Abstract
Several pathogenic human herpesviruses have recently been shown to express virally encoded microRNAs in infected cells. Although the function of these microRNAs is largely unknown, they are hypothesized to play a role in mediating viral replication by down-regulating cellular mRNAs encoding antiviral factors. Here, we report the cloning and analysis of microRNAs encoded by Rhesus Monkey Rhadinovirus (RRV), an animal virus model for the pathogenic human γ-herpesvirus Kaposi’s Sarcoma-Associated Herpesvirus (KSHV). RRV expresses several microRNAs that are encoded in the same genomic location as the previously reported KSHV microRNAs, yet these microRNAs are unrelated in primary sequence. These data set the stage for the mutational ablation and phenotypic analysis of RRV mutants lacking one or more viral microRNAs.
Keywords: microRNAs, Rhesus Monkey Rhadinovirus, Kaposi’s Sarcoma-Associated Herpesvirus
Introduction
MicroRNAs (miRNAs) are a ubiquitous class of ~22 nt long regulatory RNAs that post-transcriptionally regulate the expression of numerous eukaryotic genes (Bartel, 2004). In addition to being encoded by the genomes of all known metazoan eukaryotes, miRNAs have also been identified in several DNA viruses, particularly herpesviruses (Cullen, 2006). While some viral miRNAs appear to regulate the expression of viral genes transcribed from the opposite genome strand (Pfeffer et al., 2004; Sullivan et al., 2005), the majority are believed to downregulate cellular genes that act to reduce viral replication, including genes involved in mediating innate or adaptive immune responses (Gupta et al., 2006). Examples of herpesviruses that encode miRNAs include Kaposi’s Sarcoma-Associated Herpesvirus (KSHV), which encodes 12 viral miRNAs (Cai et al., 2005; Pfeffer et al., 2005; Samols et al., 2005; Grundhoff et al., 2006), and Epstein-Barr Virus (EBV), which expresses at least 23 miRNAs in infected cells (Pfeffer et al., 2004, 2005; Cai et al., 2006; Grundhoff et al., 2006). Analysis of rhesus lymphocryptovirus (rLCV), a non-human relative of EBV that infects rhesus macaques, has so far identified 16 distinct miRNAs, 8 of which are evolutionarily conserved in EBV (Cai et al., 2006). To extend these data, we have cloned and analyzed the miRNAs expressed by Rhesus Monkey Rhadinovirus (RRV), a simian rhadinovirus, closely related to human KSHV, that is both genetically tractable and readily grown in culture (Alexander et al., 2000; DeWire et al., 2003; Bilello et al., 2006; Estep et al., 2007; O’Connor and Kedes, 2007). Here, we report the identification of primary miRNA stem-loop precursors in RRV that give rise to 11 distinct viral miRNAs. While the genomic localization of the viral miRNAs is conserved between RRV and KSHV, none of the RRV miRNAs is similar in sequence to the previously reported KSHV miRNAs.
Results
KSHV establishes latent infections in many adherent cell lines, including telomerase-immortalized microvascular endothelial cells and transformed dermal microvascular cells (Blackbourn et al., 2000; Bechtel et al., 2003). To establish an analogous endothelial cell culture system appropriate for studying RRV, rhesus monkey vascular endothelial cell cultures (MVECs) were infected with a previously described recombinant strain of RRV (Bilello et al., 2006) that expresses green fluorescent protein (GFP) and then monitored for GFP expression over time. While GFP expression was apparent by day 4 post-infection, the percentage of GFP positive cells was low (approximately 2–4%). The RRV-infected MVEC cultures were passed when confluent and the percentage of GFP-positive cells progressively increased over time to approximately 90% at day 24 post-infection. While mild cytopathology was observed, the appearance of the infected MVEC cultures was not markedly different from that seen in uninfected cultures passed in parallel. The titer of RRV-GFP in the MVEC culture supernatant at days 20 and 24 post-infection was approximately 104 pfu/ml, approximately 1% of what is observed in the supernatant of rhesus fibroblast cultures lytically-infected with RRV-GFP (Bilello et al., 2006). The infection of the MVEC cells therefore appeared to be predominantly latent, although some of the MVECs were clearly producing infectious RRV.
