Abstract
CELO (chicken embryo lethal orphan) virus is an avian adenovirus that is being developed as a gene transfer vector. Its trimeric major coat protein (942 residues, 106,709 Da) has 42% sequence identity to human adenovirus type 2 (AdH2) hexon and 45% to AdH5 hexon. For structural studies, the growth of CELO virus has been optimized, and its hexon purified and crystallized. The hexon crystals, the first non-human example, diffract to 3.9 Å resolution. Molecular replacement using the AdH5 model was used to identify the location of the CELO hexon within the unit cell. There is one hexon monomer in the asymmetric unit of the trigonal space group P321 (a = b = 157.8 Å, c = 114.2 Å, γ = 120º) and the solvent content is 67.8%. The hexons pack in a hexagonal honeycomb so that large ~100 Å diameter channels run through the entire crystal. This remarkable property of the crystals lends itself to their exploitation as a nanomaterial. Structural studies on CELO will elucidate the differences between avian and human adenoviruses and contribute to a better understanding of adenoviruses with non-human hosts.
Keywords: Avian Adenovirus, CELO, Major Coat Protein, Hexon, Crystal Packing, Nanotechnology
CELO, an avian adenovirus with useful properties
Chicken embryo lethal orphan (CELO), an avian adenovirus (also known as fowl adenovirus type 1; FAV-1), is of interest as a gene transfer vector (Francois et al., 2001) for human gene therapy, or for vaccine delivery (Kozarsky and Wilson, 1993). Non-human adenovirus vectors, such as CELO (Shashkova et al., 2005) and chimpanzee (Farina et al., 2001), are advantageous for human therapeutic use as they evade the pre-existing immune responses frequently occurring from natural adenovirus infections.
The structural organization of CELO virus is similar to that of human adenovirus (Rux and Burnett, 2004). Its icosahedral protein capsid is formed from 240 hexons and 12 penton vertex complexes (Laver et al., 1971). CELO hexon is a very stable trimeric protein with three 942-residue polypeptide chains (106,709 Da). Sequence alignment of CELO hexon with human adenovirus types 2 (AdH2) or AdH5 hexons (86% common identity) using the program Clustal W (Higgins et al., 1996) reveals similar identities of 44% and 45% (Fig. 1) (Rux et al., 2003; Xu, 2002). The structure of CELO hexon is likely to resemble the crystal structures of AdH2 (967 residues) and AdH5 hexon (951 residues) (Rux and Burnett, 2000; Rux et al., 2003). These are very similar, with each subunit containing two jellyrolls, or β-barrels, with the “viral” fold common in spherical virus coat proteins. The differences between the human and avian sequences are concentrated in the long loops at the top of the molecule (Fig. 1) and these hypervariable regions of sequence are likely to define hexon’s type-specific antigenic determinants (Crawford-Miksza and Schnurr, 1996; Rux et al., 2003). This assumption was recently verified experimentally for a chimpanzee adenovirus, AdC68 (Pichla-Gollon et al., 2006).
Figure 1.

Sequence comparison between AdH2, AdH5 and CELO hexons. Identities are shown with red backgrounds and white letters; positions where amino acids have similar physico-chemical properties, and so are conserved, have a green background and black letters. Numbers follow the absolute alignment position (including gaps), for which the secondary structures are shown as cylinders (helices) and arrows (strands). The two eight-stranded viral jellyrolls are depicted in blue (V1) and green (V2), while other secondary structure is in black. It is not known if the initial methionine in CELO hexon is removed, as for AdH2 and AdH5 hexons where the N-terminus is acetylated. The sequence alignment was performed using Clustal W (Higgins et al., 1996) and the figure was prepared with ALSCRIPT (Barton, 1993).
Infection by adenovirus occurs in two steps, with recognition by the projecting knob at the end of the trimeric fiber leading to uptake signaled by the pentameric penton base (Rux and Burnett, 2004). CELO virus is unusual in that two fibers, instead of one, are associated with each penton base. A recent structure for the penton base of AdH2 shows how the trimeric fiber overcomes its symmetry mismatch with the pentameric base to form the penton complex, and suggests how two fibers can bind to a single base in CELO (Zubieta et al., 2005). Other structural differences between CELO and human adenovirus are also intriguing. CELO virus lacks the genes for the DNA core protein polypeptide V and the minor capsid cementing protein polypeptide IX (Chiocca et al., 1996). In human adenovirus, polypeptide IX stabilizes the virion (Furcinitti et al., 1989; van Oostrum et al., 1986) so it is somewhat surprising that the CELO virion exhibits a higher heat stability than that of AdH5 (Michou et al., 1999).
