Abstract
Achieving efficient in vivo delivery of siRNA to the appropriate target cell would be a major advance in the use of RNAi in gene function studies and as a therapeutic modality. Hepatocytes, the key parenchymal cells of the liver, are a particularly attractive target cell type for siRNA delivery given their central role in several infectious and metabolic disorders. We have developed a vehicle for the delivery of siRNA to hepatocytes both in vitro and in vivo, which we have named siRNA Dynamic PolyConjugates. Key features of the Dynamic PolyConjugate technology include a membrane-active polymer, the ability to reversibly mask the activity of this polymer until it reaches the acidic environment of endosomes, and the ability to target this modified polymer and its siRNA cargo specifically to hepatocytes in vivo after simple, low-pressure i.v. injection. Using this delivery technology, we demonstrate effective knockdown of two endogenous genes in mouse liver: apolipoprotein B (apoB) and peroxisome proliferator-activated receptor alpha (ppara). Knockdown of apoB resulted in clear phenotypic changes that included a significant reduction in serum cholesterol and increased fat accumulation in the liver, consistent with the known functions of apoB. Knockdown of ppara also resulted in a phenotype consistent with its known function, although with less penetrance than observed in apoB knockdown mice. Analyses of serum liver enzyme and cytokine levels in treated mice indicated that the siRNA Dynamic PolyConjugate was nontoxic and well tolerated.
Keywords: pH labile bonds, nonviral siRNA delivery, siRNA–polymer conjugates, endosomolytic polymers
The ability of siRNA to silence specific genes has generated great interest in its use as a research tool and therapeutic agent for a wide spectrum of disorders that include cancer, infectious disease, and metabolic conditions (1–3). Effective in vivo delivery of siRNA to the appropriate target cell is an essential component of these siRNA-based applications. Accordingly, a variety of nonviral (4–14) and viral (15–17) systems are being developed for delivery of siRNA to liver, tumors, and other tissues in vivo.
In addition to their importance in many infectious and metabolic disorders (18), hepatocytes are a particularly attractive target cell type for siRNA delivery given their ability to be accessed directly by nanoparticle-sized constructs after simple intravascular injection. Initial hepatocyte delivery efforts used hydrodynamic delivery of naked siRNA to the liver (19, 20). More recent work has used viral vectors, such as AAV or lentivirus (16, 17), or synthetic systems such as cholesterol–siRNA conjugates or stable nucleic acid lipid particles (SNALPs) (21, 22). Among the nonviral approaches, SNALP technology represents a significant advance, enabling target mRNA knockdown in liver after i.v. injection of clinically relevant doses of siRNA (21). More recently, another lipid-based system termed interfering nanoparticles (iNOPs) has also demonstrated the ability to deliver siRNA in vivo (23). A key drawback of the SNALP and iNOP systems, however, is that the siRNA complexes are only passively targeted to liver. As a result, siRNAs are delivered to a significant number of nontarget cells in the liver, potentially contributing to toxicity.
Hepatocyte targeting after administration into a peripheral vein requires that the delivery vehicle avoid nonspecific interactions en route to the target cell, which is commonly accomplished by the attachment of polyethylene glycol (PEG) (24) or other hydrophilic, noninteractive agents. Upon reaching the liver, the vehicle must then exit the intravascular space to access hepatocytes. Because of the open, fenestrated nature of the hepatic vasculature, particles <100 nm in diameter can readily exit hepatic vessels and interact with liver parenchymal cells (25). However, avoiding uptake and subsequent activation of Kupffer cells, the resident immune cells of the liver, are likely essential to avoid toxicity (26). As an example, Kupffer cell uptake of adenoviral vectors is the main cause of liver toxicity observed when these vectors are used for delivery (27). Galactose-derived ligands, which are recognized by the asialoglycoprotein receptor (ASGPr), can be used to specifically target hepatocytes (28). Certain galactose-containing ligands enable hepatocyte uptake and avoidance of Kupffer cells if properly displayed on the delivery vehicle (29, 30).
Once attached to the surface of hepatocytes, siRNA-containing complexes can enter the cells via receptor-mediated endocytosis. The siRNAs must then escape from endosomes to elicit RNAi. To accomplish efficient endosomal escape, we developed a strategy that relies on the selective activation of a latent endosomolytic agent in the acidic environment of the endosome (31). Selective activation ensures that deleterious interactions with other membranes the agent encounters before endocytosis are prevented. In our strategy, amine groups on the endosomolytic agent are modified with a maleic anhydride, creating acid-labile maleamate bonds (32). These bonds are cleaved within the acidic environment of the endosome, unmasking the agent's amines and activating its endosomolytic capabilities (31). The endosomolytic agent used in the present study is an amphipathic poly(vinyl ether) we previously developed termed PBAVE, which is composed of butyl and amino vinyl ethers (33).
In this study, we use a bifunctional maleamate linkage to reversibly attach the shielding agent PEG and the hepatocyte targeting ligand N-acetylgalactosamine (NAG) to PBAVE. The siRNA cargo itself is attached to PBAVE through a reversible disulfide linkage, which prevents displacement of the siRNA from the polymer en route to the target cell. We have named this delivery vehicle an siRNA Dynamic PolyConjugate, to indicate the fact that the siRNA, shielding agents, and targeting ligands are reversibly conjugated to a polymer whose endosomolytic properties are triggered by its chemical environment.
Results
Formulation of the siRNA Polyconjugate and Cellular Delivery.
