Abstract
The hypothesis of fetal origins of adult disease posits that early developmental exposures involve epigenetic modifications, such as DNA methylation, that influence adult disease susceptibility. In utero or neonatal exposure to bisphenol A (BPA), a high-production-volume chemical used in the manufacture of polycarbonate plastic, is associated with higher body weight, increased breast and prostate cancer, and altered reproductive function. This study shows that maternal exposure to this endocrine-active compound shifted the coat color distribution of viable yellow agouti (Avy) mouse offspring toward yellow by decreasing CpG (cytosine-guanine dinucleotide) methylation in an intracisternal A particle retrotransposon upstream of the Agouti gene. CpG methylation also was decreased at another metastable locus, the CDK5 activator-binding protein (CabpIAP). DNA methylation at the Avy locus was similar in tissues from the three germ layers, providing evidence that epigenetic patterning during early stem cell development is sensitive to BPA exposure. Moreover, maternal dietary supplementation, with either methyl donors like folic acid or the phytoestrogen genistein, negated the DNA hypomethylating effect of BPA. Thus, we present compelling evidence that early developmental exposure to BPA can change offspring phenotype by stably altering the epigenome, an effect that can be counteracted by maternal dietary supplements.
Keywords: DNA methylation, environmental epigenomics, viable yellow agouti, fetal origins of adult disease
There is now significant evidence that the risk of many chronic adult diseases and disorders results from exposure to environmental factors early in development (1, 2). Moreover, it seems that there is a link between what we are exposed to in utero and disease formation in adulthood that involves epigenetic modifications such as DNA methylation of transposable elements and cis-acting, imprinting regulatory elements (3). Many xenobiotics, ubiquitously present in the environment, have estrogenic properties and function as endocrine disruptors; however, their potential to modify the epigenome remains largely unexplored (4). The epigenome is particularly susceptible to dysregulation during gestation, neonatal development, puberty, and old age. Nevertheless, it is most vulnerable to environmental exposures during embryogenesis because the elaborate DNA methylation and chromatin patterning required for normal tissue development is programmed during early development.
Most regions of the mammalian genome exhibit little variability among individuals in tissue-specific DNA methylation levels. In contrast, DNA methylation is determined stochastically at some transposable element insertion sites. This potentially can affect the expression of neighboring genes, resulting in the formation of loci with metastable epialleles (3). Cellular epigenetic mosaicism and individual phenotypic variability then can occur even in genetically identical individuals. These sites are also particularly vulnerable to environmentally induced epigenetic alterations (5–7).
The Agouti gene in the viable yellow agouti (Avy) mouse (8) is the most extensively studied metastable epiallele, an allele that is expressed variably in genetically identical individuals because of epigenetic modifications established during early development (9). The wild-type murine Agouti gene encodes a paracrine signaling molecule that produces either black eumelanin (a) or yellow phaeomelanin (A). Both A and a transcripts are initiated from a hair cycle-specific promoter in exon 2. Transient A expression in hair follicles during a specific stage of hair growth results in a subapical yellow band on each black hair shaft, causing the brown (agouti) coat color of wild-type mice (8). The Avy allele resulted from the insertion of a murine intracisternal A particle (IAP) retrotransposon into the 5′ end of the Agouti gene (6, 8). A cryptic promoter in the proximal end of the Avy IAP promotes constitutive ectopic Agouti transcription, leading to yellow fur, obesity, diabetes, and tumorigenesis (10, 11). Methylation of cytosines in cytosine-guanine (CpG) dinucleotide sites in and near the Avy IAP correlates inversely with ectopic Agouti expression and varies dramatically among individual isogenic Avy/a mice. This results in a wide variation in coat color, ranging from yellow (unmethylated) to pseudoagouti (methylated).
The present study uses this model to evaluate how the fetal epigenome is affected by maternal exposure to the estrogenic xenobiotic chemical bisphenol A (BPA). BPA is a high production volume chemical used in the manufacture of polycarbonate plastic and epoxy resins. It is present in many commonly used products including food and beverage containers, baby bottles, and dental composites. The detection of BPA in 95% of human urine samples (12) clearly attests to the widespread use of BPA and widespread human exposure to BPA. Rodent studies have associated pre- or perinatal BPA exposure with higher body weight, increased breast and prostate cancer, altered reproductive function, and other chronic health effects (reviewed in ref. 13). BPA also enters the placenta and accumulates in fetuses after rodent maternal oral exposure (14). Herein, we report the effect of maternal BPA exposure alone or in combination with nutritional supplements on the epigenome of the offspring.
