Abstract
TRF2 (telomeric repeat binding factor 2) is an essential component of the telomeric cap, where it forms and stabilizes the T-loop junctions. TRF2 forms the T-loops by stimulating strand invasion of the 3′ overhang into duplex DNA. TRF2 also has been shown to localize to nontelomeric DNA double-strand breaks, but its functional role in DNA repair has not been examined. Here, we present evidence that TRF2 is involved in homologous recombination (HR) repair of nontelomeric double-strand breaks. Depletion of TRF2 strongly inhibited HR and delayed the formation of Rad51 foci after γ-irradiation, whereas overexpression of TRF2 stimulated HR. Depletion of TRF2 had no effect on nonhomologous end-joining, and overexpression of TRF2 inhibited nonhomologous end-joining. We propose, based on our results and on the ability of TRF2 to mediate strand invasion, that TRF2 plays an essential role in HR by facilitating the formation of early recombination intermediates.
Keywords: DNA repair, telomeres
TRF2 (telomeric repeat binding factor 2) was first identified as a major telomere binding protein that shields chromosome ends from degradation and from end-to-end fusions (1–3). Mammalian telomeres form large duplex loops in which the single-stranded 3′ end is embedded within the double-stranded DNA (4). TRF2 generates T-loop structures and recently was shown to bind to DNA junctions (4, 5). The mechanism by which TRF2 facilitates folding of telomeric DNA into T-loops involves binding of TRF2 complexes to DNA and untwisting the neighboring DNA, thereby favoring strand invasion (6).
In addition to evidence of TRF2's role in telomere maintenance, there are multiple pieces of evidence that implicate TRF2 in double-strand break (DSB) repair of nontelomeric DNA (7–9). TRF2 localizes to DSB sites at the early stages of cellular response to DSBs, appearing in the first few seconds after DSB induction and leaving as DSBs are being processed (7). TRF2 is phosphorylated by ATM in response to DNA damage (8). The phosphorylated form of TRF2 is not bound to telomeric DNA and is localized to DNA damage sites (8). In turn, overexpression of TRF2 inhibits autophosphorylation of ATM and induction of DNA damage response (9).
DSBs are repaired by two major pathways: homologous recombination (HR) and nonhomologous end-joining (NHEJ). During HR, the missing information is copied into the DSB site from a homologous sequence, whereas NHEJ joins the broken ends without homology. Several proteins involved in DSB repair interact with TRF2, such as the MRE11/Rad50/NBS1 complex, Ku70, WRN, and BLM (10–12). Thus, TRF2 is phosphorylated in response to DNA damage, localizes to DNA breaks, and interacts with DSB repair proteins, but its functional role in DSB repair is unknown. Here, we examined the role of TRF2 in DSB repair by HR and NHEJ.
Results
Reporter Cell Lines for Detection of NHEJ and HR.
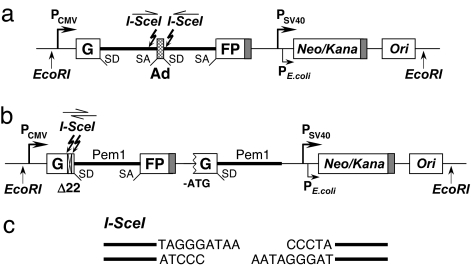
To study the role of TRF2 in NHEJ and HR, we used an in vivo assay that measures the repair of an induced chromosomal break. The reporter cassette for detection of NHEJ previously was described (13). It contains the GFP gene with an artificially engineered 3-kb intron from the Pem1 gene (GFP–Pem1) (Fig. 1a). The Pem1 intron contains adenoviral exon flanked by recognition sequences for I-SceI endonuclease in inverted orientation. Digestion with I-SceI generates DSB with incompatible DNA ends (Fig. 1c). Unrearranged NHEJ cassette is GFP-negative because adenoviral exon disrupts the GFP ORF. Upon induction of DSBs by expression of I-SceI, the adenoviral exon is removed, NHEJ reconstitutes the functional GFP gene, and green cells can be scored by flow cytometry. The HR reporter was built on the basis of GFP–PemI used for the NHEJ reporter. In the HR reporter (Fig. 1b), the first exon of the GFP–Pem1 contains a 22-bp deletion combined with insertion of three restriction sites: I-SceI–HindIII–I-SceI for induction of DSB. The deletion ensures that GFP cannot be reconstituted by an NHEJ event. The two I-SceI sites are in inverted orientation; therefore, I-SceI digestion leaves incompatible ends. The first copy of the GFP–Pem1 is followed by the promoter-less/ATG-less first exon of GFP–Pem1. The intact construct is GFP-negative. Upon induction of DSB by I-SceI digestion, the functional GFP gene will be reconstituted by intramolecular or intermolecular gene conversion between the two mutated copies of the first GFP–PemI exon. The second copy of the GFP gene is lacking the first ATG codon and the second exon, so crossing over or single-strand annealing will not restore the GFP activity. The design allows exclusive detection of gene conversion, which is the predominant HR pathway in mammalian cells (14).