At day 24 post-infection, total RNA was isolated from the RRV infected MVECs and used as a source for the isolation and reverse transcription of small RNAs of 18 nt to 26 nt in size, as previously described (Cai et al., 2005). The resultant cDNAs were concatamerized, cloned and sequenced. This cDNA cloning approach is predicted to clone all cellular and viral miRNAs as well as fragments of cellular mRNAs and non-coding RNAs that fall into this size range. The 19 to 24 bp long cDNA clones that were obtained were identified by computational analysis by comparison to the RRV genome sequence as well as to databases covering cellular miRNAs, noncoding RNAs and mRNAs. However, as the cells analyzed were of rhesus macaque origin, not all cloned sequences obtained could be clearly annotated. Nevertheless, this approach will identify all cloned cDNAs that represent RRV miRNAs.
A complete listing of all the cellular miRNAs cDNA cloned from the MVECs is provided in Supplementary Table 1 while the genomic origin of all the cDNA clones obtained is indicated in Supplementary Figure 1. Analysis of the 684 19–24 bp long cDNAs obtained from the RRV infected MVECs revealed 185 cellular miRNAs of which let-7 (82 cDNA clones), miR-125b-2 (26 clones), miR-221 (24 clones) and miR-100 (14 clones) were the most prevalent. We also obtained 138 cDNA clones of mRNA origin, 131 cDNA clones of rRNA origin and 20 cDNA clones of tRNA origin. A total of 102 rhesus cDNA clones were not identified by this analysis but were clearly not of RRV origin. The remaining 108 cDNA clones were all derived from the infecting RRV virus and represent 11 different RRV-derived miRNAs (Table 1). As expected, the viral miRNAs obtained ranged from 19 to 24 nt in length, with the majority 21 to 23 nt long.
Table 1.
Identity and genomic location of RRV miRNAs recovered from latently RRV-infected MVECs
| Name | Sequence 5’ to 3’ | Length (nt) | Hits | Position |
|---|---|---|---|---|
| miR-rR1-1-5p
miR-rR1-1-3p |
CGAUCGCACCUUUGGCCGGCCGG
GGCCACCGAGGAUGCGGUC |
23
19 |
27
1 |
116367:116345
116335:116317 |
| miR-rR1-2 | (A)UACGGCGCUGCACGGUUGGA | 20–21 | 3 | 114357:114337 |
| miR-rR1-3 | CCCGAUGAGCAGUUAGUCC | 19 | 1 | 112762:112744 |
| miR-rR1-4-5p
miR-rR1-4-3p |
UGGGGAGGGCGGUCAGCGCGC(G)
CUCGUUAACCGCCCUCCCGAGA |
21–22
22 |
39
6 |
112317:112296
112267:112246 |
| miR-rR1-5 | CCGGAACCCAAAGACACGUGCCCG | 24 | 1 | 111544:111521 |
| miR-rR1-6-5p
miR-rR1-6-3p |
CGCGGAAAGGUGUGCACAUCGU(A)
CGAUGUACGCCCUUUCGCAGU |
22–23
21 |
19
2 |
111236:111214
111201:111181 |
| miR-rR1-7-5p
miR-rR1-7-3p |
(U)GGAGAGCAGUUAACGUGCGUUC
CGCACGUCGAUUGCUCUCUAG |
22–23
21 |
3
6 |
111123:111101
111083:111063 |
This table shows the sequence, length, cloning frequency and genomic position of the RRV miRNAs cloned in this analysis. Coordinates are given relative to the 26–95 isolate of RRV (accession number AF210726). Terminal variation in the length of miRNA cDNA clones is indicated by parentheses around the variable nucleotide. Where two miRNAs appear to derive from a single stem-loop precursor RNA, the more 5’ miRNA is indicated by the suffix-5p and the more 3’ by -3p.
MiRNAs are initially transcribed as part of one arm of an RNA hairpin that in turn forms part of a longer capped, polyadenylated primary miRNA transcript (Bartel, 2004). Computer analysis of the RRV sequences flanking the cloned RRV miRNAs revealed that all but one of these candidate RRV miRNAs could indeed be folded into the imperfect RNA hairpins that are characteristic of primary miRNA transcripts (Fig. 1). However, one candidate RRV miRNA, i.e. miR-rR1-3, was not readily folded into the expected RNA hairpin structure. This RRV miRNA will be further discussed below.
Fig. 1.
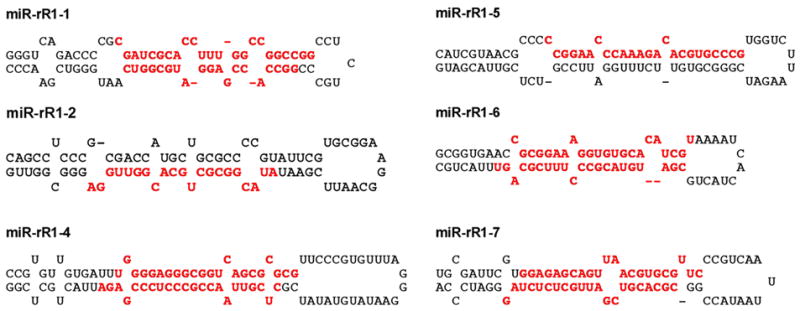
Predicted RNA folding of the primary miRNA stem-loop structures that give rise to ten of the eleven RRV miRNAs listed in Table 1. Cloned RRV miRNAs are indicated in red.