Results
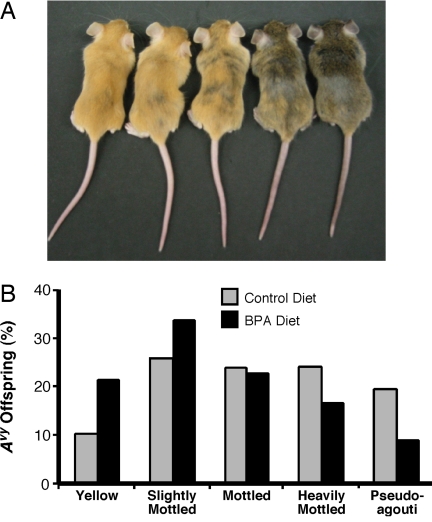
To evaluate the effects of maternal BPA exposure on the fetal epigenome, female a/a mice received a phytoestrogen-free AIN-93G diet (n = 16 litters, 120 total offspring, 60 Avy/a offspring) or a phytoestrogen-free AIN-93G diet combined with 50 mg of BPA/kg (n = 17 litters, 124 total offspring, 73 Avy/a offspring) two weeks before mating with Avy/a males and throughout gestation and lactation. Maternal dietary BPA did not significantly (P > 0.25) influence litter size, survival, wean weight, genotypic ratio, or sex ratio (data not shown). In contrast, maternal BPA significantly shifted the coat color distribution of genetically identical d22 Avy/a offspring toward the yellow coat color phenotype (χ2 P = 0.007) (Fig. 1 A and B). Twenty-one percent of offspring developmentally exposed to BPA were classified as yellow compared with 10% of control offspring. Furthermore, only 9.6% of BPA offspring were classified as pseudoagouti, compared with 18.3% of control animals.
Fig. 1.
Maternal BPA exposure shifts offspring coat color distribution toward yellow. (A) Genetically identical Avy/a offspring representing the five coat color phenotypes. (B) Coat color distribution of Avy/a offspring born to 16 control (n = 60) and 17 BPA-exposed (n = 73) litters (50-mg BPA/kg diet).
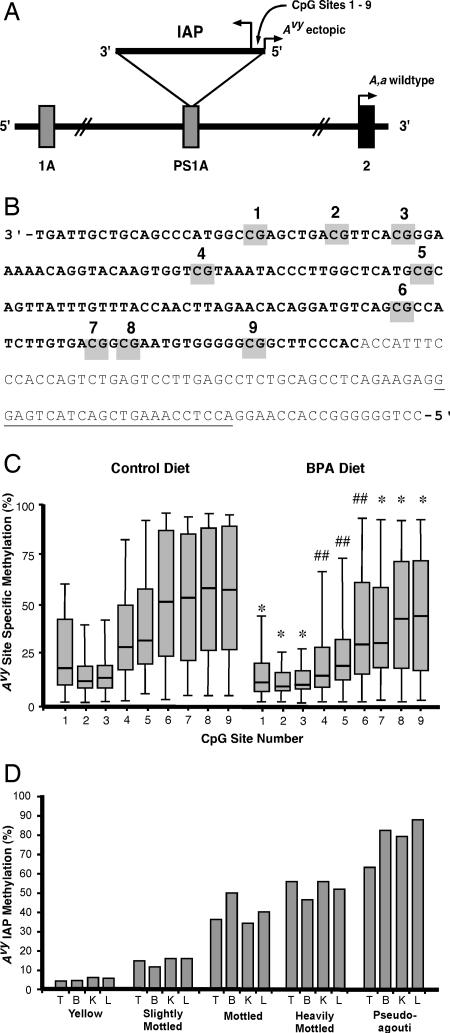
To assess BPA's effect on the epigenome of the offspring, DNA methylation at nine CpG sites in the cryptic promoter region of the Avy IAP (Fig. 2 A and B) (5) was measured by bisulfite treatment and sequencing (15). BPA-exposed offspring exhibited 27 ± 2.8% (n = 73) methylation across the nine sites compared with 39 ± 2.6% (n = 60) methylation in control offspring. Thus, BPA-exposed offspring showed a significantly (P = 0.004) decreased average percentage of cells methylated at these nine sites relative to that in control offspring. Analysis of individual CpG sites revealed significantly (P < 0.05) lower methylation in the BPA-exposed offspring at all nine sites (Fig. 2C). Moreover, for sites 4, 5, and 6, the statistical significance was an order of magnitude greater (P = 0.003, 0.004, and 0.005, respectively) than that of the other CpG sites, indicating that this region may be particularly important in modifying chromatin structure and Agouti gene expression.
Fig. 2.
Maternal BPA exposure reduces DNA methylation at nine CpG sites within the Avy IAP. (A) The Avy allele contains a contra-oriented IAP insertion within pseudoexon 1A (PS1A) of the Agouti gene. A cryptic promoter (short arrowhead labeled “Avy ectopic”) drives constitutive ectopic Agouti expression. Transcription of the Agouti gene normally initiates from a developmentally regulated hair cycle-specific promoter in exon 2 (short arrowhead labeled “A, a wild type”). (B) The IAP sequence containing nine CpG sites (gray boxes) is located between the cryptic Agouti promoter and the IAP promoter and the downstream 3′ genomic sequence. Bold text represents IAP sequence, and nonbold text represents genomic sequence. The location of the bisulfite-converted genomic reverse primer for amplifying the Avy IAP is underlined. (C) Box plots representing the percentage of cells methylated at CpG sites 1–9 in control (n = 60) and BPA-exposed (n = 73) Avy/a offspring (diet group t test; ##, P < 0.01; *, P < 0.05). (D) Average methylation across CpG sites 1–9 in d22 tissues derived from ectodermal (brain and tail), mesodermal (kidney), and endodermal (liver) tissues from BPA-exposed Avy/a offspring (n = 10) representing the five coat color phenotypes is highly correlated (Pearson's r > 0.9 and P < 0.05 for each correlation). T, tail; B, brain; K, kidney; L, liver.