Structural studies on CELO virus are aimed at understanding the architecture and stability of its virion and so aid its development as a gene transfer vector. The CELO hexon has now been purified and crystallized. Its structure will be used for combined imaging with electron microscopy reconstructions to make a quasi-atomic model of the virion, as done for AdH2 (Stewart et al., 1991; Stewart et al., 1993), AdH5 (Fabry et al., 2005) and bacteriophage PRD1 (San Martín et al., 2001; San Martín et al., 2002). Structural studies on PRD1 have shown that its coat protein is similar to that of adenovirus (Benson et al., 1999) and led to the hypothesis that double-barrel trimers are a structural marker for a common viral lineage that embraces a wide range of apparently unrelated viruses (Bamford et al., 2002; Benson et al., 2004). Hexon crystal structures have been important in comparative studies of adenoviruses by guiding the alignment of multiple hexon sequences (Rux et al., 2003). Structural information on CELO hexon and the viral epitopes is particularly important for CELO’s development as a vaccine carrier as it will aid in the construction of a series of variants for revaccination by modification of the viral coat. This strategy is being used successfully for chimpanzee adenovirus (Pichla-Gollon et al., 2006).
The CELO hexon crystals themselves are of interest as they could be engineered to create defined nanomaterials, as has been done with crystals of cowpea mosaic virus (Falkner et al., 2005). Viral coat proteins are particularly useful nanoscale building blocks (Douglas et al., 2002) as they form highly-symmetrical capsids that can be manipulated to possess predictable physical and chemical properties (Wang et al., 2002). Even larger assemblies can be formed. For example, human hexon forms two-dimensional arrays as well as virions. Its potential for making different stable inter-molecular contacts is shown by its formation of two types of array: continuous; or with a regular pattern of holes (van Oostrum et al., 1986). The high physical stability of CELO virions (Cotten et al., 1993), which is advantageous for its use as a vector, also makes it attractive for use as a bio-nanomaterial. A further practical and economic advantage is that CELO virus can be grown efficiently and inexpensively in embryonated chicken eggs (Laver et al., 1971).
CELO growth in chicken eggs; hexon purification and crystallization
For structural studies, several experimental variables were optimized to maximize the viral yield (Xu, 2002). The detailed protocols used for viral growth, and the purification of virions and hexons, are available online as supplementary material. In brief, CELO virus (Phelps strain, serotype FAV-1) was kindly supplied by Dr. Matthew Cotten (Research Institute of Molecular Pathology, Vienna, Austria). Nine-day-old embryonated chicken eggs (B&E Eggs, Stevens PA) were injected with 50 μl of virus solution containing 2 × 109 virions ml−1. After 3 days incubation, the eggs were refrigerated overnight before the allantoic fluid surrounding the embryo was harvested. Electron microscopy was used routinely for quality control. This revealed the presence of ~200 Å diameter particles in the virus stock solution obtained from Dr. Cotten, as well as one purchased from ATCC (Catalog No. VR-432). Their size and morphology corresponded to avian adenovirus-associated virus (AAAV), a common contaminant (Yates et al., 1973). AAAV is a defective parvovirus that only grows in the presence of the helper adenovirus (Atchison et al., 1965).
The CELO virions and hexon were purified by a modification of the scheme developed in our laboratory for AdH2 and AdH5 (Rux and Burnett, 2006). After removing cellular debris and lipids from the allantoic fluid, the virions were pelleted by ultra-centrifugation in a pre-cooled SW28 rotor at 27,000 rpm for 1 hr at 5 °C. Hexon was then separated from the other viral proteins in the supernatant by ammonium sulfate precipitation, followed by two anion exchange columns (Q-Sepharose Fast, and Bio-Rad Q10). Hexon fractions were concentrated to 2 ml before further purification on Hi-Load Superdex 200 or 75 gel filtration columns. The purified protein was verified by SDS-PAGE and N-terminal protein sequencing.
For crystallization, the purified protein was concentrated to ~10 mg ml−1 and exchanged with a Centricon PM-10 micro-concentrator into hexon storage buffer (10 mM sodium phosphate, pH 7.0, 0.02% sodium azide) (Rux and Burnett, 2006). Equilateral triangular plates (typically 0.06 × 0.3 × 0.3 mm3) crystallized from sitting-drops at 22 °C containing 5 μl protein and 5 μl crystallization solution (7% isopropanol, 13% PEG-4000, 0.01 M glycine and 0.1 M HEPES at pH 7.5). A sharp decrease in resolution occurred on freezing the crystals with cryoprotectants, such as 15% isopropanol and 20% PEG-4000. Diffraction data were collected with an RAXIS-IV detector (1° oscillation, 12 min exposure) at room temperature from a single crystal in a quartz capillary using a Rigaku II rotating copper anode X-ray generator.
The diffraction data (Table 1) were processed with the programs Denzo and SCALEPACK (Otwinowski and Minor, 1997) to indicate the primitive trigonal space group P321 (a = b =157.8 Å, c=114.2 Å, γ=120º). One CELO hexon monomer lies in the asymmetric unit, consistent with a trimeric molecule along the threefold crystallographic axis. The solvent content is 67.8%, with a Vm of 3.85 Å3 Da−1 (Matthews, 1968). The data reduction statistics are consistent with decay of the unfrozen crystals (Rsym=20.5%). Initially, the known structure of AdH5 hexon (PDB code 1P30) (Rux et al., 2003) was used as the search model for molecular replacement with the programs CNS (Brünger et al., 1998) or AMoRe in CCP4 (Collaborative Computational Project Number 4, 1994). The rotation function contained one strong peak with a correlation coefficient (0.20) significantly higher than background (0.15–0.17). The corresponding orientation in the translation function again gave a single prominent peak (0.45) above the noise (0.22–0.27). Rigid body refinement using AMoRe decreased the Rfactor from 43.6% to 42.8%.