Of the 11 RRV miRNAs obtained, 8 derive from 4 stem-loop precursors and therefore represent either two miRNAs of different function or a functional miRNA and a “passenger”, i.e. a non-functional, strand. In either case, these 2 short RNAs should anneal to form an imperfect stem in the primary precursor, leaving ~2 nt 3’ overhangs at each end (Bartel, 2004). As can be seen in Fig. 1, this was generally the case. Moreover, the structure of these RRV primary miRNA precursors, consisting of the central miRNA duplex, a basal ~10 bp stem extension and a large,≥10 nt terminal loop, is consistent with earlier work that has defined structural features present in primary miRNA hairpins that are recognized by the cellular RNA processing enzyme Drosha and its co-factor DGCR8 (Zeng et al., 2005).
To further confirm the validity of the RRV miRNAs listed in Table 1, and to see whether both strands of miR-rR1-1, miR-rR1-4, miR-rR1-6 and miR-rR1-7 could indeed be detected in RRV infected cells, we performed primer extension analyses to detect all 11 of the viral miRNAs listed in Table 1. The primers used were designed to be 5 or 6 nucleotides shorter, at the 3’ end, than the full-length viral miRNA sequences listed in Table 1. The RNA preparation used was derived from a human cell line, 293, that had been infected at high multiplicity with the RRV-GFP virus. Infection of 293 cells with RRV results in a mixture of cells that become lytically and latently infected (DeWire and Damania, 2005). This experiment allowed us to analyze RRV miRNA expression in a second, distinct RRV-infected cell line.
As shown in Fig. 2, this primer extension analysis detected all 11 of the RRV miRNAs in infected 293 cells that had been previously cloned from infected MVEC cells, although some signals, e.g. for miR-rR1-1-3p, were quite faint. This latter result is consistent with the fact that miR-rR1-1-3p was only cloned once from infected MVEC cells while the complimentary miR-rR1-1-5p miRNA was cloned 27 times (Table 1) and is also more readily detected (Fig. 2). This suggests that miR-rR1-1-5p is the active viral miRNA while miR-rR1-1-3p is likely the passenger strand. For other RRV miRNAs, e.g. miR-rR1-4, both the 5p and 3p strand were cloned multiple times (Table 1) and gave rise to robust primer extension signals (Fig. 2), so it is currently unclear whether only the more highly expressed miR-rR1-4-5p is physiologically relevant or whether both strands are active miRNAs. Importantly, the miR-rR1-3 miRNA, which was not readily folded into the expected primary miRNA hairpin structure, was also readily detected in RRV-infected 293 cells (Fig. 2). We therefore conclude that this candidate miRNA is, in fact, an authentic RRV miRNA. It is therefore likely that it does indeed form part of an RNA hairpin in the primary miRNA precursor even though we have not yet been able to deduce a convincing hairpin structure.
Fig. 2.
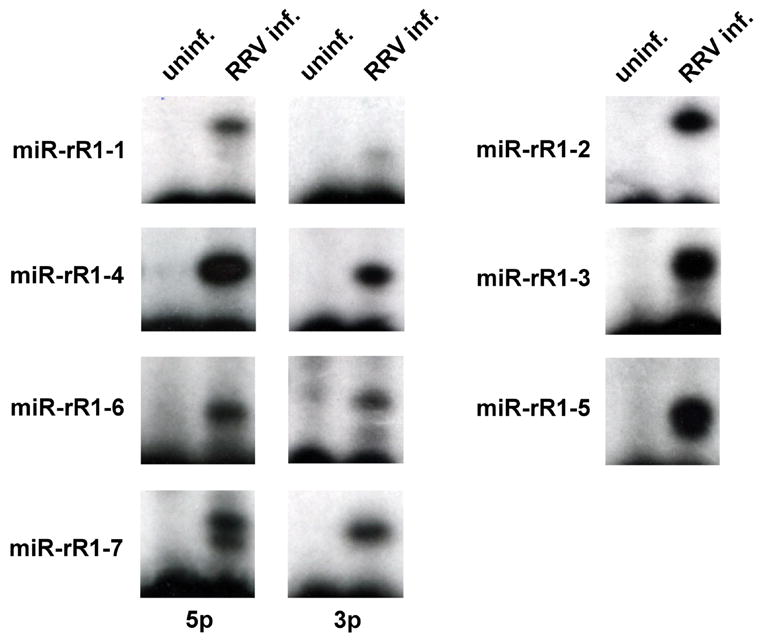
Detection of RRV miRNAs using primer extension analysis. The RNA samples analyzed were derived from control 293 cells or from 293 cells that were infected with RRV at high multiplicity and that were beginning to enter lytic replication. The primers used were all designed to be 5 or 6 nt shorter, at the 3’ end, than the mature RRV miRNAs listed in Table 1. In the case of miR-rR1-7-5p, we detected two primer extension products, differing by 1 nt in length, as predicted in Table 1. All other miRNAs gave rise to a single extension product, with the possible exception of miR-rR1-4.