The relationship between BPA dietary exposure, IAP methylation, and offspring coat color also was assessed by mediational regression analysis (16). This statistical approach showed that, although BPA diet significantly (P = 0.02) influences coat color, this association markedly decreased (P = 0.7) when average Avy CpG methylation of sites 1–9 was included in the model. Importantly, the inclusion of the methylation data in the regression model nullified the relationship between BPA diet and coat color. This demonstrates that the methylation at the Avy IAP principally mediates the effect of BPA exposure (P = 0.004) on Avy/a coat color.
Methylation levels in d22 tail tissues from a randomly chosen subset (n = 10) of BPA-exposed animals was correlated highly to methylation levels in d22 tissues derived from the ectoderm (brain), mesoderm (kidney), and endoderm (liver) (Pearson's r > 0.9 and P < 0.05 for each correlation) (Fig. 2D). The low variability in methylation among tissues from the different germ layers relative to the high variability between individual animals indicates that the Avy methylation patterns resulting from BPA exposure are established before germ layer differentiation in the embryonic stem cells.
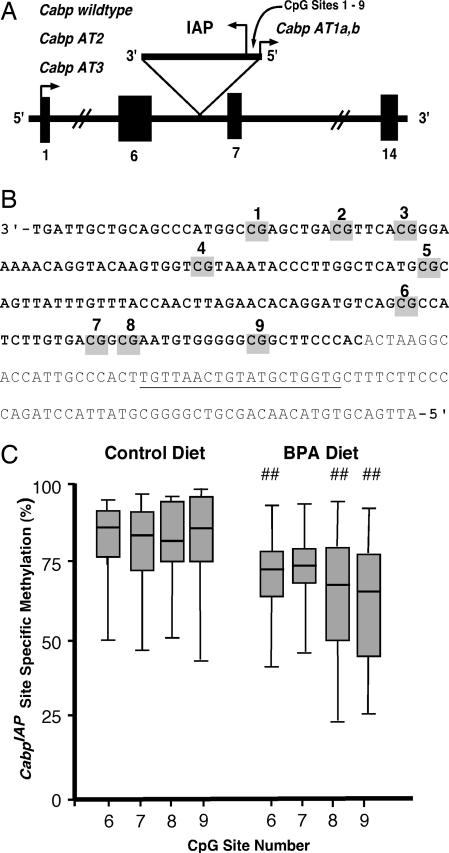
To investigate whether maternal dietary BPA induces hypomethylation at additional loci with metastable epialleles, we measured CpG methylation patterns of the CabpIAP gene. The CabpIAP metastable epiallele (17) resulted from the insertion of an IAP retrotransposon into intron 6 of the murine CDK5 activator-binding protein (Cabp), a gene responsible for CDK5 kinase inhibition (Fig. 3 A and B). Variable expressivity in genetically identical individuals is associated with differential methylation of the 5′ IAP, which results in a number of short, aberrant transcripts (Fig. 3A) (17). Bisulfite sequencing of the 5′ CabpIAP in kidney tissue, where expression is high, revealed hypomethylation in BPA-exposed d22 Avy/a and a/a offspring (P = 0.003; n = 39 control offspring and n = 39 BPA-exposed offspring). Site-specific analysis showed methylation differences only at sites 6–9 (P = 0.006, 0.09, 0.001, and 0.0001, respectively) (Fig. 3C). Average methylation at sites 6–9 was 65.8 ± 2.2% (n = 39) in BPA-exposed offspring compared with 78.8 ± 2.0% (n = 39) in control offspring (P = 0.0001). Thus, maternal dietary BPA exposure promotes DNA hypomethylation at multiple murine loci with metastable epialleles.
Fig. 3.
Maternal BPA exposure decreases offspring methylation at the CabpIAP metastable epiallele. (A) The CabpIAP metastable epiallele (17) contains a contra-oriented IAP insertion within intron 6 of the murine CDK5 activator-binding protein (Cabp) gene, resulting in short aberrant transcripts originating from the 5′ LTR of the IAP (short arrowhead labeled “Cabp AT1a,b”). Short aberrant transcripts also originate at the normal transcription start site (short arrowhead labeled “Cabp wild type”) because of premature truncation upstream of the IAP insert (Cabp AT2 and AT3). Normal Cabp transcription covers 14 exons, resulting in a 2-kb transcript. (B) The IAP sequence containing nine CpG sites located between the cryptic Cabp promoter and the IAP promoter (bold text) and the downstream 3′ genomic sequence (nonbold text). The location of the bisulfite-converted genomic reverse primer for amplifying the 5′ CabpIAP locus is underlined. (C) Box plots representing the percentage of cells methylated at CpG sites 6–9 in control (n = 39) and BPA-exposed (n = 39) Avy/a offspring (diet group t test; ##, P < 0.01).