Table 1. Data collection statistics.
| Data processing | |
|---|---|
| Space Group | P321 |
| Unit Cell | a = b = 157.8 Å, c = 114.2 Å, γ = 120.0° |
| Molecules per asymmetric unit | 1 subunit (942 residues) of a hexon trimer |
| Resolution range (Å) (outer shell) | 50 − 3.90 (3.97 − 3.90)c |
| Total reflections | 67,452 |
| Unique reflections | 14,636 |
| Completeness (%) | 95.7 (94.4) |
| Redundancya | 4.6 (4.2) |
| Rsym (%)b | 20.5 (47.8) |
| Average I/σ(I) | 7.1 (2.5) |
Redundancy is the ratio of the number of measured reflections to the unique reflections.
Rsym=Σh |Ih−<Ih>|/ΣIh, where Ih is the intensity of each individual reflection, and <Ih> is the average intensity for the multiple observations of symmetry-related reflections.
Values in parentheses refer to the highest resolution shell.
As further refinement was difficult at the current resolution, the molecular replacement solution was checked using the program MOLREP in CCP4 (Collaborative Computational Project Number 4, 1994). Three different search models were used: AdH5 hexon (PDB code 1P30) (Rux et al., 2003); AdH2 hexon (PDB code 1P2Z) (Rux et al., 2003); and a CELO hexon model, based upon the AdH5 structure, produced with the program JACKAL (http://trantor.bioc.columbia.edu/programs/jackal/index.html). Each model gave a single solution and all three solutions agreed. The correlation coefficients were 0.40 for AdH2, 0.41 for AdH5, and 0.36 for the CELO model hexon. Maps were generated with the model phases, averaged and solvent flattened in CNS (Brünger et al., 1998), and used to assess how well the CELO model fit the electron density. Density for the protein chain was evident in the base of the molecule, especially for the two jellyrolls. In the tower, the molecular envelope could be discerned but the density was discontinuous and so the peptide chain could not be traced. A total of 227 residues in the tower loops and other poorly-fitting regions were removed, and refinement carried out in CNS (Brünger et al., 1998) using the phase information from the averaged, solvent-flattened map as a restraint. The Rwork improved from 47.3% to 37.2% and the Rfree from 46.7% to 41.6%, but essentially the jellyrolls are the only well-determined part of the model. As various efforts at further refinement were fruitless, significant improvements in both resolution and diffraction quality will be necessary to obtain additional structural information. The structure factors and molecular replacement solution have been deposited in the Protein Data Bank (2INY).
Large channels run throughout the CELO hexon crystal
The remarkable crystal packing of CELO hexon, the first non-human example (Xu, 2002), is quite different than that observed for human hexons. AdH2 and AdH5 hexon crystals both have the same cubic space group, P213. In this unit cell, three of the four hexon trimers are related by the threefold crystallographic symmetry axis along the body diagonal. The fourth molecule lies in the center of the other three to give a tetrahedral arrangement that maximizes the separation of the charged towers (Athappilly et al., 1994) (Fig. 2). CELO hexon also crystallizes with coincident molecular and crystallographic threefold axes to give one monomer in its P321 asymmetric unit (Fig. 3). Nevertheless, the inter-hexon contacts are quite different and the main feature of the packing is that the hexons form layers. Even so, the trimers are not in continuous sheets, as in the virion facets, but are organized as a honeycomb (Fig. 4). Their arrangement is reminiscent of the “single-walled holey” sheets formed on EM grids by AdH2 hexons from purified samples or disrupted virions (van Oostrum et al., 1986). A notable difference is that six CELO hexons alternate in an up-down staggered arrangement around the ring, whereas the corresponding hexons in the AdH2 p6 holey sheet are parallel.
Figure 2.