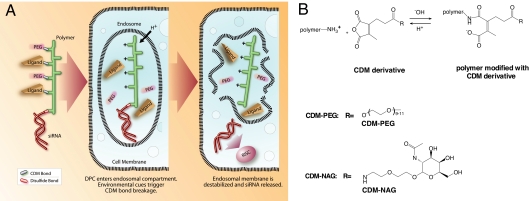
The formulation of the siRNA polyconjugate and the principles of polyconjugate-mediated siRNA delivery are shown in Fig. 1. The polyconjugate itself is constructed by first linking the siRNA payload to the PBAVE polymer through a disulfide linkage (Fig. 1A). The amount of conjugated siRNA from this reaction is typically 70–90% of the input. The siRNA–polymer conjugate is then reversibly modified with maleic anhydride derivatives synthesized from carboxy dimethylmaleic anhydride (CDM) (31) containing PEG or NAG groups (Fig. 1B). Modification with PEG reduces nonspecific interactions and allows hepatocyte targeting via the NAG ligand. The resulting siRNA polyconjugate is negatively charged, soluble, and nonaggregating under physiological conditions. The size of the siRNA polyconjugate is 10 ± 2 nm as measured by particle sizing, making it substantially smaller than the SNALP or iNOP siRNA complexes (21, 34).
Fig. 1.
Critical components of the siRNA polyconjugate and the proposed mechanism of siRNA delivery. (A) Schematic showing the siRNA Dynamic PolyConjugate, its cellular uptake, disassembly in the low pH environment of the endosome, and release of the siRNA into the cytoplasm of the target cell. CDM, carboxylated dimethyl maleic acid. (B) Mechanism of pH-sensitive CDM chemistry and the structures of the CDM derivatives used in this study. Depicted is the reaction of CDM with free tertiary amines on the polymer, which is reversible under acidic conditions.
After simple i.v. injection, the siRNA polyconjugate is designed to engage the ASGPr on hepatocytes and be taken into the cell via endocytosis (Fig. 1A). As the endosome matures, the decrease in pH induces release of the CDM-PEG and CDM-NAG groups, unmasking the positively charged amine groups on the PBAVE polymer. This release results in the activation of the endosomolytic capability of PBAVE and release of the siRNA into the reducing environment of the cytoplasm. Once there, the siRNA cargo is cleaved from the polymer, allowing the siRNA to engage RISC and induce RNAi.
Activity of the siRNA Polyconjugate in Tissue Culture.
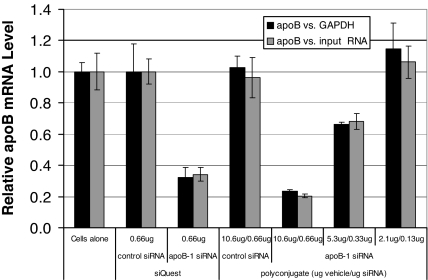
It would appear obvious that a delivery agent designed for in vivo use should also have transfection activity in culture. Therefore, we tested the ability of the polyconjugate to deliver siRNA and knock down target gene expression in mouse primary hepatocytes. We chose to target the mouse apolipoprotein B (apoB) gene, a hepatocyte-expressed gene involved in cholesterol transport. All siRNAs used in this report contained modifications designed to increase resistance to nucleases and suppress off-target effects (22, 35). We found that transfection of primary hepatocytes with apoB-1 siRNA polyconjugate was highly effective, resulting in nearly 80% knockdown of apoB mRNA (Fig. 2). The level of target gene knockdown was comparable to that in cells transfected with apoB-1 siRNA by using siQUEST, a commercially available in vitro siRNA transfection agent. As expected, decreasing the amount of siRNA polyconjugate added to the cells led to progressively decreased apoB knockdown.
Fig. 2.
siRNA polyconjugates can be used to transfect siRNA in mouse primary hepatocytes. Shown is RT-qPCR analysis of apoB mRNA knockdown in primary hepatocytes. Cells were transfected with the indicated amounts of siRNA by using a commercially available transfection reagent (siQUEST) or with serial dilutions of apoB-1 siRNA polyconjugate. Twenty-four hours after transfection, relative apoB mRNA levels were measured versus GAPDH mRNA levels or versus the amount of input RNA in the RT-qPCR and then normalized to the values in untreated cells (cells alone). Data are shown as mean ± SD.
Effective siRNA delivery using Dynamic PolyConjugate technology required that the PEG shielding agent be linked to the polymer through a reversible linkage. Attachment of PEG to the polymer backbone through a nonhydrolysable amide linkage, in place of a reversible CDM linkage, completely abolished knockdown of apoB expression [supporting information (SI) Fig. 7]. These results are consistent with previous studies of irreversible modifications of amphipathic polycations (31) and highlight the necessity for the use of reversible modifications to achieve endosomolysis.
Targeting of the siRNA Polyconjugate to Hepatocytes in Vivo.
Previous studies have shown that attachment of galactose or NAG facilitates hepatocyte targeting of a variety of uncharged, water-soluble polymers (36–38). We attached NAG to the polymer–siRNA backbone through a CDM linkage to determine whether we could target the siRNA polyconjugate to liver hepatocytes after simple i.v. injection into the tail vein of mice.