Consistent with the data shown in Table 1, miR-rR1-7-5p was found to give rise to 2 primer extension products. However, miR-rR1-2, which like miR-rR1-7-5p also gave rise to 2 cDNA clones differing by 1 nucleotide at the 5’ end (Table 1), only gave a single primer extension product (Fig. 2). This may imply that only the longer, 21 nt version of miR-rR1-2 is actually expressed in vivo and that the single, 20 nt cDNA sequence obtained had lost one 5’ nucleotide during RNA isolation or cDNA synthesis. These data also suggested that miR-rR1-5, which was cloned only once from RRV infected MVECs yet was readily detected in RRV infected 293 cells, also exists in two forms differing by 1 nt at the 5’ end. In any event, these primer extension data clearly validate the existence of all of the RRV miRNAs listed in Table 1.
An interesting question is whether these RRV miRNAs are induced during lytic RRV replication. To address this question, we took advantage of the previous finding that RRV can be maintained in a latent state in infected 293 cells by treatment with the antiviral drug ganciclovir but enters lytic replication when cultured in the absence of this inhibitor. We therefore prepared total RNA from uninfected 293 cells, 293 cells infected with RRV and maintained in the presence of ganciclovir and finally 293 cells infected with RRV and maintained in the absence of drug. To confirm the state of the viral infection, we performed RT-PCR analysis using primers specific for the RRV ORF71 mRNA, which should be expressed during both latent and lytic infection, and the RRV ORF50/RTA mRNA, which encodes a viral protein seen exclusively in lytically infected cells. As shown in Fig. 3A, the ORF71 mRNA was indeed detected in RRV-infected 293 cells regardless of drug treatment, while the ORF50/RTA mRNA was only detected in RRV-infected 293 cells cultured in the absence of ganciclovir. Primer extension analyses for three of the RRV miRNAs (miR-rR1-2, miR-rR1-3 and miR-rR1-5) showed that all three miRNAs were detectable in latently RRV infected 293 cells but were strongly activated in 293 cells undergoing lytic replication. No increase was observed when a cellular miRNA, human miR-16, was analyzed in parallel. We therefore conclude that RRV miRNA expression is indeed activated during lytic virus replication.