The Avy mouse has been used previously to primarily assess the effect of maternal nutritional supplements on the epigenome of the offspring (5–7, 18). Those studies showed that methyl donors such as folic acid (6, 7, 18) or the phytoestrogen genistein (5) act very early in development to shift the coat color distribution toward the methylated pseudoagouti phenotype. To determine if these maternal nutritional supplements counteract the hypomethylation effect of BPA exposure, the diets of female a/a mice exposed to BPA (50 mg/kg diet) were supplemented nutritionally. Female a/a mice received a BPA diet supplemented with either methyl donors (4.3 mg of folic acid/kg diet; 0.53 mg of vitamin B12/kg diet; 5 g of betaine/kg diet; 7.97 g of choline chloride/kg diet) or genistein (250 mg/kg diet) 2 weeks before mating with Avy/a males and throughout gestation and lactation. Neither of these combination diets significantly (P > 0.25) affected litter size, litter survival, wean weight, genotypic ratio, or sex ratio (data not shown).
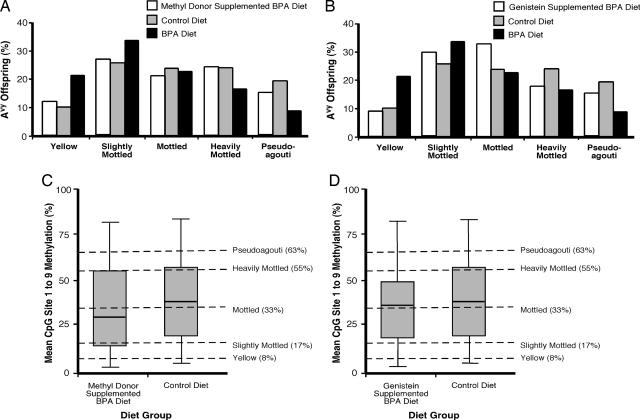
Maternal supplementation with methyl donors (n = 14 litters, 95 total offspring, 54 Avy/a offspring; χ2 P = 0.96) or genistein (n = 13 litters, 81 total offspring, 39 Avy/a offspring; χ2 P = 0.97) restored the coat color distribution in BPA-exposed offspring to that observed in control litters (Fig. 4 A and B). Ten to 13% of control, BPA-exposed/methyl donor-supplemented, or BPA-exposed/genistein-supplemented offspring were classified as yellow compared with 21% of offspring exposed to BPA alone. Furthermore, 15–18.3% of control, BPA-exposed/methyl donor-supplemented, or BPA-exposed/genistein-supplemented offspring were classified as pseudoagouti compared with only 9.6% of BPA-exposed offspring.
Fig. 4.
Maternal nutritional supplementation counteracts BPA-induced DNA hypomethylation and the shift in coat color distribution to yellow. (A) Coat color distribution of Avy/a offspring born to 14 BPA-exposed/methyl donor-supplemented mothers (n = 54), 16 control mothers (n = 60), and 17 BPA-exposed mothers (n = 73). (B) Coat color distribution of Avy/a offspring born to 13 BPA-exposed/genistein-supplemented mothers (n = 39), 16 control mothers (n = 60), and 17 BPA-exposed mothers (n = 73). (C) Box plots representing the percentage of cells methylated at CpG sites 1–9 in BPA-exposed/methyl donor-supplemented (n = 54) and control (n = 60) offspring (P = 0.25). (D) Box plots representing the percentage of cells methylated at CpG sites 1–9 in BPA-exposed/genistein-supplemented (n = 39) and control (n = 60) offspring (P = 0.46).
Maternal nutritional supplementation likewise negated the BPA-induced DNA hypomethylation in the offspring (Fig. 4 C and D). CpG methylation at the Avy IAP of BPA-exposed/methyl donor-supplemented offspring was not statistically (P = 0.25) different from that of control offspring (Fig. 4C), indicating that maternal nutritional supplementation with methyl donors counteracted the hypomethylating effect of BPA. Interestingly, genistein, at a level comparable with that consumed by humans with high soy diets, also counteracted the BPA-induced hypomethylation (P = 0.46) even though it is not a methyl-donating compound (Fig. 4D). Thus, interventions as subtle as maternal nutritional supplementation with methyl donors or genistein can nullify the deleterious effects of an estrogenic endocrine disruptor on the epigenome and can change the adult phenotype of the offspring.
Discussion
The standard control coat color distribution of Avy mice results from variable expressivity controlled by stochastic DNA methylation of an IAP retrotransposon, producing genetically identical individuals with widely varying phenotypes. The inheritance of offspring coat color, metabolic disorders, and obesity in the Avy mouse has been attributed for a long time to maternal genotype and strain (19, 20). Recently, however, the Avy model has been exploited to study phenotypic variability after maternal nutritional supplementation. Wolff et al. (7) demonstrated that maternal dietary supplementation with extra folic acid, vitamin B12, choline, and betaine shifts the coat color distribution of offspring toward the pseudoagouti phenotype. Waterland et al. (6) further demonstrated that the shift in coat color after maternal methyl donor supplementation was caused by increased methylation near the Avy IAP retrotransposon.