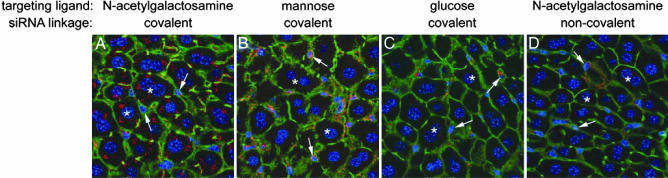
Confocal micrographs of liver sections taken from mice 1 h after injection with polyconjugate containing a Cy3-labeled, 21-mer double-stranded DNA (dsDNA) mimic of siRNA are shown in Fig. 3. When NAG was used as the targeting ligand on the polyconjugate, we observed preferential accumulation of the Cy3-labeled dsDNA in hepatocytes and only minimal association with nonparenchymal cells in liver sinusoids (Fig. 3A). Distribution was nearly homogenous throughout the different zones and of the liver acinus (SI Fig. 8). Inspection of other organs revealed minor Cy3- fluorescence in spleen and kidney, with levels estimated to be at least 20-fold lower than in liver (data not shown). Replacement of CDM-NAG on the polyconjugate with CDM-glucose resulted in markedly reduced hepatocyte uptake (Fig. 3C) (data not shown), which is consistent with the lower affinity of the ASGPr on hepatocytes for glucose (39). Significantly, attachment of mannose to the polyconjugate instead of NAG redirected the polyconjugate to nonparenchymal cells in the liver including sinusoidal endothelial and Kupffer cells, which possess mannose receptors, and away from hepatocytes (Fig. 4B) (40). These results are evidence that active, hepatocyte-specific targeting of the polyconjugate to hepatocytes is afforded by attachment of NAG. They also suggest that siRNA polyconjugates can be directed to other cell types simply by attaching the appropriate ligand.
Fig. 3.
Targeted delivery of oligonucleotide polyconjugates to liver hepatocytes in mice. Shown are confocal images of liver sections from mice injected i.v. with polyconjugate covalently linked through a CDM linkage to the targeting ligand NAG (A and D) or to mannose (B) or glucose (C) as controls. Cy3-labeled 21-mer dsDNA (red) was covalently attached to polyconjugate through a disulfide linkage (A–C) or was present in a noncovalent complex with polyconjugate (D). Livers were harvested 1 h after injection, fixed, and counterstained with ToPro-3 to visualize nuclei (blue) and Alexa 488 phalloidin to visualize cell outlines (green). Each image comprised a flattened projection of 11 optical images (0.4 μm each) to represent combined fluorescence signals from a 4-μm-thick section. Asterisks mark representative hepatocytes, and arrows indicate representative nonparenchymal cells.
Fig. 4.
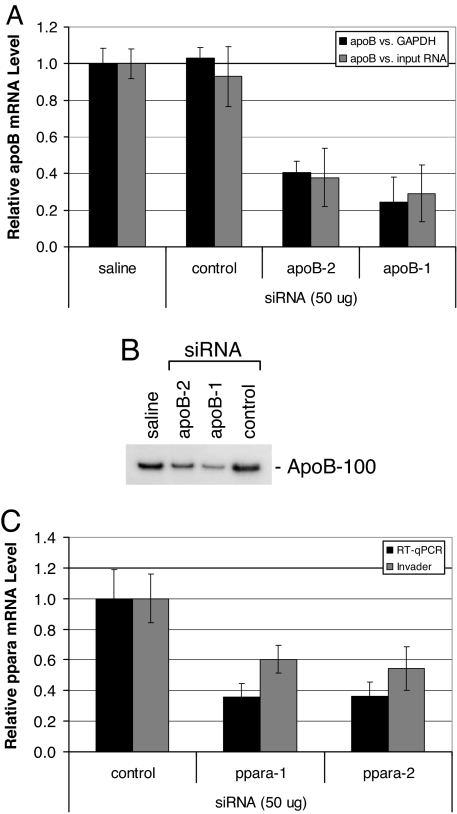
Knockdown of target gene expression in livers of mice after i.v. injection of siRNA polyconjugates. (A) Reduction of apoB mRNA levels in liver after treatment with apoB siRNA polyconjugates. RT-qPCR analyses of liver apoB levels relative to GAPDH mRNA or total input RNA were performed 2 days after injection of apoB-1, apoB-2, or control siRNA polyconjugate (800 μg of polymer, 50 μg of siRNA). Shown are the data normalized to mice receiving saline alone. n = 5, data are shown as mean ± SD. (B) Serum levels of apoB-100 protein are reduced in apoB siRNA polyconjugate-treated mice 2 days after injection. An equal volume of serum from individual mice was pooled for each group (n = 5) and subjected to Western blot analysis. (C) Reduction of ppara mRNA levels in liver after i.v. injection of ppara siRNA polyconjugates. RT-qPCR and Invader analyses of liver ppara mRNA levels relative to GAPDH or ubiquitin mRNA, respectively, 2 days after injection of ppara-1, ppara-2, or control siRNA polyconjugate (800 μg of polymer, 50 μg of siRNA). Shown are the data normalized to mice receiving saline alone. n = 5, data are shown as mean ± SD.
We also tested whether covalent attachment of the siRNA to the polyconjugate is required for efficient siRNA uptake in hepatocytes in mice. Our preliminary in vitro studies indicated that noncovalently complexed siRNA is rapidly displaced from all but the most charge-dense polycations in the presence of serum or physiological concentrations of salt (data not shown). Injection of a polyconjugate formulation in which the polymer and oligonucleotide were electrostatically complexed, but not covalently linked, resulted in much reduced hepatocyte accumulation (Fig. 3D). These results suggest that 21-mer double-stranded oligonucleotides such as siRNA are rapidly displaced from the delivery vehicle and that covalent attachment of the siRNA to the delivery vehicle is necessary for efficient delivery to the target organ.