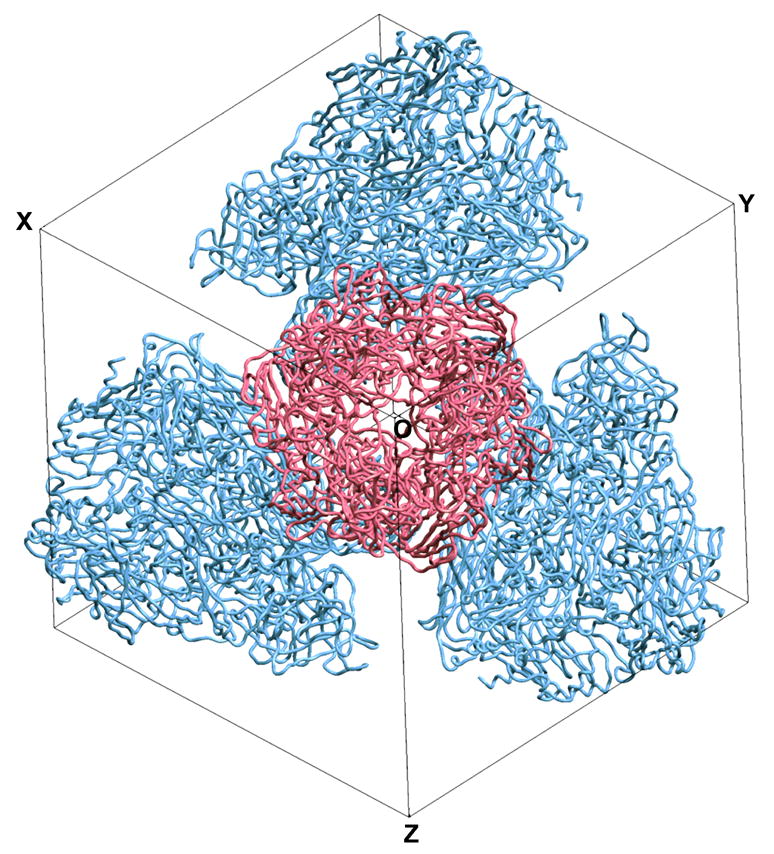
Crystal packing in the P213 unit cell of AdH5 hexon. The stereoview is down the threefold axis of the central molecule (red), which is aligned along the body diagonal of the cubic unit cell. The red molecule interacts with the bodies of the three blue molecules through its base (Rux and Burnett, 2000). Figure produced with SETOR (Evans, 1993).
Figure 3.
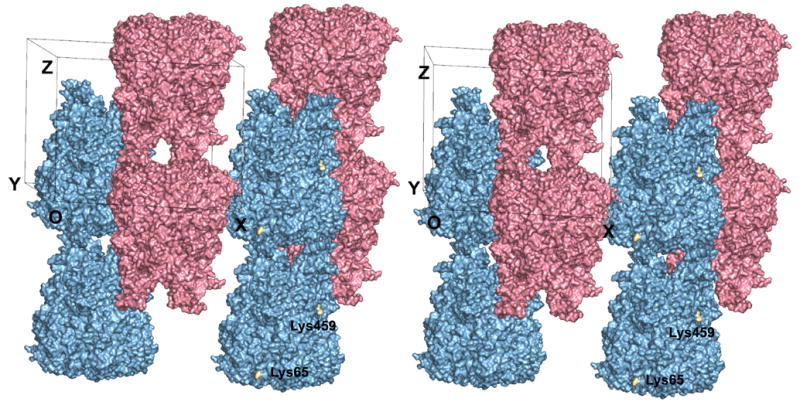
Crystal packing in the P321 unit cell of CELO hexon. The stereoview is almost along the crystallographic twofold axis. There are two hexon trimers in the unit cell (pink and blue), with their molecular axes aligned along the crystallographic threefold axes. These form anti-parallel columns of hexons throughout the crystal. The crystal has alternating layers of hexons with the same color that interlock with each other. Lys65 and Lys459, which lie on the surface of the channel through the crystal, are highlighted in gold and labeled. The CELO hexon model was produced by JACKAL. All residues are included, except those at the tip of the DE1 loop (residues 135–169) that are disordered in the AdH5 and AdH2 hexon crystal structures and so were unconstrained in the molecular modeling. Figure produced with PyMol (DeLano, 2002).
Figure 4.
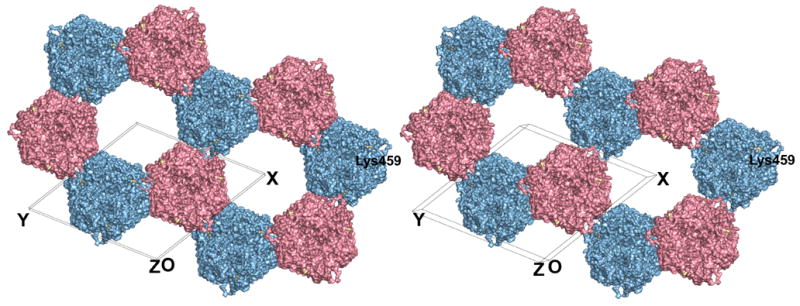
Honeycomb structure in CELO hexon crystals. The stereoview, parallel to the crystallographic threefold axes, shows the different columns of hexon in pink and blue, as in Fig. 3. Adjacent hexons point in opposite directions, with pink molecules up and blue ones down. Six anti-parallel columns surround each of the open channels running throughout the crystal. Lys459 is highlighted in gold and labeled. Figure produced by PyMol (DeLano, 2002).