Fig. 1.
Reporter cassettes for detection of NHEJ and HR. (a) Reporter cassette for detection of NHEJ. The cassette consists of the GFP gene under CMV promoter with an engineered intron from the rat Pem1 gene, interrupted by an adenoviral exon (Ad). The adenoviral exon is flanked by I-SceI recognition sites in inverted orientation for induction of DSBs. In this construct, the GFP gene is inactive; however, upon induction of a DSB and successful NHEJ, the construct becomes GFP+. The construct also contains the hybrid neomycin/kanamycin-resistance gene and bacterial origin of replication. The cassette is flanked by EcoRI sites to facilitate rescue of reporter cassettes from genomic DNA. SD, splice donor; SA, splice acceptor. Shaded rectangles indicate polyadenylation sites. (b) Reporter cassette for detection of HR. The cassette consists of two mutated copies of GFP-Pem1. In the first copy of GFP-Pem1, the first GFP exon contains a deletion of 22 nt and an insertion of two I-SceI recognition sites in inverted orientation. The 22-nt deletion ensures that GFP cannot be reconstituted by an NHEJ event. The second copy of GFP-Pem1 is lacking the ATG and the second exon of GFP. Upon induction of DSBs by I-SceI, gene conversion events reconstitute active GFP gene. (c) Incompatible DNA ends generated by digestion of two inverted I-SceI sites.
The NHEJ or HR reporter cassettes were integrated stably on a chromosome of HCA2 normal human fibroblasts immortalized by expression of human telomerase reverse transcriptase (HCA2–hTERT). The cell lines containing single unrearranged copies of the reporter cassettes were identified by Southern blot.
TRF2 Overexpression Inhibits NHEJ and Stimulates HR.
To examine the effect of TRF2 overexpression on NHEJ, the plasmid encoding the I-SceI endonuclease was contransfected with a TRF2 expression vector (pCMV–TRF2) or a control plasmid expressing the hypoxanthine phosphoribosyltransferase (HPRT) gene (pHPRT–CAG32) and a DsRed-expressing plasmid (pDsRed2-N1) into NHEJ reporter cell lines by using Amaxa Nucleofector (Amaxa, Gaithersburg, MD). Expression of I-SceI and TRF2 was verified by Western blot (Fig. 2 b and c). I-SceI expression was high on day 1 after transfection and subsequently declined, suggesting that the induction of DSBs occurs on day 1. TRF2 showed strong expression on day 1, and the levels slowly declined over the next 4 days. Thus, the highest levels of TRF2 were present during DSB induction. NHEJ of I-SceI-induced DSBs results in the appearance of GFP+ cells. To quantify NHEJ events, the cells were examined by flow cytometry on day 4 after transfection to allow for maximum GFP expression. To normalize for the efficiency of transfection, the ratio of GFP+ to DsRed+ cells was used as a measure of NHEJ efficiency. TRF2 overexpression inhibited NHEJ approximately 2-fold (Fig. 3a).
Fig. 2.
Overexpression of TRF2 proteins in HCA2–hTERT cells. (a) Schematic diagram of the full-length TRF2 and the deletion mutants TRF2ΔBΔM, TRF2ΔB, and TRF2ΔM. The numbers correspond to amino acid positions. (b) Western blot analysis of TRF2 overexpression. HCA2–hTERT cells were transfected with full-length TRF2, TRF2ΔBΔM, TRF2ΔB, or TRF2ΔM using Amaxa electroporation. The cells were harvested at the indicated time after transfection. In each lane, 50 μg of total protein was loaded and hybridized with anti-TRF2 antibodies (see Materials and Methods). Equal loading was verified by hybridization with anti-Actin antibodies. (c) Expression of I-SceI in HCA2–hTERT cells. Cells were transfected with I-SceI-expressing plasmid and harvested at the indicated time, and 50 μg of total protein was analyzed by Western blot with anti-HA antibodies. Equal loading was verified by hybridization with anti-actin antibodies.
