Abstract
Background
The power of microarray analysis can be realized only if data is systematically archived and linked to biological annotations as well as analysis algorithms.
Description
The Longhorn Array Database (LAD) is a MIAME compliant microarray database that operates on PostgreSQL and Linux. It is a fully open source version of the Stanford Microarray Database (SMD), one of the largest microarray databases. LAD is available at http://www.longhornarraydatabase.org
Conclusions
Our development of LAD provides a simple, free, open, reliable and proven solution for storage and analysis of two-color microarray data.
Background
Microarray experiments in all forms produce a great quantity of data. In addition to the primary microarray data, details about the biological samples that were analyzed must be correctly recorded and archived. This information has to be linked to biological annotations for the genes on the array in order for any experiment to have immediate or historical comparative value.
Many relational databases developed for this purpose have recently become available. ArrayDB, BASE, GeneX, MADAM and MIDAS are all examples of recent development efforts mainly from academic sources [1-4]. Most of these solutions have the advantage that the source code is available and hence in principle are open to being modified by users, but many of them still have some shortcomings. Some of these offerings store numerical data but lack the ability to simultaneously archive and visualize primary array images. Many attempt to support both two-color and Affymetrix data while not fully supporting the intricacies of storing and analyzing either. Others are simply not proven to scale to thousands of experiments or are built upon technologies that do not guarantee the integrity of the data. Finally, some solutions do not provide for a complete web-browser accessible implementation. GeneX, for example, is designed to rely upon a suite of both web-based and client applications for a variety of import and analysis features [1]. This architectural decision expands its graphical feature potential; however, it simultaneously reduces its geographical and web distributed user capabilities important to many research environments.
One of the most successful, proven, and heavily utilized microarray databases is the Stanford Microarray Database (SMD – http://genome-www.stanford.edu/microarray) [5]. SMD currently archives more than 34,500 microarray experiments including 4500 from more than a hundred different publications and supports approximately 700 users [6]. It has a great breadth of features that include data filtering, data analysis, visualization toolsets, and constantly updated biological annotations for many organisms. Additionally, SMD features a strict hierarchical user and group model of user accounts such that experiments can be collaboratively shared or protected as desired by individual researchers.
SMD's source code has been freely available for some time, which theoretically allows any researcher to install their own SMD server within their research environment. SMD in this form, however, is based on proprietary hardware and software infrastructure that would require a significant capital expenditure from any laboratory that wished to operate such a server. SMD has been primarily developed, maintained, and supported on the Sun Solaris operating system [6]. Solaris is tailored for Sun hardware and processors which are incompatible with and not nearly as widespread as Intel computer systems are in research environments.
Additionally, SMD was designed and written to utilize the Oracle relational database management system. In fact, the Solaris operating system was chosen to host SMD specifically because updates and bug fixes for Oracle usually appear first for Solaris relative to other operating system environments [7].
The cost of initial investment and long-term ownership of these technologies is significantly higher, however, than alternative open-source technology choices. Additionally, not only is Oracle expensive, it is a very demanding database in terms of the expertise required of professional database administrators to maintain it as a piece of software infrastructure.
Given the numerous strengths and proven nature of SMD, we wanted to adapt it to run on a free, open-source, widely available and powerful operating system and relational database. We chose the combination of Linux and PostgreSQL to replace Solaris and Oracle. We have named our open source version of SMD the Longhorn Array Database (LAD). The LAD code-base as well as detailed installation instructions are available at http://www.longhornarraydatabase.org.
Construction and content
The Linux translation
We adopted a two-step strategy to accomplish the open source port. We first ported SMD from Solaris to Linux while still using Oracle as the relational database. This allowed us to test and ensure that the application subset of the port operated exactly as it should.
We introduced only the required targeted changes throughout the entire source tree of SMD so that it would run under Linux. This was done so as to not alter or disrupt its original algorithmic design.
Oracle to PostgreSQL
Once LAD was fully operational on Linux (powered by Oracle) we undertook its migration to an open-source relational database that could support all the features that LAD required, such as support for transactions, foreign-key integrity constraints, indexes, and sequences. The Linux-supported, open-source relational database that met these requirements was PostgreSQL http://www.postgresql.org.
PostgreSQL is an advanced open-source object-relational database management system that supports nearly all SQL constructs. These constructs include transactions, triggers, stored procedures, subselects, and user-defined types and functions. The use of such features is generally considered to be critical for ensuring data integrity. MySQL, for example, has only very recently attained this feature set, and microarray databases that use MySQL as their engine do not make use of these transactional features [3].