A prominent feature of CELO hexon crystals is that alternate layers interlock, so columns of hexons making top-to-base interactions alternate with columns inverted by twofold symmetry running in the opposite direction (Fig. 3). These two columns lie along the two crystallographic threefold axes in the unit cell. The up-down interlocking occurs because each hexon in one column is partially buried by three neighboring hexons in columns pointing in the opposite direction. Thus, each hexon has top-to-base interactions vertically with the next one in the same column, and base-to-base interactions horizontally with hexons in the three surrounding inverted columns. In particular, the base-to-base contacts mainly occur through one facet of the pseudo-hexagonal base, which is formed from the two viral jellyrolls in one hexon monomer. The top-to-base contacts juxtapose the hypervariable loops of one trimer with the pseudo-hexagonal base of another. Despite this potential for stabilization, these loops must be flexible as their electron density is much more diffuse than that for the jellyrolls. This feature may be the origin of the poor diffraction as it could lead to long-range disorder in the columns. The electron density is best defined for the two jellyrolls in the base, which has the most sequence similarity across species and is also the best-defined part in the AdH2 and AdH5 crystal structures. The hexon tops in one column do not contact hexons in an adjacent column. The base-to-base packing produces a hexagonal honeycomb with large ~100 Å diameter channels running through the entire crystal (Fig. 4). The channel itself has a pseudo-hexagonal shape, with each face formed by the jellyrolls from adjacent hexon subunits in one trimeric hexon. This remarkable open lattice provides a symmetrical array that might be useful for nanotechnology.
The dissimilar crystal packing of CELO and human adenovirus hexons most probably reflects sequence differences in the towers and six additional residues at the CELO N-terminus. In the P213 crystals for AdH2 and AdH5 hexons, the N-termini lie in a region of inter-molecular contacts. There is insufficient room at this position for the six additional residues in CELO hexon’s longer N-terminus, so these would prevent crystallization in P213. Although AdH2, AdH5 and CELO hexon have very similar isoelectric points (~5), CELO crystallizes at pH 7.5. In contrast, AdH2 and AdH5 hexons both crystallize at pH 3.2, and crystal formation is very sensitive to the exact value (Rux and Burnett, 2006). This is particularly true for AdH2 hexon, which has a striking acidic stretch of residues (133–161) containing 16 glutamates and 4 aspartates. This stretch, which lies in the extended DE1 loop in the tower at the top of the molecule, has a counterpart in AdH5 (Rux and Burnett, 2000) but not in CELO. In AdH2 and AdH5 hexon crystals, the acidic stretches project into solvent and are disordered in the crystal structure. The same lack of charged towers occurs in a chimpanzee adenovirus hexon (AdC68), and this too forms non-cubic crystals (Xue and Burnett, 2006). In the orthorhombic C222 crystals of AdC68 hexon, the molecules form closely packed sheets of antiparallel hexon ribbons so that their interactions mimic those in the viral capsid. While all AdC68 hexons in a sheet have the same vertical orientation, the sheets stack so that hexons form columns with alternating top-to-top and base-to-base contacts. Thus, although CELO hexon is like AdC68 in employing its pseudo-hexagonal shape to form planar arrays, the interactions still do not resemble those in the viral capsid.
Architectural implications for the CELO virion
An intriguing structural difference between CELO and human adenovirus is that the CELO virion lacks the minor protein polypeptide IX (14 kDa) (Chiocca et al., 1996). Combined difference imaging with structures from electron microscopy and X-ray crystallography shows that polypeptide IX lies at four related regions of local threefold symmetry on the facet exterior (Fabry et al., 2005; Furcinitti et al., 1989; Stewart et al., 1993). Polypeptide IX acts as a “cementing” protein to stabilize the interactions of the central nine hexons in the 12-hexon facet (Furcinitti et al., 1989; van Oostrum et al., 1986). These are released as the “group-of-nine hexons” (GON) when human virions dissociate (Prage et al., 1970). Virions form in the absence of polypeptide IX (Colby and Shenk, 1981), but polypeptide IX is indispensable for complete DNA packaging (Ghosh-Choudhury et al., 1987).
CELO virions lack polypeptide IX and do not yield GONs upon disruption (Yates et al., 1973), but they are more stable than those for AdH5 (Michou et al., 1999). The cementing function provided by polypeptide IX in human adenovirus must be compensated for by another structural feature in CELO, such as a somewhat different hexon-hexon interaction. An interesting speculation is that the additional six residues at the CELO hexon N-terminus, which would be located at the edge of the base, could be interacting with the N-termini of neighboring hexon trimers to stabilize the virion. Such stabilizing interactions are quite common in virions, and occur in the bacteriophage PRD1, which is related to adenovirus (Bamford et al., 2002; Benson et al., 1999; Benson et al., 2004). These issues are being explored in a parallel comparative cryo-EM study of adenovirus made possible by the purification of CELO virions (Scheres et al., 2005; Dr. Carmen San Martín and RMB, unpublished results).
CELO hexon crystals as a potential nanobiomaterial
The exploitation of biological assembly mechanisms to fabricate hybrid systems is an attractive avenue for nanotechnology (Katz and Willner, 2005). Proteins that associate into sheets, tubes, ribbons, helices and spheres can be combined with nanoparticles or nanorods to make defined structures. Cavity or channel-containing proteins, such as ferritin (Harrison and Arosio, 1996), or pore-containing bacterial cell surface layers (S-layers) (Schuster et al., 2005) can be used for nanoelectronics (Katz and Willner, 2005).