In this study, the Avy model has been used to examine the effects of maternal exposure on the fetal epigenome from a xenobiotic contaminant rather than from a nutritional agent. We observed a statistically significant shift in offspring coat color phenotype toward yellow among genetically identical individuals whose mothers were exposed to BPA. The shift in offspring coat color was mediated by decreased methylation at nine CpG sites located immediately upstream of the Avy IAP cryptic promoter. Moreover, the particularly robust reduction in methylation at CpG sites 4–6 indicates that DNA methylation at specific sites may act in concert with underlying chromatin structure and histone modifications to affect variable Agouti gene expression. DNA methylation at the Avy locus was similar in tissues from the three germ layers, providing evidence that epigenetic patterning during early stem cell development is sensitive to BPA exposure. We also observed a statistically significant decrease in methylation at the CabpIAP metastable epiallele in BPA-exposed offspring, indicating that BPA-induced DNA hypomethylation is not locus-dependent.
In this study, maternal dosages of BPA (≈10 mg per kg of body weight per day) were designed to be an order of magnitude lower than the dietary administered maximum nontoxic threshold in rodents (200 mg per kg of body weight per day) (21). Although it is difficult to quantify human BPA exposure because of multiple exposure routes and rapid metabolism (22), human urinary analysis detected a median BPA level of 1.3 parts per billion (12). Thus, although maternal exposure to BPA in this investigation is likely higher than typical human exposure, it produced no significant effects on reproductive outcomes, litter size, or offspring health.
Evidence supporting epigenetic dysregulation as a mode of action of exogenous estrogenic compounds is mounting. Methylation studies conducted by Li et al. (23, 24) with the estrogenic pharmaceutical agent diethylstilbestrol showed hypomethylation in two critical DNA control regions in mice exposed in utero or in the perinatal period. Recently, neonatal estradiol and BPA exposure were associated with altered epigenetic patterning of the Phosphodiesterase type 4 variant 4 (Pde4d4) gene as animals age (25). We now show that a maternal dietary exposure to BPA also markedly alters the adult phenotype of the offspring by hypomethylating the epigenome.
In contrast to findings with BPA and other exogenous estrogens (23–25), in vivo developmental (5) or adult (26) dietary exposure to the plant phytoestrogen genistein induces gene hypermethylation. Using the Avy model, we previously have shown that maternal genistein supplementation shifts offspring coat color distribution toward pseudoagouti (5) by also hypermethylating the epigenome. Interestingly, Fang et al. (27, 28) observed partial reversal of DNA hypermethylation of p16INK4a, RARβ, and MGMT in cancer cells as well as DNA methyltransferase inhibition after treatment with genistein. Although tumor-suppressor gene silencing by DNA methylation occurs frequently in cancer cells, genome-wide DNA hypomethylation coupled with genome instability are the earliest events to occur in the genesis of cancer (29, 30). Given the marked dysregulation of the epigenome in cancer cells, it is not unexpected that a difference in epigenetic response to genistein would be observed in differentiating cells during normal development in vivo, compared with cancer cells in vitro.
Unlike exogenous estrogens, which are typically associated with enhanced risk of carcinogenesis (31) and increased body weight (32), genistein exposure has been linked to chemoprevention in rodent studies (33), decreased cancer in the Asian population (34), and reduced adipocyte deposition in mice (35). Genistein inhibits tyrosine kinase (36), scavenges free radicals (37), and exhibits mixed estrogenic and anti-estrogenic properties (38) depending on timing, dose, and tissue. Thus, its biological activities result from the activation of both estrogen receptor and nonestrogen receptor pathways. To assess the importance of these pathways in altering DNA methylation and chromatin structure in the offspring after maternal exposure to estrogenic agents like genistein and BPA, studies using estrogen receptor knockout (ERKOα and ERKOβ) mice (39) on an Avy background need to be conducted.
In this study, we made the important discovery that maternal nutritional supplementation with methyl donors or genistein counteracts BPA-induced hypomethylation, resulting in a control coat color distribution in the BPA-exposed offspring. BPA is ubiquitously present in the human population, so these findings hold promise for reducing disease susceptibility by public health nutrition interventions. Exposure to endocrine active compounds like BPA also is linked to epigenetic alterations and disease pathologies that are inherited through the germ line, even in the absence of continued exposure (40, 41). The epigenetic effects of maternal methyl-donor supplementation on coat color distribution in Avy offspring also have been shown to be inherited transgenerationally (42). Thus, the ability to counteract negative environmental toxicant effects, such as DNA hypomethylation, via in utero or possibly even adult nutritional supplementation, has the potential to protect human health in the present and future generations; however, the effectiveness of this preventive approach would be expected to be inversely related to the level of toxicant exposure.