Fig. 3.
Effect of TRF2 overexpression on NHEJ (a) and HR (b). HCA2–hTERT cells containing NHEJ or HR reporter cassettes were cotransfected with the plasmids encoding the full-length or truncated TRF2 (5 μg), I-SceI (5 μg), and pDsRed2-N1 (0.1 μg). I-SceI induces DSBs in the GFP-based reporter cassettes (Fig. 1), and successful repair by NHEJ for NHEJ reporter or HR for HR reporter results in the appearance of GFP+ cells. To quantify NHEJ or HR events, the cells were analyzed by flow cytometry 4 days after transfection. The DsRed was used to normalize transfection efficiency. The ratio of GFP+ to DsRed+ cells was used as a measure of NHEJ or HR. The experiments were repeated at least five times. Error bars indicate SD, and asterisks indicate the treatments significantly different from controls (P < 0.01, Student's t test).
We then analyzed the sequences at the novel junctions. The NHEJ reporter cassette contains the bacterial origin of replication and the kanamycin resistance gene (Fig. 1), which enables rescue of the reporter cassettes in Escherichia coli. The reporter cassette and the E. coli sequences are flanked by EcoRI sites to facilitate the plasmid rescue (Fig. 1). Genomic DNA was digested by EcoRI, and the fragments were circularized and transformed into competent E. coli cells. Unrearranged reporter cassettes were eliminated by digestion with PstI and ScaI enzymes. Reporter cassettes that underwent NHEJ were rescued from the cells transfected with TRF2 or with the control plasmid and were sequenced. The junctions contained deletions of 3 to 29 bp, and two clones contained insertions of 163 and 522 bp at the junction [supporting information (SI) Fig. 6]. These sequences are similar to previously reported NHEJ events in mammalian cells (13).
To determine which TRF2 domains are required for repression of NHEJ, we generated constructs lacking the Myb domain (pCMV–TRF2ΔM), basic domain (pCMV–TRF2ΔB), or both (pCMV–TRF2ΔBΔM) (Fig. 2a). Truncated forms of TRF2 were expressed at comparable levels to the wild-type TRF2 (Fig. 2b). Overexpression of the TRF2ΔM, TRF2ΔB, or TRF2ΔBΔM had no significant effect on the efficiency of NHEJ, indicating that both Myb and the basic domains are required for the repression of NHEJ (Fig. 3a).
The effect of TRF2 overexpression on HR was analyzed in the same manner as for NHEJ. HR reporter cell lines were cotransfected with the expression vectors for I-SceI, TRF2 or a control plasmid pHPRT–CAG32, and DsRed2-N1. Gene conversion events in the reporter cassette reconstitute the wild-type GFP sequence within the first exon of GFP. This was confirmed by rescuing the reporter cassettes in E. coli and sequencing. The frequency of GFP+ cells corresponding to HR events was analyzed by flow cytometry. To normalize for the efficiency of transfection, the ratio of GFP+ to DsRed+ cells was used as a measure of HR. Overexpression of the wild-type TRF2 caused ≈40% increase in the HR efficiency (Fig. 3b). Overexpression of TRF2ΔBΔM or TRF2ΔB had no effect on HR, whereas overexpression of TRF2 lacking Myb domain increased the efficiency of HR by >2-fold (Fig. 3b). These results indicate that TRF2 is involved in the regulation of both NHEJ and HR pathways of DSB repair at a chromosomal DNA break.
TRF2 Knockdown Does Not Affect NHEJ and Inhibits HR.