Viruses have many suitable properties for nanotechnology. The self-assembly properties of their coat proteins are ideal for fabrication. Viral capsids, which are highly regular and relatively abundant, are a particularly promising source of material for catalysis, molecular entrapment, and novel optical and magnetic applications. A good example is cowpea mosaic virus (CPMV) (Lomonossoff et al., 1991), whose structure is known (Lin et al., 1999). Mutant CPMV virions, genetically altered to express a cysteine residue on the viral surface, selectively bind small molecules and show promise as a programmable platform for organic chemical reactions (Wang et al., 2002).
Similarly, the self-assembled nanoscale arrays in CELO hexon crystals, with ~100 Å large channels running throughout the assembly, provide a three-dimensional template. This could be harnessed to generate nanocircuitry by incorporating metals or semiconductors. The channel is large enough to accommodate nanoparticles of gold or silver. Two lysine residues, Lys65 and Lys459, on the inner surface of the hexon nanotubes (Figs. 3, 4) could be exploited to bind gold nanoparticles and produce conductive nanowires. Moreover, bioengineering could be used to modify the residues lining the nanotubes, or to improve the crystalline order by modifying the loops. For example, the cysteine thiol groups could be used to anchor nanoparticles, foreign proteins, or peptides.
The highly ordered lattice of nanotubes in the CELO hexon crystal lattice, like other self-assembled architecture, affords precise atomic level control over the spatial distribution of attached ligands to provide an ordered multivalent material. An obvious immediate application is to use the crystals as a template for growing bundles of metallic nanowires. It will be of great interest to see the practical uses developed by the fascinating new field of biomaterials as it exploits viral proteins such as CELO hexon.
Supplementary Material
Acknowledgments
We thank Drs. John J. Rux and Susan L. Pichla-Gollon for helpful advice and discussion on virus and hexon purification. We are grateful to Drs. Donald L. Ewert, Walter Gerhard, and Andrew J. Caton for their help in teaching us the intricacies of viral growth in eggs. We are obliged to Dr. Matthew Cotten (MRC Laboratories, Banjul, The Gambia, West Africa) for his kind supply of CELO virus and useful suggestions for its purification. We are indebted to Dr. Carmen San Martín for performing the electron microscopy and her interest in the project. We thank Christie A. Cheever, a rotation graduate student at the University of Pennsylvania, for help with CELO virus purification. The research was supported by grants to RMB from the National Institutes of Health (AI 17270) and the National Science Foundation (MCB 95-07102). Other support was provided by the Wistar Cancer Center (CA 10815) and the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Atchison RW, Casto BC, Hammon W. Adenovirus-associated defective virus particles. Science. 1965;149:754–756. doi: 10.1126/science.149.3685.754. [DOI] [PubMed] [Google Scholar]
- Athappilly FK, Murali R, Rux JJ, Cai Z, Burnett RM. The refined crystal structure of hexon, the major coat protein of adenovirus type 2, at 2.9 Å resolution. J Mol Biol. 1994;242:430–455. doi: 10.1006/jmbi.1994.1593. [DOI] [PubMed] [Google Scholar]
- Bamford DH, Burnett RM, Stuart DI. Evolution of viral structure. Theor Popul Biol. 2002;61:461–470. doi: 10.1006/tpbi.2002.1591. [DOI] [PubMed] [Google Scholar]
- Barton GJ. ALSCRIPT: A tool to format multiple sequence alignments. Protein Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- Benson SD, Bamford JKH, Bamford DH, Burnett RM. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell. 1999;98:825–833. doi: 10.1016/s0092-8674(00)81516-0. [DOI] [PubMed] [Google Scholar]
- Benson SD, Bamford JKH, Bamford DH, Burnett RM. Does common architecture reveal a viral lineage spanning all three domains of life? Mol Cell. 2004;16:673–685. doi: 10.1016/j.molcel.2004.11.016. [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chiocca S, Kurzbauer R, Schaffner G, Baker A, Mautner V, Cotten M. The complete DNA sequence and genomic organization of the avian adenovirus CELO. J Virol. 1996;70:2939–2949. doi: 10.1128/jvi.70.5.2939-2949.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colby WW, Shenk T. Adenovirus type 5 virions can be assembled in vivo in the absence of detectable polypeptide IX. J Virol. 1981;39:977–980. doi: 10.1128/jvi.39.3.977-980.