The growing interest in the developmental basis of adult disease combined with the presence of BPA in products commonly encountered by pregnant women and newborns has led to attempts in Canada, California, Maryland, and Minnesota to ban its use and has led to the European Union's proposed adoption of the precautionary principle, which requires manufacturers to demonstrate chemical safety before use. Furthermore, a 2005 review of the BPA literature (43) shows that >90% of government-funded, low dose BPA studies report adverse health effects, supporting a reevaluation of human health risks associated with its exposure. This stands in stark contrast to a 2004 industry-sponsored review (44) concluding that the evidence supporting low dose effects of BPA is extremely weak. The results of our study strongly support the inclusion of epigenetic effects of xenobiotic chemicals like BPA into the risk assessment process, as well as the investigation of nutritional supplementation as a parental preventive approach to counteracting environmental influences on the epigenome.
Methods
Animals and Diets.
Avy mice were obtained from a colony that has been maintained with sibling mating and forced heterozygosity for the Avy allele for over 220 generations, resulting in a genetically invariant background (6). The Avy allele is passed through the paternal lineage to avoid bias associated with maternal transmission, in which its epigenotype is not completely reset (10). This study was approved by the Duke University Institutional Animal Care and Use Committee.
Virgin a/a females, 8–10 weeks of age, were assigned to receive one of four diets: (i) modified AIN-93G diet (diet 95092 with 7% corn oil substituted for 7% soybean oil; Harlan Teklad, Madison, WI); (ii) modified AIN-93G diet supplemented with 50 mg/kg diet of BPA (diet 06156; Harlan Teklad); (iii) modified AIN-93G diet supplemented with 50 mg/kg diet of BPA and 250 mg/kg diet of genistein (diet 06309; Harlan Teklad); or (iv) modified AIN-93G diet supplemented with 50 mg/kg diet of BPA and methyl donor compounds (diet 06310; Harlan Teklad), including 4.3 mg of folic acid/kg diet, 0.53 mg of vitamin B12/kg diet, 5 g of betaine/kg diet, and 7.97 g of choline chloride/kg diet). Harland Teklad supplied all diet ingredients except BPA (Sigma-Aldrich, St. Louis, MO), genistein (Indofine Chemical Company, Hillsborough, NJ), and betaine (Spectrum Chemicals, Gardena, CA). Dosages of BPA were designed to be an order of magnitude lower than the dietary–administered, maximum, nontoxic threshold in rodents (200 mg per kg of body weight per day) (21). Dosages of genistein (5) and methyl donors (6) were those previously used to study the effects of nutritional factors on the fetal epigenome. Diets were provided 2 weeks before mating with Avy/a males and throughout pregnancy and lactation. At day 22, all offspring were weighed, photographed, and rated for coat color phenotype before being killed. Tail, brain, liver, and kidney tissues were collected for analysis from BPA and control offspring. Tail tissue was collected from the coexposure offspring.
A single observer visually classified d22 Avy/a offspring coat color phenotype into one of five categories based on proportion of brown: yellow fur (<5% brown), slightly mottled (between 5 and 50% brown), mottled (∼50% brown), heavily mottled (between 50 and 95% brown), and pseudoagouti (>95% brown).
DNA Methylation Assay.
For all Avy/a offspring, total DNA was isolated from d22 tail using buffer ATL, proteinase K, and Rnase A (Qiagen Inc., Valencia, CA) followed by phenol–chloroform extraction and ethanol precipitation. In a subset of Avy/a offspring, total DNA also was isolated from d22 brain, liver, and kidney tissue. In addition, total DNA was isolated from d22 tail in 50% of a/a offspring, as described above.
Sodium bisulfite modification of DNA was performed by using a protocol adapted from Grunau et al. (15) as described (5, 6), except that deamination was carried out at 55°C for 5 h instead of 4 h. Regions of interest were amplified from bisulfite-modified DNA in 50-μl PCRs using 1.5 units of Platinum TaqDNA polymerase (Invitrogen, Carlsbad, CA), 15 pmol of primers, 1.5 mM MgCl2, and 10 mM dNTPs (94°C × 2 min; 94°C × 30 sec, 55°C × 30 sec, and 72°C × 60 sec for 40 cycles; 72°C × 9 min). For the Avy allele, we used forward primer IAPF3 (5′ ATT TTT AGG AAA AGA GAG TAA GAA GTA AG 3′) and reverse primer IAPR4 (5′ TAA TTC CTA AAA ATT TCA ACT AAT AAC TCC 3′) (336-bp product) (5). For the CabpIAP gene, we used forward primer CABP_F (5′ GGT TAG GAA GAA TAT TAT AGA TTA 3′) and reverse primer CABP_R2 (5′ CAC CAA CAT ACA ATT AAC A 3′) (408-bp product).