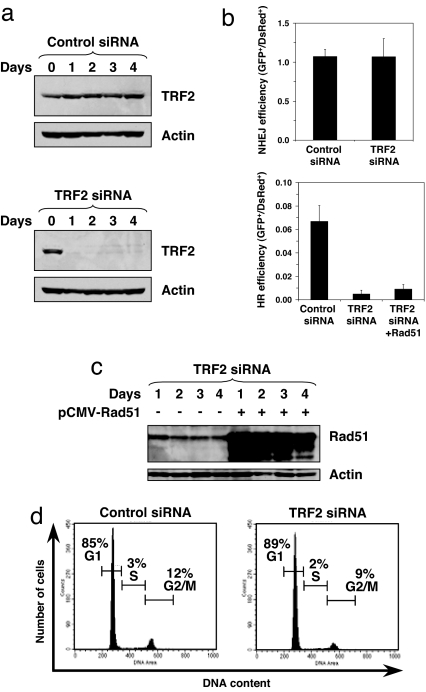
To further understand the role of TRF2 in NHEJ and HR, we examined the effect of knockdown of TRF2. NHEJ and HR reporter cell lines were transfected with siRNA to TRF2 or control siRNA twice with a 3-day interval. Single siRNA transfection led to only partial reduction of TRF2 levels, and two consecutive siRNA transfections eliminated most of the TRF2 protein (Fig. 4a). Three days after the second siRNA transfection, cells were transfected with expression vector for I-SceI to induce DSBs. Four days after I-SceI transfection, the cells were analyzed by flow cytometry. Knockdown of TRF2 had no effect on the efficiency of NHEJ but suppressed HR almost entirely (Fig. 4b). The lack of a corresponding increase in NHEJ efficiency in TRF2-depleted cells is likely explained by a low frequency of HR relative to NHEJ. Alternatively, the cells with unrepaired DSBs may enter senescence, which is a typical response of normal human fibroblasts to unrepaired DSBs (15).
Fig. 4.
Effect of TRF2 depletion on NHEJ and HR. (a) Depletion of TRF2 by siRNA. HCA2–hTERT cells containing NHEJ or HR reporter cassettes were transfected with 100 pmol of anti-TRF2 or control siRNAs (SI00742630 and 1022076; Qiagen, Valencia, CA). The siRNA transfection was repeated twice with 3-day intervals. The amount of TRF2 was analyzed by Western blot at the indicated time after the second siRNA transfection. Each lane contains 50 μg of whole-cell protein extract. Actin was used as a loading control. (b) NHEJ and HR efficiency in TRF2-depleted cells. Reporter cell lines were transfected with I-SceI plasmid and DsRed2-N1 on day 3 after the second siRNA transfection. The ratio of GFP+ to DsRed+ cells serves as a measure of NHEJ or HR efficiency. The experiments were repeated four times. Error bars indicate SD. (c) Western blot showing Rad51 overexpression. Cells were transfected with a plasmid encoding Rad51 cDNA under CMV promoter. (d) Cell cycle distribution of TRF2-depleted cells on day 1 after I-SceI transfection. Cell cycle distribution was determined by PI staining and FACS analysis.
TRF2 depletion is known to cause the formation of DNA damage foci at telomeres and may potentially indirectly affect HR by depleting the available Rad51 pools. To test this possibility, we overexpressed Rad51 cDNA in TRF2 siRNA-depleted cells (Fig. 4c). Overexpression of Rad51 did not rescue HR deficiency in TRF2-depleted cells (Fig. 4b), which is evidence against this possibility.
Because HR is most active in the G2 stage of the cell cycle, we compared cell cycle distribution in cells transfected with the control and TRF2 siRNAs. Cell cycle distribution was analyzed on day 1 after I-SceI transfection when DSB repair is expected to take place (Fig. 4d). There was a very minor change in the cell cycle profile. The number of G2/M cells dropped from 12% in the cells transfected with the control siRNA to 9% in the cells transfected with TRF2 siRNA. At the same time, the HR efficiency dropped 14-fold (from 0.066 in the control to 0.0048 in TRF2-depleted cells). If the change in the HR frequency in TRF2-depleted cells was due to the drop in the number of cells in G2/M, then the expected HR frequency after TRF2 depletion would be 0.05. This expected HR frequency falls outside of the 95% confidence interval of our observed HR frequency (0.0015–0.0081). Therefore, we conclude that the observed inhibition of HR cannot be explained by cell cycle arrest elicited by TRF2 depletion. HR also was suppressed by 60% in the cells subjected to single TRF2 siRNA transfection, which did not affect cell growth (data not shown). These results indicate that TRF2 is required for DSB repair by HR.
TRF2 Knockdown Results in Impaired Rad51, Irradiation-Induced Foci.