The LAD database schema was re-created in PostgreSQL, and Oracle-specific SQL code, constructs, and syntax in the SMD tree were translated to a more standards-compliant SQL set of statements so that they would execute correctly with PostgreSQL. We optimized the indexing of certain table structures and profiled query execution of involved joins to ensure that operations would complete in acceptable timeframes. The end result was the LAD source code which interoperated with PostgreSQL with the same efficiency with which it worked with Oracle. While Oracle is a very powerful relational database, it is expensive to license and operate. The ability to install and run LAD with a feature-rich open-source relational database substantially decreased its required initial cost of investment.
Additionally, the use of PostgreSQL greatly reduces the level of complexity required to run a production microarray database due to the ease with which it can be installed and maintained. This opens up the possibility of a larger community of developers becoming involved with a proven array data warehouse. If researchers are simply interested in developing new array analysis routines, LAD now provides the incentive and the proven means to develop and test novel plug-ins. It is possible to use LAD for the development of query and visualization tools that directly interact with the database. In essence users are not restricted to LAD tools for analysis and may only utilize its intrinsic data loading capabilities while analyzing microarray data via custom-developed algorithms and interfaces.
Utility and Discussion
Linux implementation toolset
LAD is designed to be utilized through any modern, Javascript-enabled web browser (Figure 1). We have found that multiple browsers including but not limited to Microsoft Internet Explorer, Netscape, Galeon, and Mozilla interact with LAD in a completely functional manner.
Figure 1.
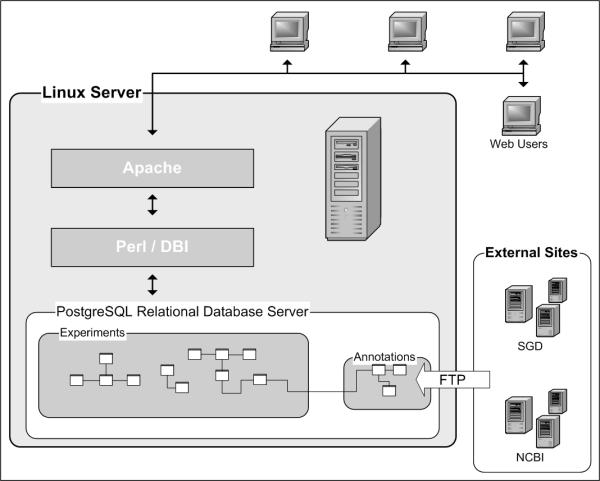
Architecture of LAD deployment. LAD is accessed exclusively through a web browser. Additionally, it relies completely on the open-source technologies Linux and PostgreSQL for operation. Organism annotations are kept up to date via a scripted linkage to external databases. The Saccharomyces Genome Database (SGD – http://www.yeastgenome.org) and the National Center for Biotechnology Information (NCBI – http://www.ncbi.nlm.nih.gov/) are shown as examples.
We have tested LAD upon RedHat 7.3, Mandrake 8.2, Mandrake 9.0, SUSE 8.0, and we are now running our production server on Mandrake Linux 9.1. All of these Linux distributions included PostgreSQL versions 7.2 or 7.3 which were utilized without modification or customization other than basic configuration options.
We have found that compatibility with any specific Linux distribution is most dependent on the version of the Apache HTTP server that is bundled with it. LAD requires Apache version 1.x as opposed to the more recent Apache 2.x releases. This is due to the way Apache 1.x handles and correctly interprets many SMD implemented non-parsed header declarations. It is likely that LAD could be made compatible with Apache 2.x. We have decided to continue to utilize Apache 1.x for the near future.
LAD was initially constructed using Perl version 5.6 but is fully compatible with Perl version 5.8. In addition to the base Perl distribution, LAD requires specialized modules such as GD, CGI, and DBI to interact with microarray images, control screen flow and layout, and communicate with the relational database respectively. All of these peripheral modules are freely available and regularly updated on the Comprehensive Perl Archive Network (CPAN) at http://www.cpan.org.
For its clustering and image manipulation functions LAD relies on many native programs that must be compiled from source code to run on the targeted operating system. We modified the source code of these programs such that they compiled and executed correctly on the Linux platform.
LAD also requires several system libraries such as libgd, libjpg, libtiff, libxpm, libfreetype, zlib, libpng, and the graphics manipulation packages ImageMagick and netpbm. In particular, libgd needs to be patched to support GIF file images (the native format of LAD). Detailed instructions for this are available at the libgd author's website http://www.rhyme.com.au/gd/.