Avy and CabpIAP PCR products were resolved by electrophoresis on a 2% agarose gel, excised, gel-extracted (GenElute, Sigma Chemical Co., St. Louis, MO), and sequenced manually (Thermo Sequenase Radiolabeled Terminator Cycle Sequencing kit; USB Corporation, Cleveland, OH) according to manufacturer's instructions (95°C × 30 sec, 55°C × 30 sec, and 72°C × 60 sec for 35 cycles) by using forward sequencing primer IAPF5 (5′ ATT ATT TTT TGA TTG TTG TAG TTT ATG G 3′). Sequencing products were resolved using PAGE. Percentage of cells methylated at the nine CpG sites in the Avy and CabpIAP IAP regions was quantified by phosphor imaging as follows: percentage of cells methylated = 100 × [(C intensity)/(C intensity + T intensity)] (45). For the Avy allele, the nine CpG sites studied are located at nucleotide positions 206, 214, 220, 244, 265, 306, 319, 322, and 334 of GenBank accession number AF540972. For the CabpIAP allele, CpG sites 5–9 are located at nucleotide positions 3, 44, 57, 60, and 72 of GenBank accession number BB842254.
Statistical Analysis.
Diet group comparisons of the proportion of offspring in each of the five coat color classes were performed by χ2 analysis. Average IAP CpG methylation and site-specific CpG methylation between the control and BPA groups were analyzed by two-sample hypothesis testing of means and ANOVA, Bonferroni-corrected for multiple comparisons by using STATA version 8.0 software (College Station, TX). Relationships among diet supplementation, Avy IAP methylation, and coat color were analyzed by mediational regression analysis (16).
Acknowledgments
We thank R. Das, K. Maloney, and J. Weidman for technical assistance and D. Skaar for critical reading of the manuscript. This work was supported by National Institutes of Health Grants ES015165, ES13053, and T32ES07031 and by Department of Energy Grant DE-FG02-05ER64101.
Abbreviations
- Avy
viable yellow agouti
- BPA
bisphenol A
- IAP
intracisternal A particle
- Cabp
CDK5 activator-binding protein.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Bateson P, Barker D, Clutton-Brock T, Deb D, D'Udine B, Foley RA, Gluckman P, Godfrey K, Kirkwood T, Lahr MM, et al. Nature. 2004;430:419–421. doi: 10.1038/nature02725. [DOI] [PubMed] [Google Scholar]
- 2.McMillen IC, Robinson JS. Physiol Rev. 2005;85:571–633. doi: 10.1152/physrev.00053.2003. [DOI] [PubMed] [Google Scholar]
- 3.Jirtle RL, Skinner MK. Nat Rev Genet. 2007;8:253–262. doi: 10.1038/nrg2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crews D, McLachlan JA. Endocrinology. 2006;147:4–10. doi: 10.1210/en.2005-1122. [DOI] [PubMed] [Google Scholar]
- 5.Dolinoy DC, Wiedman J, Waterland R, Jirtle RL. Environ Health Perspect. 2006;114:567–572. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waterland R, Jirtle R. Mol Cell Biol. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolff GL, Kodell RL, Moore SR, Cooney CA. FASEB J. 1998;12:949–957. [PubMed] [Google Scholar]
- 8.Duhl D, Vrieling H, Miller K, Wolff G, Barsh G. Nat Genet. 1994;8:59–65. doi: 10.1038/ng0994-59. [DOI] [PubMed] [Google Scholar]
- 9.Rakyan VK, Blewitt ME, Druker R, Preis JI, Whitelaw E. Trends Genet. 2002;18:348–351. doi: 10.1016/s0168-9525(02)02709-9. [DOI] [PubMed] [Google Scholar]
- 10.Morgan H, Sutherland H, Martin D, Whitelaw E. Nat Genet. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 11.Miltenberger R, Mynatt R, Wilkinson J, Woychik R. J Nutr. 1997;127:1902S–1907S. doi: 10.1093/jn/127.9.1902S. [DOI] [PubMed] [Google Scholar]
- 12.Calafat A, Kuklenyik Z, Reidy J, Caudill S, Ekong J, Needham L. Environ Health Perspect. 2005;113:391–395. doi: 10.1289/ehp.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maffini MV, Rubin BS, Sonnenschein C, Soto AM. Mol Cell Endocrinol. 2006;254–255:179–186. doi: 10.1016/j.mce.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 14.Takahashi O, Oishi S. Environ Health Perspect. 2000;108:931–935. doi: 10.1289/ehp.00108931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grunau C, Clark S, Rosenthal A. Nucleic Acids Res. 2001;29:E65. doi: 10.1093/nar/29.13.e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baron R, Kenny D. J Pers Soc Psychol. 1986;51:1173–1182. doi: 10.1037//0022-3514.51.6.1173. [DOI] [PubMed] [Google Scholar]