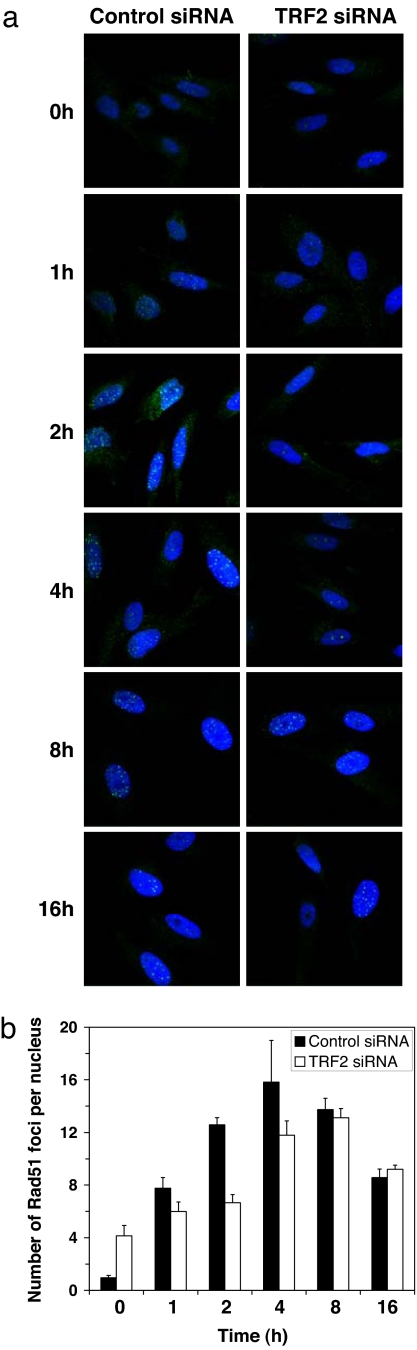
We found that TRF2 is required for HR and that TRF2 was shown to be recruited to DSBs at an early stage (7), so we hypothesized that TRF2 might control the formation of Rad51-ionizing, radiation-induced foci. We analyzed the formation of Rad51 foci at various time points after exposure to 8 Gy of γ-irradiation (Fig. 5). HCA2–hTERT cells were transfected with siRNA to TRF2 or control siRNA as described above. Three days after the second siRNA transfection, cells were γ-irradiated and fixed at various time points after irradiation. Nonirradiated cells transfected with the control siRNA contained on average one focus per nucleus. The number of Rad51 foci increased rapidly within the first hour after irradiation and reached its maximum at 4 h after irradiation (Fig. 5b). In TRF2-depleted cells, the initial number of foci was four, which is in agreement with previous reports that show that depletion of TRF2 generates DNA damage signal at telomeres (16). After irradiation of TRF2-depleted cells, the number of Rad51 foci remained almost unchanged within the first 2 h, and increased to 11.8 foci per nucleus at 4 h. The number of Rad51 foci in TRF2-depleted cells reached its maximum only at 8 h after irradiation. Thus, depletion of TRF2 delayed Rad51 foci formation by ≈4 h. This result shows that the effect of TRF2 on HR is associated with formation of Rad51 foci, which further supports the involvement of TRF2 in HR.
Fig. 5.
Effect of TRF2 depletion on formation of Rad51 foci after γ-irradiation. (a) Representative Rad51 foci in cells transfected with control or TRF2 siRNA. Cells were exposed to 8 Gy of irradiation and fixed for immunohistochemical analysis of Rad51 at the indicated times. Green is immunofluorescent staining of Rad51; blue is DNA stained with DAPI. (b) Numbers of Rad51 foci per nucleus in control siRNA or TRF2 siRNA-treated cells. The experiments were repeated three times, and 180 nuclei were counted per each experimental point. Error bars indicate SD.
Discussion
This report demonstrates that TRF2 is involved in DSB repair of nontelomeric DNA. Our results show that TRF2 represses NHEJ and is required for HR. Both N-terminal and C-terminal TRF2 domains are required for repression of NHEJ. Overexpression of the full-length TRF2 and C-terminally truncated TRF2 (TRF2ΔM) stimulate HR at nontelomeric DSB. Depletion of TRF2 does not affect NHEJ but strongly inhibits HR and delays the formation of Rad51 foci after irradiation. These results suggest that TRF2 plays a functional role in HR and may inhibit NHEJ by directing DSB repair toward HR pathway.
The inhibitory effect of TRF2 on NHEJ is not surprising because it also blocks NHEJ of telomeric DNA, but the requirement of TRF2 for HR is an unexpected finding, because it contrasts with the role of TRF2 at telomeres, where it inhibits sister chromatid exchanges and T-loop recombination (17, 18).