Knockdown of Target Genes in Liver of Mice by Using siRNA Polyconjugates.
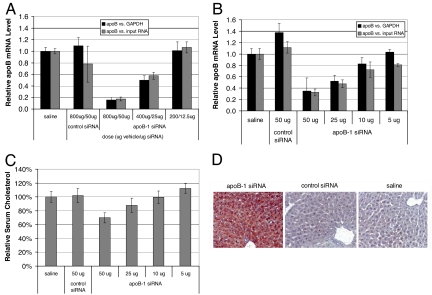
To assess the ability of the polyconjugate to deliver siRNA and knockdown target gene expression in vivo, polyconjugate containing apoB-1 siRNA (800 μg of polymer, 50 μg of siRNA) was delivered to C57BL/6 mice by using a single simple i.v. injection. Livers from injected mice were harvested 2 days after injection and assayed for apoB mRNA levels by using reverse transcriptase quantitative PCR (RT-qPCR). The apoB mRNA levels were measured relative to the level of the housekeeping GAPDH mRNA and micrograms of total input RNA, to reduce the possibility that any differences observed in relative apoB mRNA levels were due to nonspecific effects on housekeeping-gene expression. As shown in Fig. 4A, mice treated with apoB-1 siRNA polyconjugate had significantly reduced apoB mRNA levels compared with mice receiving polyconjugate containing a non-apoB control siRNA or mice injected with saline only (n = 5, P < 0.00001). Specifically, the mean apoB mRNA level in mice receiving apoB-1 siRNA polyconjugate was reduced by 76 ± 14% compared with the saline treated group relative to GAPDH mRNA levels, whereas apoB mRNA levels in mice injected with the control siRNA were unaffected. Similar results were obtained if apoB mRNA levels were measured relative to total RNA.
To confirm the specificity of the apoB knockdown, a separate group of mice was treated with an siRNA targeting a different region of the apoB mRNA. Mice receiving apoB-2 siRNA polyconjugate also exhibited a significant reduction in apoB mRNA levels (60 ± 6% reduction, n = 5, P < 0.00001). Western blot analysis of apoB-100 protein levels in serum reflected the reduction in liver apoB mRNA levels in mice receiving either apoB-1 or apoB-2 siRNAs (Fig. 4B). ApoB mRNA expression was not reduced in the jejunum, another tissue that expresses the apoB gene, suggesting that the polyconjugate does not target this tissue (data not shown).
We also prepared and tested polyconjugates containing siRNAs targeting peroxisome proliferator-activated receptor alpha (ppara), a gene important in controlling fatty acid metabolism in liver where it is expressed solely in hepatocytes (41, 42). Polyconjugate-mediated delivery of two different siRNAs targeting ppara resulted in significant knockdown of ppara mRNA levels in liver (Fig. 4C). Relative to mice receiving a control siRNA, ppara mRNA levels in mice receiving ppara-1 siRNA were reduced by between 40 ± 9% and 64 ± 9%, as determined by Invader or RT-qPCR assays, respectively. A similar reduction in ppara mRNA levels was observed in mice injected with polyconjugate containing ppara-2 siRNA, which targets a separate region of the ppara mRNA sequence. Injection of polyconjugates prepared without the hepatocyte-targeting ligand NAG resulted in no ppara knockdown (SI Fig. 9). This is consistent with the results obtained from siRNA tracking studies shown in Fig. 3, which revealed that the presence of NAG on the polyconjugate was necessary for hepatocyte uptake.
The potential toxicity of the siRNA polyconjugate was assessed by measuring serum levels of liver enzymes and cytokines (SI Table 1). Slight elevations of ALT and AST levels were detected in mice receiving control siRNA or apoB-1 siRNA polyconjugates as compared with saline-treated mice, 48 h after injection. However, the increased levels were not significant (P > 0.05), and histological examination of liver sections did not reveal signs of liver toxicity (data not shown). Similarly, analysis of TNF-α and IL-6 levels in serum by using ELISA revealed that both were slightly elevated 6 h after injection of polyconjugate but returned to baseline by 48 h. The increases observed at 6 h would not be expected to cause significant immune stimulation and are at least four orders of magnitude lower than those observed upon stimulation with lipopolysaccharide (43, 44) and one to three orders of magnitude lower than after injection of adenovirus (27, 45). No significant differences in serum levels of INF-α were detected at any of the time points, except for a slight increase at 6 h after injection of apoB-1 siRNA polyconjugate. These results indicate that the targeted siRNA polyconjugate is well tolerated.
Dose–Response and Phenotypic Analyses of Mice Receiving apoB siRNA Polyconjugate.
We investigated dose–response using two different experimental strategies: by decreasing the amount of siRNA polyconjugate delivered to the mice by serial dilutions of the formulation and by holding the amount of polymer constant but decreasing the amount of siRNA conjugated to it. Injection of simple serial dilutions of the apoB-1 siRNA polyconjugate into mice led to a progressive decrease in the amount of knockdown of liver apoB mRNA (Fig. 5A). At the highest injected dose (800 μg of polymer, 50 μg of siRNA, i.e., 2.5 mg/kg), apoB mRNA levels in the liver were reduced 84 ± 5% relative to GAPDH mRNA on day 2 after injection compared with mice injected with saline only. Similar results were obtained when apoB mRNA levels were measured relative to total RNA. Injection of 2-fold less siRNA polyconjugate (400 μg of polymer, 25 of μg siRNA) resulted in a 50 ± 8% reduction in relative apoB mRNA levels. Injection of 4-fold less resulted in no apoB knockdown as compared with the saline control group. Holding the amount of polymer constant but decreasing the amount of apoB-1 siRNA conjugated to it led to quantitatively similar results at each siRNA dose (Fig. 5B). This finding suggests that the amount of endosomolytic polymer present in the delivery vehicle is not the limiting factor for the knockdown observed, but rather it is the amount, or potency, of the siRNA conjugated to it.
