutoglou E. Spatial genome organization in the formation of chromosomal translocations. Semin Cancer Biol. 2007;17:80–90. doi: 10.1016/j.semcancer.2006.10.008. (Epub 2006 Oct 26) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalitsis P, Griffiths B, Choo AKH. Mouse telocentric sequences reveal a high rate of homogenization and possible role in Robertsonian translocation. Proc Natl Acad Sci USA. 2006;103:8786–8791. doi: 10.1073/pnas.0600250103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hecht F, Morgan R, Hecht BK. Robertsonian chromosome recombinations are rare in cancer. Cancer Genet Cytogenet. 1988;35:79–81. doi: 10.1016/0165-4608(88)90125-2. [DOI] [PubMed] [Google Scholar]
- 48.Slijepcevic P. Telomeres and mechanisms of Robertsonian fusions. Chromosoma. 1998;107:136–140. doi: 10.1007/s004120050289. [DOI] [PubMed] [Google Scholar]
- 49.Mai S, Fluri M, Siwarski D, Huppi K. Genomic instability in MycER-activated Rat1A-MycER cells. Chromosome Res. 1996;4:365–371. doi: 10.1007/BF02257272. [DOI] [PubMed] [Google Scholar]
- 50.Felsher DW, Bishop JM. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc Natl Acad Sci USA. 1999;96:3940–3944. doi: 10.1073/pnas.96.7.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCormack SJ, Weaver Z, Deming S, Natarajan G, Torri J, Johnson MD, Liyanage M, Ried T, Dickson RB. Myc/p53 interactions in transgenic mouse mammary development, tumorigenesis and chromosomal instability. Oncogene. 1998;16:2755–2766. doi: 10.1038/sj.onc.1201804. [DOI] [PubMed] [Google Scholar]
- 52.Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, Wahl GM. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene- induced genetic instability. Mol Cell. 2002;9:1031–1044. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- 53.Karlsson A, Deb-Basu D, Cherry A, Turner S, Ford J, Felsher DW. Defective double-strand DNA break repair and chromosomal translocations by MYC overexpression. Proc Natl Acad Sci USA. 2003;100:9974–9979. doi: 10.1073/pnas.1732638100. (Epub 2003 Aug 8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ray S, Atkuri KR, Deb-Basu D, Adler AS, Chang HY, Herzenberg LA, Felsher DW. MYC can induce DNA breaks in vivo and in vitro independent of reactive oxygen species. Cancer Res. 2006;66:6598–6605. doi: 10.1158/0008-5472.CAN-05-3115. [DOI] [PubMed] [Google Scholar]
- 55.Hande P, Slijepcevic P, Silver A, Bouffler S, van Buul P, Bryant P, Lansdorp P. Elongated telomeres in scid mice. Genomics. 1999;56:221–223. doi: 10.1006/geno.1998.5668. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.