Despite the inhibitory role of TRF2 in HR at telomeres, multiple evidence indicates the potential role of TRF2 in HR of nontelomeric DNA. TRF2 mediates formation of T-loops at telomeres (4, 19), which resemble the structure of Holliday junctions. The basic domain of TRF2 binds to Holliday junctions in a sequence-unspecific manner (5), suggesting that TRF2 may participate in HR of nontelomeric DNA. Furthermore, a recent biochemical study has shown that TRF2 mediates the invasion of 3′ overhangs into duplex DNA (6). TRF2 complexes are able to generate positive supercoiling, thereby untwisting the neighboring DNA and facilitating strand invasion (6). TRF2 does not behave like RecA in strand invasion, as it does not require ATP and does not catalyze strand exchange. Importantly, TRF2 was able to mediate strand invasion of both telomeric and nontelomeric sequences (6), which suggests that it can facilitate strand invasion during repair of DSBs.
The N-terminal domain of TRF2 has DNA binding activity, which is unspecific to telomeric sequence (5, 6). We show that the TRF2 mutant (TRF2ΔM) that contains this unspecific DNA-binding domain and lacks the C-terminal domain stimulates HR when overexpressed. Thus, it is likely that the N-terminal domain of TRF2 is involved in strand invasion during HR repair of DSBs, and the C-terminal domain plays a regulatory role by limiting the HR activity of TRF2.
Depletion of TRF2 delays Rad51 foci formation after irradiation by ≈4 h. TRF2 may recruit directly Rad51 or, alternatively, may act earlier in the pathway. Multiple proteins facilitate Rad51 recruitment to DSBs in mammalian cells; among them are Rad52, Rad51 paralogs, and BRCA2 and possibly others (20, 21). TRF2 may interact with these proteins and facilitate further recruitment of Rad51. Why is the formation of Rad51 foci delayed, and the DSBs are not processed by NHEJ pathway? The exact interaction between HR and NHEJ during DSB repair is not fully understood. Recent studies have shown that both NHEJ and HR factors are recruited to the sites of DSBs (22, 23). The NHEJ factors are recruited first, and Rad51 is recruited relatively late (within 1 h after DSB induction). This finding opposes previous models of competition between HR and NHEJ factors for binding to DSBs and suggests that, based on the configuration of DNA ends or other factors, certain breaks are preferentially processed by NHEJ, and others are preferentially processed by HR. In the absence of TRF2, the breaks that would normally be repaired by HR stay unrepaired for a longer period. However, in a normal situation, TRF2 is recruited to the site of DSB very early, within 2 sec of irradiation (7). Thus, it is possible that TRF2 arrives early, marks certain DSBs for repair by HR, and facilitates strand invasion by Rad51.
In summary, our study has assigned a functional role to TRF2 in HR repair of genomic DSBs. Based on our results, immunolocalization studies, and the biochemical properties of TRF2, we propose that TRF2 binds rapidly to DSB sites, “prepares the ground” for Rad51, and facilitates strand invasion by Rad51.
Materials and Methods
Cell Culture.
The hTERT-immortalized HCA2 cell line (15) and its derivatives were cultured in Eagle's minimum essential media with Eagle's salts (Gibco) containing 15% FCS (Gibco) and 1% penicillin and streptomycin (Gibco) in 5% CO2/3% O2 at 37°C.
Transfections.
Cells were split 2 days before transfection at the density of 5 × 105 cells per plate. Plasmids or siRNA were transfected into HCA2–hTERT cells by an Amaxa Nucleofector machine using the Human Dermal Fibroblast Nucleofector kit and U20 program.
Construction of Cell Reporter Lines Containing Cassettes for Analysis of the Efficiency of NHEJ and HR.
The linearized reporter cassettes (0.5 μg) were transfected into HCA2–hTERT cells by Amaxa electroporation. One day after transfection, cells were placed on selection with G418 at 1 mg/ml. G418-resistant colonies were picked 7–10 days later. The clones were screened by Southern blot for intact reporter cassettes and the number of integrated copies was analyzed. Single-copy integrants were selected for further study.
TRF2 Constructs.