Fig. 5.
apoB-1 siRNA polyconjugate dose–response and phenotypes of treated mice. (A) Knockdown of apoB mRNA after injection of serial dilutions of the apoB-1 siRNA polyconjugate. The indicated dose of apoB-1 or control siRNA polyconjugate was injected intravenously in mice. The livers were harvested 2 days later, and the relative levels of apoB mRNA to those of GAPDH mRNA or total input RNA were measured by RT-qPCR. Data are normalized to mice receiving saline alone. n = 5, data are shown as mean ± SD. (B) Reducing the amount of apoB-1 siRNA attached to the polyconjugate decreases apoB knockdown. Mice were injected with polyconjugate (800 μg of polymer) covalently attached to the indicated amount of apoB-1 or control siRNA. The livers were harvested and relative apoB mRNA levels were determined as in A. n = 5, data are shown as mean ± SD. (C) Serum cholesterol is reduced in a siRNA dose-dependent manner in mice treated with apoB-1 siRNA polyconjugate. Serum from mice in B was collected after a brief fast (4 h) and analyzed for total cholesterol. Values were normalized to mice receiving saline only. n = 5, data are shown as mean ± SD. (D) Knockdown of apoB results in increased hepatic lipid content. Liver sections were taken from briefly fasted mice 2 days after injection of apoB-1 or control siRNA polyconjugate (800 μg of polymer, 50 μg of siRNA), or saline only. Sections were fixed, and lipids were detected by staining with oil red.
A hallmark of apoB deficiency is decreased serum cholesterol levels due to impairment of VLDL assembly and cholesterol transport from the liver (46). To determine whether the level of apoB knockdown shown in Fig. 5B was sufficient to elicit a physiological response in these mice, we measured their total serum cholesterol levels. At the highest delivered siRNA dose (800 μg of polymer, 50 μg), we observed a significant decrease in mean serum cholesterol levels (30 ± 7%, n = 5, P < 0.001) relative to mice receiving a control siRNA or saline only (Fig. 5C). Similar results were obtained in animals treated with apoB-2 siRNA polyconjugate (data not shown). Decreasing the amount of siRNA attached to the polyconjugate led to a progressive decrease in the amount of cholesterol lowering observed, consistent with decreased apoB mRNA knockdown measured in these animals (Fig. 5B).
Impairment of VLDL assembly in the liver and the resultant decrease in VLDL export might also be expected to alter hepatic triglyceride levels because triglycerides are also incorporated into VLDL particles (47). Indeed, transgenic mice expressing a truncated form of apoB found in patients with familial hypobetalipoproteinemia, also display a reduced capacity to transport hepatic triglycerides (48). To assess the effects of apoB knockdown on triglyceride transport, we performed oil red staining of liver sections obtained from mice injected with apoB-1 siRNA polyconjugate. Inspection of the liver sections revealed dramatically increased hepatic lipid content compared with control mice (Fig. 5D). Decreased serum triglyceride levels were also detected in these mice, providing further evidence for diminished hepatic triglyceride export capacity (data not shown). Together, these results indicate that simple i.v. injection of apoB-1 siRNA polyconjugate results in a knockdown of expression of apoB in the liver with expected phenotypic effects.
Longevity and Phenotypic Effect of apoB Knockdown.
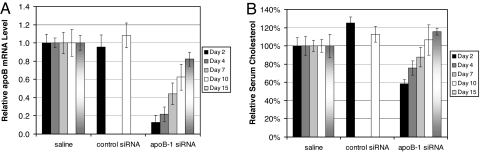
We performed a time course experiment to determine the duration of apoB knockdown and cholesterol lowering in mice after injection of a single dose of apoB-1 siRNA polyconjugate. Consistent with our results described in the previous sections, injection of apoB-1 siRNA polyconjugate (800 μg of polymer, 50 μg of siRNA) resulted in a reduction of mean apoB mRNA levels by 87 ± 8% on day 2 relative to control mice (Fig. 6A). The reduction in apoB expression was accompanied by a 42 ± 5% reduction in total serum cholesterol levels (Fig. 6B). Decreases in apoB mRNA expression remained significant through day 10 and had returned to near control levels by day 15. Reduction in serum cholesterol remained significant through day 4 (n = 5, P < 0.01) and did not fully recover to control levels until day 10. These results indicate that sustained apoB knockdown and lowered serum cholesterol levels can be attained after a single i.v. injection of siRNA polyconjugate, and that the phenotype can be reversed as apoB expression returns to normal.
Fig. 6.
Longevity of apoB knockdown and the phenotypic response in mice. (A) Duration of apoB mRNA knockdown in liver. RT-qPCR analyses of liver apoB levels relative to GAPDH mRNA or total input RNA were performed at the indicated time after injection of apoB-1 or control siRNA polyconjugate (800 μg of polymer, 50 μg of siRNA). Shown are the data normalized to mice receiving saline alone. n = 5, data are shown as mean ± SD. (B) Duration of serum cholesterol reduction. Serum from mice in A was collected after a 4-h fast and analyzed for total cholesterol. Values were normalized to mice receiving saline only. Mice treated with control siRNA polyconjugate were only assayed on days 2 and 10 after injection. n = 5, data are shown as mean ± SD.