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Cotten M, Wagner E, Zatloukal K, Birnstiel ML. Chicken adenovirus (CELO virus) particles augment receptor-mediated DNA delivery to mammalian cells and yield exceptional levels of stable transformants. J Virol. 1993;67:3777–3785. doi: 10.1128/jvi.67.7.3777-3785.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford-Miksza L, Schnurr DP. Analysis of 15 adenovirus hexon proteins reveals the location and structure of seven hypervariable regions containing serotype-specific residues. J Virol. 1996;70:1836–1844. doi: 10.1128/jvi.70.3.1836-1844.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics Systems. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- Douglas T, Strable E, Willits D, Aitouchen A, Libera M, Young M. Protein engineering of a viral cage for constrained nanomaterials synthesis. Adv Mater. 2002;14:415–418. [Google Scholar]
- Evans SV. SETOR: Hardware-lighted three-dimensional solid model representations of macromolecules. J Mol Graph. 1993;11:134–138. doi: 10.1016/0263-7855(93)87009-t. [DOI] [PubMed] [Google Scholar]
- Fabry CMS, Rosa-Calatrava M, Conway JF, Zubieta C, Cusack S, Ruigrok RWH, Schoehn G. A quasi-atomic model of human adenovirus type 5 capsid. EMBO J. 2005;24:1645–1654. doi: 10.1038/sj.emboj.7600653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falkner JC, Turner ME, Bosworth JK, Trentler TJ, Johnson JE, Lin T, Colvin VL. Virus crystals as nanocomposite scaffolds. J Am Chem Soc. 2005;127:5274–5275. doi: 10.1021/ja044496m. [DOI] [PubMed] [Google Scholar]
- Farina SF, Gao GP, Xiang ZQ, Rux JJ, Burnett RM, Alvira MR, Marsh J, Ertl HCJ, Wilson JM. Replication-defective vector based on a chimpanzee adenovirus. J Virol. 2001;75:11603–11613. doi: 10.1128/JVI.75.23.11603-11613.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francois A, Eterradossi N, Delmas B, Payet V, Langlois P. Construction of avian adenovirus CELO recombinant in cosmids. J Virol. 2001;75:5288–5301. doi: 10.1128/JVI.75.11.5288-5301.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furcinitti PS, van Oostrum J, Burnett RM. Adenovirus polypeptide IX revealed as capsid cement by difference images from electron microscopy and crystallography. EMBO J. 1989;8:3563–3570. doi: 10.1002/j.1460-2075.1989.tb08528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh-Choudhury G, Haj-Ahmad Y, Graham FL. Protein IX, a minor component of the human adenovirus capsid, is essential for the packaging of full length genomes. EMBO J. 1987;6:1733–1739. doi: 10.1002/j.1460-2075.1987.tb02425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison PM, Arosio P. The ferritins: molecular properties, iron storage function and cellular regulation. Biochim Biophys Acta. 1996;1275:161–203. doi: 10.1016/0005-2728(96)00022-9. [DOI] [PubMed] [Google Scholar]
- Higgins DG, Thompson JD, Gibson TJ. Using CLUSTAL for multiple sequence alignments. Meth Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- Katz E, Willner I. Integrated nanoparticle-biomolecule hybrid systems: Synthesis, properties, and applications. Angew Chem Int Ed. 2005;43:6042–6108. doi: 10.1002/anie.200400651. [DOI] [PubMed] [Google Scholar]
- Kozarsky KF, Wilson JM. Gene therapy: Adenovirus vectors. Curr Opin Genet Dev. 1993;3:499–503. doi: 10.1016/0959-437x(93)90126-a. [DOI] [PubMed] [Google Scholar]
- Laver WG, Younghusband HB, Wrigley NG. Purification and properties of chick embryo lethal orphan virus (an avian adenovirus) Virology. 1971;45:598–614. doi: 10.1016/0042-6822(71)90175-9. [DOI] [PubMed] [Google Scholar]
- Lin T, Chen Z, Usha R, Stauffacher CV, Dai JB, Schmidt T, Johnson JE. The refined crystal structure of cowpea mosaic virus at 2.8 Å resolution. Virology. 1999;265:20–34. doi: 10.1006/viro.1999.0038. [DOI] [PubMed] [Google Scholar]
- Lomonossoff G, Shanks M, Holness C, Maule A, Evans D, Chen Z, Stauffacher C, Johnson JE. Comovirus capsid proteins: Synthesis, structure, and evolutionary implications. In: Smith CJ, editor. Biochemistry and Molecular Biology of Plant-Pathogen Interactions. Clarendon; Oxford: 1991. pp. 76–91. [Google Scholar]
- Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- Michou AI, Lehrmann H, Saltik M, Cotten M. Mutational analysis of the avian adenovirus CELO, which provides a basis for gene delivery vectors. J Virol. 1999;73:1399–1410. doi: 10.1128/jvi.73.2.1399-1410.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pichla-Gollon SL, Drinker M, Zhou X, Xue F, Rux JJ, Gao G-P, Wilson JM, Ertl HCJ, Burnett RM, Bergelson JM. Major neutralizing epitope identified for a chimpanzee adenovirus-based vaccine vector. 2006 Submitted for publication. [Google Scholar]