p16–1 plasmid containing TRF2 cDNA was kindly provided by Titia de Lange (The Rockefeller University, New York, NY). To construct pCMV–TRF2, the TRF2 cDNA from p16-1 was cloned into EcoRI site of pDsRed2-N1 (Clontech), and DsRed cDNA was removed by NotI/BamH I digestion. To construct pCMV-TRF2ΔBΔM, the TRF2 fragment was amplified with primers (5′-CGCGCGGCCGCCGAGGCACGGCTGGAAGAGGCAGTC and 5′-CTTGGGCCCTTATTATTCTTCTACAGTCCACTTCTGC) using a GC-rich PCR kit (Roche, Gipf-Oberfrick, Switzerland), the primers containing the NotI and ApaI restriction sites. The amplified fragment was cloned with NotI and ApaI restriction sites into a vector containing CMV promoter and N-terminal FLAG epitope. The sequences of TRF2 and TRF2ΔBΔM in the pCMV-TRF2 and pCMV-TRF2ΔBΔM were confirmed by sequencing. Both clones were found to contain a deletion of 3 nt at positions 1303–1305 in TRF2 ORF. To repair this mutation, we amplified a TRF2 cDNA fragment by RT-PCR from RNA extracted from normal human fibroblasts and replaced the PstI/SalI fragment in the pCMV-TRF2 and pCMV-TRF2ΔBΔM with the PstI/SalI fragment from cDNA. The resulting plasmids were sequenced, and their sequences were identical to the TRF2 sequence in The National Center for Biotechnology Information. The pCMV-TRF2ΔM plasmid was constructed by cloning the PstI/SalI fragment of pCMV-TRF2ΔBΔM into PstI/SalI-digested pCMV-TRF2. The pCMV-TRF2ΔB was derived cloning the HindIII/PstI fragment of pCMV-TRF2ΔBΔM into HindIII/PstI-digested pCMV-TRF2.
Antibodies for Western Blots.
TRF2 was detected with ab4182 antibodies (Abcam, Cambridge, MA) to the central domain of TRF2. These antibodies can recognize all of the three TRF2 constructs when overexpressed; however, their affinity is too low to detect endogenous TRF2. Therefore, for siRNA experiments, endogenous TRF2 was detected by using antiTRF2 antibodies 2645 (Cell Signaling Technology, Beverly, MA) to the C-terminal domain of TRF2. I-SceI was detected with anti-HA antibody 2367 (Cell Signaling Technology).
FACS Analysis.
Cells were harvested, washed in 1× PBS, and kept on ice before FACS analysis. FACS analysis was performed on FACSCalibur instrument (BD Biosciences, Franklin Lakes, NJ) using red-versus-green fluorescence plot as described in Seluanov et al. (13). The gating was determined by analyzing cells transfected with pEGFP-N1 (Clontech, Mountain View, CA), pDsRed2-N1 (Clontech), and a control plasmid (pHPRT-CAG32). A minimum of 20,000 cells per sample were analyzed. Data were analyzed with CellQuest software.
Plasmids Rescue.
The reporter cassettes in the HR and NHEJ cell lines are flanked by EcoRI sites and contain the kanamycin resistance gene and the bacterial origin of replication (Fig. 1). Genomic DNA (10 μg) was digested with EcoRI enzyme overnight. Digested DNA was ligated overnight at 16°C with New England Biolabs (Ipswich, MA) DNA ligase and was purified with phenol/chloroform. The purified DNA was digested overnight with PstI/ScaI enzymes, which cut inside adenoviral exon to remove unrearranged constructs. The DNA was purified with phenol/chloroform and transfected into Stbl2-competent cells (Invitrogen, Carlsbad, CA). Colonies were characterized by digestion and sequenced.
Immunohistochemistry.
HCA2–hTERT cells were transfected twice with control or with TRF2 siRNA, and 1 day after second transfection, the cells were transferred on fibronectin-pretreated culture slides (BD Falcon, Franklin Lakes, NJ) at 2 × 104 cells per slide. Two days after plating, cells were treated with 8 Gy of γ-irradiation using a Shepherd irradiator (6,000-Cu 137Cs source). One, 2, 4, 8, and 16 h after irradiation, cells were fixed with 4% freshly prepared paraformaldehyde for 15 min at room temperature, washed twice briefly with ice cold PBS, and permeabilized with 0.25% Triton X-100 for 10 min at room temperature. Cells were washed three times for 5 min with PBS at room temperature and blocked with goat serum (1:100) at room temperature for 1 h. Then the cells were incubated with primary antibody to Rad51 (1:500, PC130; Calbiochem, San Diego, CA) overnight at 4°C, washed three times with PBS, and incubated with FITC-conjugated anti-rabbit secondary antibody (1:200) for 1 h at room temperature in the dark. Cells were washed for 10 min three times, stained with 1 μg/ml DAPI for 2 min at room temperature in the dark, and washed briefly three times with PBS. The slides were covered with VECTASHIELD mounting media (Vector Laboratories, Burlingame, CA). The images were taken using a Leica (Deerfield, IL) confocal microscope TCS SP.