Discussion
In this study, we demonstrated the ability to use Dynamic PolyConjugate technology to deliver siRNA to hepatocytes in cells in culture and in mice. In mice, siRNA polyconjugates were used to elicit knockdown of two different genes, apoB and ppara. Maximal knockdown of 80–90% was achieved in vivo with a 2.5 mg/kg dose of apoB siRNA, a result that is similar to what has been achieved by using the SNALP delivery system (21, 22). Time-course studies also indicate a similar knockdown longevity, with an apparent half-life of between 7 and 10 days. On the basis of serum liver enzyme analyses, cytokine assays, and liver histology, liver toxicity was not observed. The ability of the siRNA polyconjugate to target hepatocytes and avoid Kupffer cell uptake is a likely explanation for the lack of toxicity.
Delivery of apoB siRNA caused a phenotypic effect that was manifested by lowered serum cholesterol and ApoB protein levels, and by a fatty liver. A single dose of siRNA polyconjugate resulted in decreased cholesterol levels for 1 week to 10 days. Once the serum cholesterol levels had recovered, we were able to re-administer the apoB siRNA and reduce the serum cholesterol levels a second time (data not shown). The presence of fatty liver was not commented upon in prior reports in which apoB siRNA was delivered in vivo, but a study in which antisense apoB oligonucleotides were used indicated that apoB knockdown did not cause fatty liver (10, 21, 22, 49). However, fatty livers are observed in transgenic mice possessing a mutant apoB gene associated with familial hypobetalipoproteinemia in humans (48). The possibility that apoB knockdown causes fatty liver should be taken into account if apoB is to be considered a therapeutic target in the treatment of hypercholesterolemia in humans. On the other hand, knockdown of apoB may provide a useful mouse model to study the acute effects of fatty liver, an important human clinical problem (50).
The ability to generate a knockdown phenotype after delivery of siRNA polyconjugates was not limited to treatment with apoB siRNA. Mice treated with siRNA polyconjugates targeting ppara also displayed the gene-appropriate phenotype, characterized by a significant increase in serum triglycerides after delivery of ppara siRNA polyconjugate, a phenotype that is consistent with known ppara function (data not shown) (41). However, this phenotype was less reproducible than the apoB knockdown phenotype. It is possible that the level of knockdown achieved against this target was not always sufficient to yield consistent phenotypic effects. The use of more potent siRNAs may help in this regard.
A key feature of Dynamic PolyConjugate technology is the use of targeting ligands to direct the polyconjugate to a specific cell type. We present evidence in this report that hepatocyte-specific targeting is achieved by using polyconjugates containing the NAG ligand and that substituting mannose for NAG results in redirection of the polyconjugate to nonparenchymal liver cells and away from hepatocytes. The targetability of the Dynamic PolyConjugates makes it unique among other recently developed nanotechnologies such as SNALP and iNOP for delivery of siRNA to hepatocytes in liver. Rather than using ligand-mediated targeting, these latter technologies instead rely on the inherent ability of the liver to clear foreign particles of a certain size. One drawback of this approach is that it can result in uptake by cell types in liver other than hepatocytes. Indeed, an intense signal from fluorescently labeled siRNA is observed in Kupffer cells when delivered by using SNALPs (51), a situation that would have the potential to induce Kupffer-cell-mediated liver toxicity. In addition, the size of the SNALP or iNOP siRNA complexes is much greater than the siRNA polyconjugate (21, 34), further increasing the risk of inducing Kupffer cell activation (52). The larger size of the SNALP and iNOP siRNA complexes would also restrict their use to vascular endothelial cells or tissues such as liver with blood vessel fenestrations of >50 nm. The small size of the particles prepared by using Dynamic PolyConjugate technology should allow more flexibility in targeting other cell types.
In summary, Dynamic PolyConjugate technology seamlessly incorporates several design features including an endosomolytic polymeric carrier that is shielded by reversible covalent modifications, reversibly attached cellular receptor ligands, and labile conjugation of siRNA. This is a modular platform system in which these different elements could be adapted to enable siRNA delivery for a wide variety of purposes. Selective targeting is an important characteristic of the Dynamic PolyConjugate technology, and we anticipate that other ligands could be easily incorporated into this system to enable siRNA-mediated knockdown of genes in other tissues and cell types.
Methods: Polyconjugate Synthesis and Formulation
SATA-modified siRNAs were synthesized by reaction of 5′ amine-modified siRNA with 1 weight equivalents (wt eq) of N-succinimidyl-S-acetylthioacetate (SATA) reagent (Pierce) and 0.36 wt eq of NaHCO3 in water at 4°C for 16 h. The modified siRNAs were then precipitated by the addition of 9 vol of ethanol and incubation at −80°C for 2 h. The precipitate was resuspended in 1× siRNA buffer (Dharmacon) and quantified by measuring absorbance at the 260-nm wavelength.
PBAVE was synthesized according to published procedures (33). PBAVE (30 mg/ml in 5 mM TAPS, pH 9) was modified by addition of 1.5 wt % SMPT (Pierce). After a 1-h incubation, 0.8 mg of SMPT-PBAVE was added to 400 μl of isotonic glucose solution containing 5 mM TAPS (pH 9). To this solution was added 50 μg of SATA-modified siRNA. For the dose–response experiments where [PBAVE] was constant, different amounts of siRNA were added. The mixture was then incubated for 16 h. To the solution was then added 5.6 mg of Hepes free base followed by a mixture of 3.7 mg of CDM-NAG and 1.9 mg of CDM-PEG. The solution was then incubated for at least 1 h at room temperature before injection.