- Prage L, Pettersson U, Höglund S, Lonberg-Holm K, Philipson L. Structural proteins of adenoviruses. IV Sequential degradation of the adenovirus type 2 virion. Virology. 1970;42:341–358. doi: 10.1016/0042-6822(70)90278-3. [DOI] [PubMed] [Google Scholar]
- Rux JJ, Burnett RM. Type-specific epitope locations revealed by X-ray crystallographic study of adenovirus type 5 hexon. Mol Ther. 2000;1:18–30. doi: 10.1006/mthe.1999.0001. [DOI] [PubMed] [Google Scholar]
- Rux JJ, Burnett RM. Adenovirus structure. Hum Gene Ther. 2004;15:1167–1176. doi: 10.1089/hum.2004.15.1167. [DOI] [PubMed] [Google Scholar]
- Rux JJ, Burnett RM. Large scale purification and crystallization of adenovirus hexon. In: Wold WSM, Tollofson AE, editors. Adenovirus Methods and Protocols. 2. Vol. 2. Humana Press; Totowa, New Jersey: 2006. pp. 231–250. (Methods in Molecular Medicine, Vol. 131) [DOI] [PubMed] [Google Scholar]
- Rux JJ, Kuser PR, Burnett RM. Structural and phylogenetic analysis of adenovirus hexons by use of high-resolution X-ray crystallographic, molecular modeling, and sequence-based methods. J Virol. 2003;77:9553–9566. doi: 10.1128/JVI.77.17.9553-9566.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Martín C, Burnett RM, de Haas F, Heinkel R, Rutten T, Fuller SD, Butcher SJ, Bamford DH. Combined EM/X-ray imaging yields a quasi-atomic model of the adenovirus-related bacteriophage PRD1 and shows key capsid and membrane interactions. Structure. 2001;9:917–930. doi: 10.1016/s0969-2126(01)00642-6. [DOI] [PubMed] [Google Scholar]
- San Martín C, Huiskonen JT, Bamford JKH, Butcher SJ, Fuller SD, Bamford DH, Burnett RM. Minor proteins, mobile arms and membrane-capsid interactions in the bacteriophage PRD1 capsid. Nature Struct Biol. 2002;9:756–763. doi: 10.1038/nsb837. [DOI] [PubMed] [Google Scholar]
- Scheres SHW, Marabini R, Lanzavecchia S, Cantele F, Rutten T, Fuller SD, Carazo JM, Burnett RM, San Martín C. Classification of single-projection reconstructions for cryo-electron microscopy data of icosahedral viruses. J Struct Biol. 2005;151:79–91. doi: 10.1016/j.jsb.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Schuster B, Gyorvary E, Pum D, Sleytr UB. Nanotechnology with S-layer proteins. Meth Mol Biol. 2005;300:101–123. doi: 10.1385/1-59259-858-7:101. [DOI] [PubMed] [Google Scholar]
- Shashkova EV, Cherenova LV, Kazansky DB, Doronin K. Avian adenovirus vector CELO-TK displays anticancer activity in human cancer cells and suppresses established murine melanoma tumors. Cancer Gene Therapy. 2005;12:617–626. doi: 10.1038/sj.cgt.7700822. [DOI] [PubMed] [Google Scholar]
- Stewart PL, Burnett RM, Cyrklaff M, Fuller SD. Image reconstruction reveals the complex molecular organization of adenovirus. Cell. 1991;67:145–154. doi: 10.1016/0092-8674(91)90578-m. [DOI] [PubMed] [Google Scholar]
- Stewart PL, Fuller SD, Burnett RM. Difference imaging of adenovirus: Bridging the resolution gap between X-ray crystallography and electron microscopy. EMBO J. 1993;12:2589–2599. doi: 10.1002/j.1460-2075.1993.tb05919.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Oostrum J, Smith PR, Mohraz M, Burnett RM. Interpretation of electron micrographs of adenovirus hexon arrays using a crystallographic molecular model. J Ultrastruct Mol Struct Res. 1986;96:77–90. doi: 10.1016/0889-1605(86)90009-1. [DOI] [PubMed] [Google Scholar]
- Wang Q, Kaltgrad E, Lin T, Johnson JE, Finn MG. Natural supramolecular building blocks: Wild-type cowpea mosaic virus. Chem Biol. 2002;9:805–811. doi: 10.1016/s1074-5521(02)00165-5. [DOI] [PubMed] [Google Scholar]
- Xu L. Ph.D. thesis. University of Pennsylvania, Philadelphia; PA: 2002. A tale of two viruses with therapeutic potential: Structural studies on CELO, an avian adenovirus, and the bacteriophage PRD1. [Google Scholar]
- Xue F, Burnett RM. Capsid-like arrays in crystals of chimpanzee adenovirus hexon. J Struct Biol. 2006;154:217–221. doi: 10.1016/j.jsb.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Yates VJ, el-Mishad AM, McCormick KJ, Trentin JJ. Isolation and characterization of an avian adenovirus-associated virus. Infect Immun. 1973;7:973–980. doi: 10.1128/iai.7.6.973-980.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubieta C, Schoehn G, Chroboczek J, Cusack S. The structure of the human adenovirus 2 penton. Mol Cell. 2005;17:121–135. doi: 10.1016/j.molcel.2004.11.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