We have deployed LAD on an Intel-based dual-Xeon Dell Precision 530 workstation with 1 GB of RAM and 500 GB of hard disk space. This is a very affordable mid-tier machine for this type of application. We have found that this dual CPU machine performs significantly better for a large user group than a single CPU machine of the same caliber in that the machine can more easily facilitate several data queries and operations in parallel. However, a fully functional version of LAD can be installed on a machine with 256 MB RAM and a 10 GB hard disk. We have developed a simplified installer that allows LAD to be configured and operational within minutes after the prerequisites are installed [8].
We have also developed novel features in LAD, such as features that facilitate the analysis of time-course experiments, sharing and regeneration of analytical data sets such as hierarchical clusters, and retrieval of original microarray images, settings, and results files. Enhanced user and group management and system maintenance functions have also been implemented. We are currently hosting more than 1300 microarray experiments including both analytical and image data for each. The smallest of these experiments contains over 15,000 spots with many having nearly 50,000.
MIAME support
A critical part of microarray experimentation and subsequent data analysis is the ability to collaboratively and meaningfully share the enormous amount of data and information that describe individual experiments. Lack of standards for presenting and exchanging data on both experimental conditions and the numerical microarray data makes relative comparison of microarray experiments produced in separate research environments a near impossibility.
A standard entitled MIAME – the Minimum Information About a Microarray Experiment – has been proposed to address this problem [9]. MIAME is in essence a comprehensive specification that details the minimum annotation that should accompany the publication of any microarray data set. Subsections pertaining to experimental design, array design, sample preparation, hybridization protocols, actual quantitative results, and normalization controls are all addressed within the specification.
MIAME enjoys significant support from both the research and journal publishing communities [10]. It is therefore imperative that any long-term microarray data warehouse and analysis environment support the MIAME specification. This will ensure that as results are accumulated within the database the appropriate experimental and conditional annotations are simultaneously recorded and archived.
To make LAD MIAME compliant, we have implemented a strategy that allows for a MIAME addendum to be attached to each experiment that is submitted to the database. This information is associated with each experiment in a new table within the relational database. Subsequent recall of that experiment also recalls all MIAME annotation information. Since a large fraction of this required information will remain constant or change slightly from experiment to experiment, MIAME annotation is implemented through the use of reusable templates. This will enforce MIAME compliance without encumbering the experiment submission process.
Future plans
The open and modular nature of the LAD code base makes it possible in principle to integrate new features developed for SMD into LAD. Conversely, new features developed for LAD by us or others may possibly be integrated into the SMD source tree.
LAD currently supports Axon GenePix http://www.axon.com and ScanAlyze http://rana.lbl.gov data file formats for data upload. Depending on public interest, the LAD code base may be enhanced to support a larger variety of microarray data formats and file types. It is important to also note, however, that because LAD is available as a fully open-source implementation, its code could be adapted by researchers to process a multitude of data layouts. Additionally, simple scripts could be written to convert unsupported file formats to either of the supported GenePix or ScanAlyze file types.
Currently, LAD is able to export data for single experiments in a tab-delimited spreadsheet format. Both raw data and up-to-date gene annotation can be extracted. Further support for export to public repositories like the Gene Expression Omnibus (GEO – http://www.ncbi.nlm.nih.gov/geo) is available within the code base. Minimal effort would be required in order to provide graphical interfaces to guide the export of both single and groups of experiments in a variety of formats such as MAGE-ML [10].
The efforts we have made to comply with and support the MIAME standard are simple but effective. We plan to utilize our MIAME infrastructure by adding further utilization of its information throughout the code base. Both data viewing and analysis will receive MIAME-based filtering screens so that researchers can utilize the information included in MIAME addendums to specifically filter the experiments they wish to work with at any given time.
We plan to utilize our open source system to develop novel tools to aid the analysis and visualization of expression profiling experiments as well as microarray data from novel experimental approaches such as the use of chromatin immunoprecipitation to identify the binding of transcription factors to DNA [11], comparative genomic hybridization to measure DNA copy number changes [11] and protein microarrays [12]. Additionally we hope that the ready availability of an open database system will spur similar efforts from others.
We wish to enhance the ability of researchers to collaborate with regards to experimental results and analysis via more integrated communication tools and stored analysis results. In this way, researchers will have the ability to not only share their data with collaborators but will also have the ability to save their filtering, perusal, and scientific findings as well. In this form LAD can also serve as a platform for publishing primary microarray data in conjunction with more traditional avenues of publication.