Fig. 3.
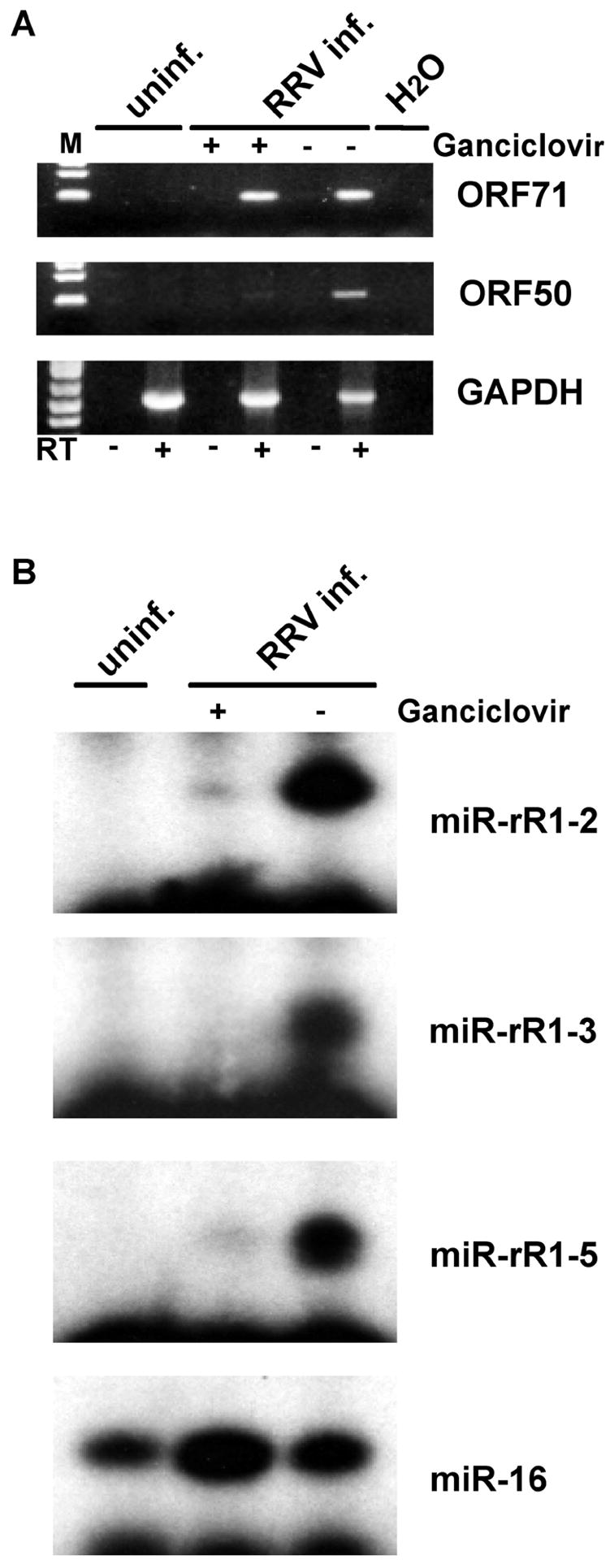
RRV miRNA expression is enhanced upon entry into the lytic phase. A) RT-PCR analysis of the expression of a latent RRV mRNA (ORF71), a lytic RRV mRNA (ORF50/RTA) and a control cellular mRNA (GAPDH) using total RNA derived from uninfected 293 cells, 293 cells maintained in the latent phase of the RRV replication cycle by treatment with ganciclovir, or lytically RRV-infected 293 cells. Reactions were controlled for contaminating cellular DNA by performing the PCR amplification without first performing a reverse transcription (RT) step. B) This primer extension analysis was performed as described in Fig. 2 using the RNA preparations described in panel A. The cellular miR-16 miRNA served as a loading control.
The 12 KSHV miRNA previously identified by ourselves and others (Cai et al., 2005; Pfeffer et al., 2005; Grundhoff et al., 2006) cluster into a small, primarily non-coding region within the KSHV genome located between the ORF69 and ORF71 (also known as the V-FLIP) genes. As shown in Fig. 4, this is also true in RRV, although RRV lacks the K12/Kaposin gene found in KSHV. In both viruses, all the miRNAs are in the same transcriptional orientation and, in latently KSHV-infected cells, it has, in fact, been shown that all the KSHV miRNAs are derived from transcripts that initiate 5’ to ORF71 and terminate transcription 3’ to the K12/Kaposin gene (Cai and Cullen, 2006). While relatively little is known about the transcriptional regulation of RRV, it is known that there are both latent and lytic promoters located 5’ to the ORF73 gene in RRV (DeWire and Damania, 2005) and it is therefore possible that these promoters give rise to transcripts that are processed to yield all the RRV miRNAs listed in Table 1 and schematically represented in Fig. 4.
Fig. 4.
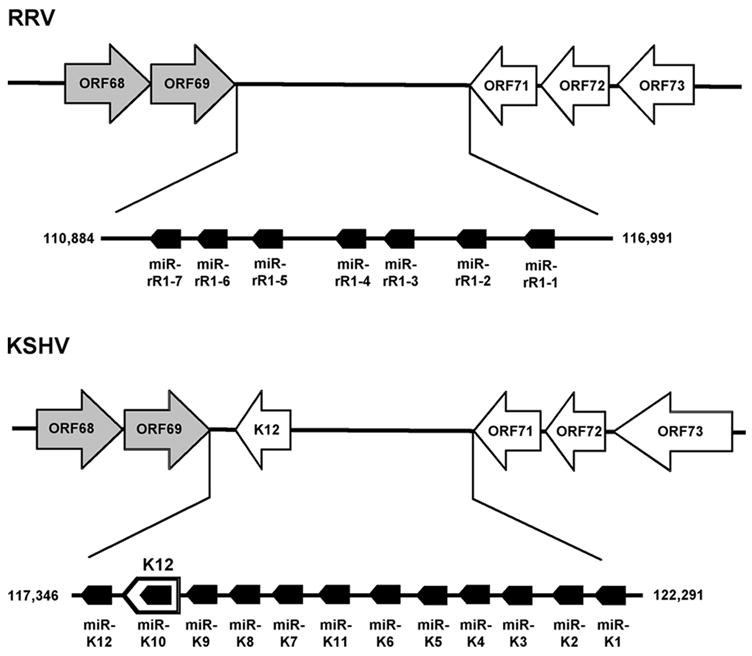
Schematic representation of the genomic localization of the miRNAs encoded by RRV and KSHV. This schematic shows the viral genes conserved between RRV and KSHV in the latency associated region, as well as the K12/Kaposin gene unique to KSHV. ORF73, ORF72, ORF71 and K12 are expressed during latent infection while ORF68 and ORF69 are lytic genes (indicated in grey). The transcriptional orientation of the viral protein coding genes and of the viral miRNAs is indicated.