Supplementary Material
Acknowledgments
We thank Dr. Titia de Lange for the kind gift of TRF2 cDNA, Dara Brown and A'Shantee O'Steen for help with construction of reported cell lines, and Steven Rosenblatt for help with plasmid rescue. This work was supported by National Institutes of Health Grant AG027237, an American Federation for Aging Research grant and a Komen Foundation grant (both to V.G.), and Ellison Medical Foundation grants (to A.S. and V.G.).
Abbreviations
- DSB
double-strand break
- HR
homologous recombination
- NHEJ
nonhomologous end-joining
- HPRT
hypoxanthine phosphoribosyltransferase
- hTERT
human telomerase reverse transcriptase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702410104/DC1.
References
- 1.Broccoli D, Smogorzewska A, Chong L, de Lange T. Nat Genet. 1997;17:231–235. doi: 10.1038/ng1097-231. [DOI] [PubMed] [Google Scholar]
- 2.Bilaud T, Brun C, Ancelin K, Koering CE, Laroche T, Gilson E. Nat Genet. 1997;17:236–239. doi: 10.1038/ng1097-236. [DOI] [PubMed] [Google Scholar]
- 3.Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- 4.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 5.Fouche N, Cesare AJ, Willcox S, Ozgur S, Compton SA, Griffith JD. J Biol Chem. 2006;281:37486–37495. doi: 10.1074/jbc.M608778200. [DOI] [PubMed] [Google Scholar]
- 6.Amiard S, Doudeau M, Pinte S, Poulet A, Lenain C, Faivre-Moskalenko C, Angelov D, Hug N, Vindigni A, Bouvet P, et al. Nat Struct Mol Biol. 2007;14:147–154. doi: 10.1038/nsmb1192. [DOI] [PubMed] [Google Scholar]
- 7.Bradshaw PS, Stavropoulos DJ, Meyn MS. Nat Genet. 2005;37:193–197. doi: 10.1038/ng1506. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka H, Mendonca MS, Bradshaw PS, Hoelz DJ, Malkas LH, Meyn MS, Gilley D. Proc Natl Acad Sci USA. 2005;102:15539–15544. doi: 10.1073/pnas.0507915102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karlseder J, Hoke K, Mirzoeva OK, Bakkenist C, Kastan MB, Petrini JH, de Lange T. PLoS Biol. 2004;2:E240. doi: 10.1371/journal.pbio.0020240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu XD, Kuster B, Mann M, Petrini JH, de Lange T. Nat Genet. 2000;25:347–352. doi: 10.1038/77139. [DOI] [PubMed] [Google Scholar]
- 11.Song K, Jung D, Jung Y, Lee SG, Lee I. FEBS Lett. 2000;481:81–85. doi: 10.1016/s0014-5793(00)01958-x. [DOI] [PubMed] [Google Scholar]
- 12.Opresko PL, von Kobbe C, Laine JP, Harrigan J, Hickson ID, Bohr VA. J Biol Chem. 2002;277:41110–41119. doi: 10.1074/jbc.M205396200. [DOI] [PubMed] [Google Scholar]
- 13.Seluanov A, Mittelman D, Pereira-Smith OM, Wilson JH, Gorbunova V. Proc Natl Acad Sci USA. 2004;101:7624–7629. doi: 10.1073/pnas.0400726101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson RD, Jasin M. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorbunova V, Seluanov A, Pereira-Smith OM. J Biol Chem. 2002;277:38540–38549. doi: 10.1074/jbc.M202671200. [DOI] [PubMed] [Google Scholar]
- 16.Celli GB, de Lange T. Nat Cell Biol. 2005;7:712–718. doi: 10.1038/ncb1275. [DOI] [PubMed] [Google Scholar]
- 17.Wang RC, Smogorzewska A, de Lange T. Cell. 2004;119:355–638. doi: 10.1016/j.cell.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 18.Celli GB, Denchi EL, de Lange T. Nat Cell Biol. 2006;8:885–890. doi: 10.1038/ncb1444. [DOI] [PubMed] [Google Scholar]
- 19.Stansel RM, de Lange T, Griffith JD. EMBO J. 2001;20:5532–5540. doi: 10.1093/emboj/20.19.5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Symington LS. Microbiol Mol Biol Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esashi F, Christ N, Gannon J, Liu Y, Hunt T, Jasin M, West SC. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 22.Kim JS, Krasieva TB, Kurumizaka H, Chen DJ, Taylor AM, Yokomori K. J Cell Biol. 2005;170:341–347. doi: 10.1083/jcb.200411083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rapp A, Greulich KO. J Cell Sci. 2004;117:4935–4945. doi: 10.1242/jcs.01355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.