Finally, we wish to extend LAD's ability to integrate and communicate with our microarray analysis platforms. We plan to add MAGE-ML export capability such that data can be readily exported from LAD into other analysis programs that conform to the MIAME standard of data representation. In this way, LAD will always serve as an excellent archival tool, analysis platform, and gateway to analysis by a world of research toolsets yet to be developed.
Conclusions
The Longhorn Array Database (LAD) is a fully open-source, MIAME compliant microarray database based on PostgreSQL and Linux.
Availability
LAD source-code is freely available to all interested users. The download and installation instructions can be found at: http://www.longhornarraydatabase.org
Authors' contributions
PK was responsible for the code modifications to port SMD to the open-source platform. GS was responsible for the initial development of SMD and provided consultation on conversion of the code base. VI provided overall supervision and funding of the project.
Acknowledgments
Acknowledgements
We thank Raul Davidovich for discussions and Adron Harris for support. This work was supported by an NIH INIA Program (Integrative Neuroscience Initiative on Alcoholism) grant AA13518. P.J.K. was supported in part by a pre-doctoral NIAAA-Alcohol Training Grant.
Contributor Information
Patrick J Killion, Email: pkillion@mail.utexas.edu.
Gavin Sherlock, Email: sherlock@genome.stanford.edu.
Vishwanath R Iyer, Email: vishy@mail.utexas.edu.
References
- Gardiner-Garden M, Littlejohn TG. A comparison of microarray databases. Brief Bioinform. 2001;2:143–158. doi: 10.1093/bib/2.2.143. [DOI] [PubMed] [Google Scholar]
- Ermolaeva O, Rastogi M, Pruitt KD, Schuler GD, Bittner ML, Chen Y, Simon R, Meltzer P, Trent JM, Boguski MS. Data management and analysis for gene expression arrays. Nat Genet. 1998;20:19–23. doi: 10.1038/1670. [DOI] [PubMed] [Google Scholar]
- Saal LH, Troein C, Vallon-Christersson J, Gruvberger S, Borg A, Peterson C. BioArray Software Environment (BASE): a platform for comprehensive management and analysis of microarray data. Genome Biol. 2002;3:SOFTWARE0003. doi: 10.1186/gb-2002-3-8-software0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazma A, Parkinson H, Sarkans U, Shojatalab M, Vilo J, Abeygunawardena N, Holloway E, Kapushesky M, Kemmeren P, Lara GG, Oezcimen A, Rocca-Serra P, Sansone SA. ArrayExpress--a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003;31:68–71. doi: 10.1093/nar/gkg091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherlock G, Hernandez-Boussard T, Kasarskis A, Binkley G, Matese JC, Dwight SS, Kaloper M, Weng S, Jin H, Ball CA, Eisen MB, Spellman PT, Brown PO, Botstein D, Cherry JM. The Stanford Microarray Database. Nucleic Acids Res. 2001;29:152–155. doi: 10.1093/nar/29.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollub J, Ball CA, Binkley G, Demeter J, Finkelstein DB, Hebert JM, Hernandez-Boussard T, Jin H, Kaloper M, Matese JC, Schroeder M, Brown PO, Botstein D, Sherlock G. The Stanford Microarray Database: data access and quality assessment tools. Nucleic Acids Res. 2003;31:94–96. doi: 10.1093/nar/gkg078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball CA, Sherlock G. LIMS, Databases, and Data Management. In: Bowtell D and Sambrook J, editor. DNA Microarrays: A Molecular Cloning Manual. CSHL Press; 2003. pp. 552–581. [Google Scholar]
- LAD LAD is available at http://www.longhornarraydatabase.org
- Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Miller M, Stewart J, Troup C, Sarkans U, Chervitz S, Bernhart D, Sherlock G, Ball C, Lepage M, Swiatek M, Marks WL, Goncalves J, Markel S, Iordan D, Shojatalab M, Pizarro A, White J, Hubley R, Deutsch E, Senger M, Aronow BJ, Robinson A, Bassett D, Stoeckert C. J., Jr., Brazma A. Design and implementation of microarray gene expression markup language (MAGE-ML) Genome Biol. 2002;3:RESEARCH0046. doi: 10.1186/gb-2002-3-9-research0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollack JR, Iyer VR. Characterizing the physical genome. Nat Genet. 2002;32 Suppl:515–521. doi: 10.1038/ng1035. [DOI] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nature. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]