Discussion
Previous analyses of several distinct γ-herpesviruses, including EBV, its rhesus counterpart rLCV, KSHV and the murine γ-herpesvirus mouse herpesvirus 68, have identified miRNAs in all of these herpesviruses (Pfeffer et al., 2004, 2005; Cai et al., 2005, 2006; Grundhoff et al., 2006). Therefore, our observation that RRV also encodes miRNAs is not a surprise. There were two principal reasons why this research was undertaken. First, in the case of the human lymphocryptovirus EBV and its rhesus counterpart rLCV, we have previously reported that the viral miRNAs are conserved in terms of genomic localization and, moreover, that 8 of the 16 miRNAs cloned from rLCV are conserved in EBV at the primary sequence level (Cai et al., 2006). We were therefore curious as to whether RRV, a rhadinovirus that is closely related to the human pathogenic herpesvirus KSHV (Alexander et al., 2000), would show a similar pattern. Second, RRV is genetically tractable in that mutations can be introduced into the RRV genome and their effect then analyzed in tissue culture and in infected rhesus macaques (Bilello et al., 2006; Estep et al., 2007; O’Connor and Kedes, 2007). Indeed, in immunocompromised rhesus macaques, RRV is pathogenic. This result is reminiscent of KSHV, which is also innocuous in immunocompetent patients but pathogenic in immunodeficient individuals, including AIDS patients.
The data reported here reveal that the miRNAs present in KSHV and RRV are indeed all located in a single cluster in each virus that is found at the same genomic location. In addition, in both KSHV and RRV, all the viral miRNAs are in the same transcriptional orientation (Fig. 4). It therefore seems likely that the transcription and processing of the RRV miRNAs occurs via essentially the same mechanism seen in KSHV. Specifically, in KSHV, miRNA expression is dependent on promoters located 5’ to the viral ORF72 and ORF73 genes and requires the read-through of a “leaky” poly-(A) addition site located 3’ to ORF71 (Cai and Cullen, 2006). This results in long transcripts that, at least in latently KSHV infected cells, are processed to give rise to all 12 KSHV miRNAs (Fig. 3). In RRV, transcription is less well understood but lytic and latent promoters located 5’ to RRV ORF73 have been identified (DeWire and Damania, 2005) and there is a consensus poly(A) addition site located immediately 3’ to RRV ORF71. Whether this RRV poly(A) addition site shares the leakiness characteristic of the equivalent site in KSHV, thereby giving rise to a single primary miRNA precursor for all the RRV miRNAs, remains to be determined.
One important difference between KSHV and RRV is that, in the case of KSHV, the majority of the viral miRNAs show at most a modest increase in expression upon entry into lytic replication (Cai et al., 2005; Pfeffer et al., 2005). The exceptions are miR-K10 and miR-K12, which are significantly induced upon entry into lytic replication due to the presence of a potent lytic promoter located immediately 5’ to the K12 ORF (Cai and Cullen, 2006). In contrast, in the case of RRV, it appears that the expression of all the viral miRNAs is strongly enhanced during lytic replication (Fig. 3). This difference likely results from the fact that RRV contains a potent lytic promoter located immediately 5’ to the ORF73 gene (DeWire and Damania, 2005) that is lacking in KSHV (Cai and Cullen, 2006). In contrast, the lytic promoter located 5’ to the K12 ORF of KSHV, as well as the K12 ORF itself, are both absent in RRV (Fig. 4).