CDM-PEG and CDM-NAG were synthesized from the acid chloride generated by using oxalyl chloride. To the acid chloride was added 1.1 molar equivalents polyethylene glycol monomethyl ether (molecular weight average of 450) to generate CDM-PEG or (aminoethoxy)ethoxy-2-(acetylamino)-2-deoxy-β-d-glucopyranoside to generate CDM-NAG. The final product was purified by using reverse-phase HPLC with a 0.1% TFA water/acetonitrile gradient.
Additional methods are available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Mark Noble, Tracie Milarch, Mavis Eldridge, and John Fuller of the Mirus animal care facility for their invaluable assistance and members of the Mirus scientific staff for critically reading the manuscript.
Abbreviations
- CDM
carboxy dimethylmaleic anhydride
- iNOP
interfering nanoparticle
- NAG
N-acetylgalactosamine
- SNALP
stable nucleic acid lipid particle.
Footnotes
Conflict of interest statement: All of the authors except J.A.W. are employees of Mirus Bio.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703778104/DC1.
References
- 1.Dykxhoorn DM, Palliser D, Lieberman J. Gene Ther. 2006;13:541–552. doi: 10.1038/sj.gt.3302703. [DOI] [PubMed] [Google Scholar]
- 2.Kim DH, Rossi JJ. Nat Rev. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 3.Behlke MA. Mol Ther. 2006;13:644–670. doi: 10.1016/j.ymthe.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, Feng Y, Palliser D, Weiner DB, Shankar P, et al. Nat Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 5.Urban-Klein B, Werth S, Abuharbeid S, Czubayko F, Aigner A. Gene Ther. 2005;12:461–466. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- 6.Schiffelers RM, Ansari A, Xu J, Zhou Q, Tang Q, Storm G, Molema G, Lu PY, Scaria PV, Woodle MC. Nucleic Acids Res. 2004;32:e149. doi: 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sioud M, Sorensen DR. Biochem Biophys Res Commun. 2003;312:1220–1225. doi: 10.1016/j.bbrc.2003.11.057. [DOI] [PubMed] [Google Scholar]
- 8.Minakuchi Y, Takeshita F, Kosaka N, Sasaki H, Yamamoto Y, Kouno M, Honma K, Nagahara S, Hanai K, Sano A, et al. Nucleic Acids Res. 2004;32:e109. doi: 10.1093/nar/gnh093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ge Q, Filip L, Bai A, Nguyen T, Eisen HN, Chen J. Proc Natl Acad Sci USA. 2004;101:8676–8681. doi: 10.1073/pnas.0402486101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, et al. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 11.Zhang W, Yang H, Kong X, Mohapatra S, San Juan-Vergara H, Hellermann G, Behera S, Singam R, Lockey RF, Mohapatra SS. Nat Med. 2005;11:56–62. doi: 10.1038/nm1174. [DOI] [PubMed] [Google Scholar]
- 12.Bitko V, Musiyenko A, Shulyayeva O, Barik S. Nat Med. 2005;11:50–55. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- 13.Hassani Z, Lemkine GF, Erbacher P, Palmier K, Alfama G, Giovannangeli C, Behr JP, Demeneix BA. J Gene Med. 2005;7:198–207. doi: 10.1002/jgm.659. [DOI] [PubMed] [Google Scholar]
- 14.Hu-Lieskovan S, Heidel JD, Bartlett DW, Davis ME, Triche TJ. Cancer Res. 2005;65:8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- 15.Xia H, Mao Q, Paulson HL, Davidson BL. Nat Biotechnol. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- 16.Tiscornia G, Singer O, Ikawa M, Verma IM. Proc Natl Acad Sci USA. 2003;100:1844–1848. doi: 10.1073/pnas.0437912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 18.Arias IM, Boyer JL, Fausto N, Jakoby WB, Schachter D, Shafritz DA, editors. The Liver, Biology and Pathobiology. New York: Raven; 1994. [Google Scholar]
- 19.McCaffrey AP, Meuse L, Pham TT, Conklin DS, Hannon GJ, Kay MA. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- 20.Lewis DL, Hagstrom JE, Loomis AG, Wolff JA, Herweijer H. Nature Genet. 2002;32:107–108. doi: 10.1038/ng944. [DOI] [PubMed] [Google Scholar]
- 21.Zimmermann TS, Lee AC, Akinc A, Bramlage B, Bumcrot D, Fedoruk MN, Harborth J, Heyes JA, Jeffs LB, John M, et al. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]
- 22.Judge AD, Bola G, Lee AC, MacLachlan I. Mol Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 23.Baigude H, McCarroll J, Yang C-S, Swain PM, Rana TM. ACS Chem Biol. 2007;2:237–241. doi: 10.1021/cb7000582. [DOI] [PubMed] [Google Scholar]