- 17.Druker R, Bruxner TJ, Lehrbach NJ, Whitelaw E. Nucleic Acids Res. 2004;32:5800–5808. doi: 10.1093/nar/gkh914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cooney CA, Dave AA, Wolff GL. J Nutr. 2002;132:2393–2400. doi: 10.1093/jn/132.8.2393S. [DOI] [PubMed] [Google Scholar]
- 19.Wolff G. Genetics. 1978;88:529–539. doi: 10.1093/genetics/88.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolff GL, Roberts DW, Galbraith DB. J Hered. 1986;77:151–158. doi: 10.1093/oxfordjournals.jhered.a110206. [DOI] [PubMed] [Google Scholar]
- 21.Takahashi O, Oishi S. Food Chem Toxicol. 2003;41:1035–1044. doi: 10.1016/s0278-6915(03)00031-0. [DOI] [PubMed] [Google Scholar]
- 22.Volkel W, Colnot T, Csanady G, Filser J, Dekant W. Chem Res Toxicol. 2002;15:1281–1287. doi: 10.1021/tx025548t. [DOI] [PubMed] [Google Scholar]
- 23.Li S, Hansman R, Newbold R, Davis B, McLachlan JA, Barrett JC. Mol Carcinog. 2003;38:78–84. doi: 10.1002/mc.10147. [DOI] [PubMed] [Google Scholar]
- 24.Li S, Washburn K, Moore R, Uno T, Teng C, Newbold R, McLachlan J, Negishi M. Cancer Res. 1997;57:4356–4359. [PubMed] [Google Scholar]
- 25.Ho S, Tang W, Belmonte de Frausto J, Prins G. Cancer Res. 2006;66:5624–5632. doi: 10.1158/0008-5472.CAN-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Day J, Bauer A, desBordes C, Zhuang Y, Kim B.-E., Newton LG, Nehra V, Forsee KM, MacDonald RS, Besch-Williford C, et al. J Nutr. 2002;132:2419–2423. doi: 10.1093/jn/132.8.2419S. [DOI] [PubMed] [Google Scholar]
- 27.Fang MZ, Chen D, Sun Y, Jin Z, Christman JK, Yang CS. Clin Cancer Res. 2005;11:7033–7041. doi: 10.1158/1078-0432.CCR-05-0406. [DOI] [PubMed] [Google Scholar]
- 28.Fang M, Chen D, Yang C. J Nutr. 2007;137:223S–228S. doi: 10.1093/jn/137.1.223S. [DOI] [PubMed] [Google Scholar]
- 29.Esteller M, Herman JG. J Pathol. 2002;196:1–7. doi: 10.1002/path.1024. [DOI] [PubMed] [Google Scholar]
- 30.Feinberg AP, Tycko B. Nat Rev Cancer. 2004;4:143–153. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 31.Prins G, Birch L, Tang W, Ho S. Reprod Toxicol. 2007;23:374–382. doi: 10.1016/j.reprotox.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newbold RR, Padilla-Banks E, Snyder RJ, Phillips TM, Jefferson WN. Reprod Toxicol. 2007;23:290–296. doi: 10.1016/j.reprotox.2006.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lamartiniere C, Cotroneo M, Fritz WA, Wang J, Mentor-Marcel R, Elgavish A. J Nutr. 2002;132:552S–558S. doi: 10.1093/jn/132.3.552S. [DOI] [PubMed] [Google Scholar]
- 34.Adlercreutz H. Environ Health Perspect. 1995;103:103–112. doi: 10.1289/ehp.95103s7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naaz A, Yellayi S, Zakroczymski MA, Bunick D, Doerge DR, Lubahn DB, Helferich WG, Cooke PS. Endocrinology. 2003;144:3315–3320. doi: 10.1210/en.2003-0076. [DOI] [PubMed] [Google Scholar]
- 36.Akiyama T, Ishida J, Nakagawa S, Ogawara H, Watanabe S, Itoh N, Shibuya M, Fukami Y. J Biol Chem. 1987;262:5592–5595. [PubMed] [Google Scholar]
- 37.Wei H, Wei L, Frenkel K, Bowen R, Barnes S. Nutr Cancer. 1993;20:1–12. doi: 10.1080/01635589309514265. [DOI] [PubMed] [Google Scholar]
- 38.Price K, Fenwick G. Food Addit Contam. 1985;2:73–106. doi: 10.1080/02652038509373531. [DOI] [PubMed] [Google Scholar]
- 39.Carpenter KD, Korach KS. Ann NY Acad Sci. 2006;1092:361–373. doi: 10.1196/annals.1365.033. [DOI] [PubMed] [Google Scholar]
- 40.Newbold RR, Padilla-Banks E, Jefferson WN. Endocrinology. 2006;147:s11–17. doi: 10.1210/en.2005-1164. [DOI] [PubMed] [Google Scholar]
- 41.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cropley JE, Suter CM, Beckman KB, Martin DIK. Proc Natl Acad Sci USA. 2006;103:17308–17312. doi: 10.1073/pnas.0607090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.vom Saal F, Hughes C. Environ Health Perspect. 2005;113:926–933. doi: 10.1289/ehp.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gray G, Cohen J, Cunha G, Hughes C, McConnell E, Rhomberg L, Sipes I, Mattison D. Hum Ecol Risk Assess. 2004;10:875–921. [Google Scholar]
- 45.Waterland RA, Lin J-R, Smith CA, Jirtle RL. Hum Mol Genet. 2006;15:705–716. doi: 10.1093/hmg/ddi484. [DOI] [PubMed] [Google Scholar]