A perhaps somewhat surprising result reported here is that we only identified 6 distinct primary miRNA stem-loops in RRV (Fig. 2) that give rise to 10 viral miRNAs (Table 1) (An additional RRV miRNA, miR-rR1-3, could not be assigned to an RNA stem-loop structure but was readily detectable in RRV infected cells, as shown in Figs. 2 and 3.). It therefore is clearly possible that we have missed other viral miRNAs that are expressed at a low level in RRV-infected MVEC cells, although we did clone several “passenger” strands for the more highly expressed viral miRNAs (Table 1). However, even if other miRNAs are located in this region of the RRV genome, computer analysis does not reveal the presence of any sequences closely similar to any of the KSHV miRNAs.
The finding that RRV has not conserved any of the miRNAs present in KSHV, while 8 out of the 16 miRNAs expressed by rLCV are conserved in EBV (Cai et al., 2006), likely reflects the greater sequence divergence between the genomes of KSHV and RRV, when compared to EBV and rLCV (Alexander et al., 2000; Cai et al., 2006). However, despite the fact that none of the KSHV miRNAs are conserved in RRV, it clearly remains possible that these viral miRNAs down-regulate the same host innate or adaptive immune response pathways and possibly even the same mRNAs. In any event, these results set the stage for the mutational inactivation of the miRNAs present in RRV and the future analysis of the effect of these mutations on the replication of RRV in culture and in vivo and on the ability of RRV to exert a pathogenic effect in immunodeficient rhesus macaques.
Materials and Methods
Maintenance of MVECs and RRV-GFP infection
Primary rhesus monkey vascular endothelial cell cultures (MVEC21150 CGB-5), kindly provided by Dr. Jay Nelson (Oregon Health Sciences University, Portland, OR), were maintained in Endothelial Cell Medium 2 (EBM-2; Cambrex BioScience, Walkersville, MD) supplemented with SingleQuots (Cambrex). When confluent, MVECs were passed using Accutase (Innovative Cell Technologies, San Diego, CA). One day after passage, MVEC cultures were infected with approximately 2 ×105 infectious units of RRV-GFP (Bilello et al., 2006) and passed when cultures became confluent.
RNA extraction, cloning and analysis
At day 24 post-infection, RNA was isolated from RRV-GFP-infected MVEC cultures using TRIzol Reagent (Invitrogen, Carlsbad, CA). After the addition of 0.2 vol. chloroform, the samples were shaken vigorously for 15 sec and incubated for 3 minutes at room temperature. Samples were then centrifuged at 12,000 ×g for 15 minutes at 4°C and the aqueous phase transferred to a fresh tube. Total RNA was precipitated by the addition of 0.5 vol. isopropanol, incubated at room temperature for 10 min and centrifuged at 12,000 ×g for 10 minutes at 4°C. The supernatant was discarded and the RNA pellet was washed in 1 vol. 75% ethanol followed by centrifugation at 7,500 ×g for 5 min at room temperature. The resultant RNA pellet was briefly dried and dissolved in ddH2O. The isolation, cDNA cloning and sequence analysis of the small, 18–25 nt long RNAs present within this preparation of RRV-infected MVEC RNA was then performed as previously described (Cai et al., 2005, 2006).
Human 293 cells were infected with the RRV-GFP virus at a multiplicity of ~1 pfu/cell in the presence of polybrene (5 μg/ml). After 24 h, the supernatant medium was replaced with fresh medium. At 72 h post-infection, all the 293 cells showed GFP expression and ~50% of the cells showed a cytopathic effect indicative of lytic RRV replication. To generate latently RRV-infected 293 cell cultures, 293 cells were infected as above. After one hour, the media was replaced by fresh media containing 500 μM ganciclovir (Calbiochem). After 72 h, essentially all the 293 cells showed GFP expression but no cytopathic effect was evident. Total RNA was then isolated from control, uninfected and from latently or lytically RRV-infected 293 cells, as described above. Primer extension analyses were performed as previously described (Cai and Cullen, 2006) using primers that were designed to be 5 or 6 nt shorter than human miR-16 and the mature, full-length RRV miRNAs listed in Table 1.
RT-PCR analysis
Two μg of total RNA isolated from uninfected 293 cells, latently RRV infected 293 cells or lytically RRV infected 293 cells was treated with DNase and was then subjected to reverse transcription using Oligo dT primers (Invitrogen/Superscript II). The resultant cDNAs were amplified with previously described primers specific for RRV ORF50/RTA (Dittmer et al., 2005), RRV ORF71 (DeWire and Damania, 2005) or human GAPDH (Cai et al., 2004). The PCR amplification was performed for 35 cycles (ORF71) or 30 cycles (ORF50 and GAPDH) at 94°C for 30s, 42°C for 30s and 72°C for 30s.