- 24.Greenwald RB, Conover CD, Choe YH. Crit Rev Ther Drug Carrier Syst. 2000;17:101–161. [PubMed] [Google Scholar]
- 25.Snoeys J, Lievens J, Wisse E, Jacobs F, Duimel H, Collen D, Frederik P, De Geest B. Gene Ther. 2007;14:604–612. doi: 10.1038/sj.gt.3302899. [DOI] [PubMed] [Google Scholar]
- 26.Roberts RA, Ganey PE, Ju C, Kamendulis LM, Rusyn I, Klaunig JE. Toxicol Sci. 2007;96:2–15. doi: 10.1093/toxsci/kfl173. [DOI] [PubMed] [Google Scholar]
- 27.Lieber A, He CY, Meuse L, Schowalter D, Kirillova I, Winther B, Kay MA. J Virol. 1997;71:8798–8807. doi: 10.1128/jvi.71.11.8798-8807.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu J, Nantz MH, Zern MA. Front Biosci. 2002;7:d717–d725. doi: 10.2741/A806. [DOI] [PubMed] [Google Scholar]
- 29.Biessen EA, Bakkeren HF, Beuting DM, Kuiper J, Van Berkel TJ. Biochem J. 1994;299(Pt 1):291–296. doi: 10.1042/bj2990291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coombs PJ, Taylor ME, Drickamer K. Glycobiology. 2006;16:1C–7C. doi: 10.1093/glycob/cwj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rozema DB, Ekena K, Lewis DL, Loomis AG, Wolff JA. Bioconjugate Chem. 2003;14:51–57. doi: 10.1021/bc0255945. [DOI] [PubMed] [Google Scholar]
- 32.Kirby AJ, Lancaster PW. J Chem Soc Perkin Trans. 1972;2:1206–1214. [Google Scholar]
- 33.Wakefield DH, Klein JJ, Wolff JA, Rozema DB. Bioconjugate Chem. 2005;16:1204–1208. doi: 10.1021/bc050067h. [DOI] [PubMed] [Google Scholar]
- 34.Dufes C, Uchegbu IF, Schatzlein AG. Adv Drug Delivery Rev. 2005;57:2177–2202. doi: 10.1016/j.addr.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 35.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, Nichols K, et al. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim EM, Jeong HJ, Park IK, Cho CS, Kim CG, Bom HS. J Nucl Med. 2005;46:141–145. [PubMed] [Google Scholar]
- 37.Watanabe Y, Liu X, Shibuya I, Akaike T. J Biomater Sci Polym Ed. 2000;11:833–848. doi: 10.1163/156856200744048. [DOI] [PubMed] [Google Scholar]
- 38.Pimm MV, Perkins AC, Duncan R, Ulbrich K. J Drug Target. 1993;1:125–131. doi: 10.3109/10611869308996068. [DOI] [PubMed] [Google Scholar]
- 39.Sarkar M, Liao J, Kabat EA, Tanabe T, Ashwell G. J Biol Chem. 1979;254:3170–3174. [PubMed] [Google Scholar]
- 40.Jansen RW, Molema G, Ching TL, Oosting R, Harms G, Moolenaar F, Hardonk MJ, Meijer DK. J Biol Chem. 1991;266:3343–3348. [PubMed] [Google Scholar]
- 41.Kersten S, Seydoux J, Peters JM, Gonzalez FJ, Desvergne B, Wahli W. J Clin Invest. 1999;103:1489–1498. doi: 10.1172/JCI6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schoonjans K, Staels B, Auwerx J. J Lipid Res. 1996;37:907–925. [PubMed] [Google Scholar]
- 43.Matsumoto M, Sakao Y, Akira S. Int Immunol. 1998;10:1825–1835. doi: 10.1093/intimm/10.12.1825. [DOI] [PubMed] [Google Scholar]
- 44.Matsuzaki J, Kuwamura M, Yamaji R, Inui H, Nakano Y. J Nutr. 2001;131:2139–2144. doi: 10.1093/jn/131.8.2139. [DOI] [PubMed] [Google Scholar]
- 45.Benihoud K, Esselin S, Descamps D, Jullienne B, Salone B, Bobe P, Bonardelle D, Connault E, Opolon P, Saggio I, et al. Gene Ther. 2007;14:533–544. doi: 10.1038/sj.gt.3302885. [DOI] [PubMed] [Google Scholar]
- 46.Burnett JR, Barrett PH. Crit Rev Clin Lab Sci. 2002;39:89–137. doi: 10.1080/10408360208951113. [DOI] [PubMed] [Google Scholar]
- 47.Gibbons GF, Wiggins D, Brown AM, Hebbachi AM. Biochem Soc Trans. 2004;32:59–64. doi: 10.1042/bst0320059. [DOI] [PubMed] [Google Scholar]
- 48.Chen Z, Fitzgerald RL, Averna MR, Schonfeld G. J Biol Chem. 2000;275:32807–32815. doi: 10.1074/jbc.M004913200. [DOI] [PubMed] [Google Scholar]
- 49.Crooke RM, Graham MJ, Lemonidis KM, Whipple CP, Koo S, Perera RJ. J Lipid Res. 2005;46:872–884. doi: 10.1194/jlr.M400492-JLR200. [DOI] [PubMed] [Google Scholar]
- 50.Schonfeld G. Annu Rev Nutr. 1995;15:23–34. doi: 10.1146/annurev.nu.15.070195.000323. [DOI] [PubMed] [Google Scholar]
- 51.Morrissey DV, Blanchard K, Shaw L, Jensen K, Lockridge JA, Dickinson B, McSwiggen JA, Vargeese C, Bowman K, Shaffer CS, et al. Hepatology. 2005;41:1349–1356. doi: 10.1002/hep.20702. [DOI] [PubMed] [Google Scholar]
- 52.Popielarski SR, Hu-Lieskovan S, French SW, Triche TJ, Davis ME. Bioconjugate Chem. 2005;16:1071–1080. doi: 10.1021/bc0501146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.