Supplementary Material
Acknowledgments
The authors thank Blossom Damania and Jay Nelson for reagents used in this research. This research was supported by grants from the National Institute of Health to BRC (AI067968) and RCD (AI063928 and RR00168).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander L, Denekamp L, Knapp A, Auerbach MR, Damania B, Desrosiers RC. The primary sequence of Rhesus monkey rhadinovirus isolate 26–95: Sequence similarities to Kaposi’s sarcoma-associated herpesvirus and Rhesus monkey rhadinovirus isolate 17577. J Virol. 2000;74:3388–3398. doi: 10.1128/jvi.74.7.3388-3398.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bechtel JT, Liang Y, Hvidding J, Ganem D. Host range of Kaposi’s sarcoma-associated herpesvirus in cultured cells. J Virol. 2003;77:6474–6481. doi: 10.1128/JVI.77.11.6474-6481.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilello JP, Morgan JS, Damania B, Lang SM, Desrosiers RC. A genetic systemic for Rhesus monkey rhadinovirus: Use of recombinant virus to quantitate antibody-mediated neutralization. J Virol. 2006;80:1549–1562. doi: 10.1128/JVI.80.3.1549-1562.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackbourn DJ, Lennette E, Klencke B, Moses A, Chandran B, Weinstrein M, Glogau RG, Witte MH, Way DL, Kutzkey T, Herndier B, Levy JA. The restricted cellular host range of human herpesvirus 8. AIDS. 2000;14:1123–1133. doi: 10.1097/00002030-200006160-00009. [DOI] [PubMed] [Google Scholar]
- Cai X, Cullen BR. Transcriptional origin of Kaposi’s sarcoma-associated herpesvirus microRNAs. J Virol. 2006;80:2234–2242. doi: 10.1128/JVI.80.5.2234-2242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc Natl Acad Sci USA. 2005;102:5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Schäfer A, Lu S, Bilello JP, Desorsiers RC, Edwards R, Raab-Traub N, Cullen BR. Epstein-Barr virus microRNAs are evolutionarily conserved and differentially expressed. PLos Pathog. 2006;2:e23. doi: 10.1371/journal.ppat.0020023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. Viruses and microRNAs. Nat Genet. 2006;38:S25–S30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- DeWire SM, Damania B. The latency-associated nuclear antigen of Rhesus monkey rhadinovirus inhibits viral replication through repression of Orf50/Rta transcriptional activation. J Virol. 2005;79:3127–3138. doi: 10.1128/JVI.79.5.3127-3138.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWire SM, Money ES, Krall SP, Damania B. Rhesus monkey rhadinovirus (RRV): Construction of a RRV-GFP recombinant virus and development of assays to assess viral replication. Virology. 2003;312:122–134. doi: 10.1016/s0042-6822(03)00195-8. [DOI] [PubMed] [Google Scholar]
- Dittmer DP, Gonzalez CM, Vahrson W, DeWire SM, Hines-Boykin R, Damania B. Whole-genome transcription profiling of rhesus monkey rhadinovirus. J Virol. 2005;79:8637–8650. doi: 10.1128/JVI.79.13.8637-8650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estep RS, Powers MF, Yen BK, Li H, Wong SW. Construction of an infectious Rhesus rhadinovirus (RRV) bacterial artificial chromosome (BAC) for the analysis of Kaposi’s sarcoma-associated herpesvirus (KSHV) related disease development. J Virol. 2007 Jan. 10; doi: 10.1128/JVI.01997-06. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundhoff A, Sullivan CS, Ganem D. A combined computational and microarray-based approach identifies novel microRNAs encoded by human gamma-herpesviruses. RNA. 2006;12:1–18. doi: 10.1261/rna.2326106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Gartner JJ, Sethupathy P, Hatzigeorgiou AG, Fraser NW. Anti-apoptotic function of a microRNA encoded by the HSV-1 latency-associated transcript. Nature. 2006;442:82–85. doi: 10.1038/nature04836. [DOI] [PubMed] [Google Scholar]
- O’Connor CM, Kedes DH. Rhesus monkey rhadinovirus: A model for the study of KSHV. Curr Top Microbiol Immunol. 2007;312:43–69. doi: 10.1007/978-3-540-34344-8_2. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander S, Grässer FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Zavolan M, Grässer FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T. Identification of virus-encoded microRNAs. Science. 2004;304:734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- Samols MA, Hu J, Skalsky RL, Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi’s sarcoma-associated herpesvirus. J Virol. 2005;79:9301–9305. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D. SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells. Nature. 2005;435:682–686. doi: 10.1038/nature03576. [DOI] [PubMed] [Google Scholar]
- Zeng Z, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


