Abstract
Adducts induced by the antitumor alkylator ecteinascidin 743 (ET-743, Yondelis, trabectedin) represent a unique challenge to the DNA repair machinery because no pathway examined to date is able to remove the ET adducts, whereas cells deficient in nucleotide excision repair show increased resistance. We here describe the processing of the initial ET adducts into cytotoxic lesions and characterize the influence of cellular repair pathways on this process. Our findings show that exposure of proliferating mammalian cells to pharmacologically relevant concentrations of ET-743 is accompanied by rapid formation of DNA double-strand breaks (DSBs), as shown by the neutral comet assay and induction of focalized phosphorylated H2AX. The ET adducts are stable and can be converted into DSBs hours after the drug has been removed. Loss of homologous recombination repair has no influence on the initial levels of DSBs but is associated with the persistence of unrepaired DSBs after ET-743 is removed, resulting in extensive chromosomal abnormalities and pronounced sensitivity to the drug. In comparison, loss of nonhomologous end-joining had only modest effect on the sensitivity. The identification of DSB formation as a key step in the processing of ET-743 lesions represents a novel mechanism of action for the drug that is in agreement with its unusual potency. Because loss of repair proteins is common in human tumors, expression levels of selected repair factors may be useful in identifying patients particularly likely to benefit, or not, from treatment with ET-743.
Keywords: cancer therapy, natural products, lesion processing, DNA repair, response prediction
Compounds forming covalent DNA adducts (hereafter named DNA alkylators) are currently included in virtually all clinical drug regiments for the treatment of both leukemias and solid tumors. Lately, this group of compounds has attracted renewed interest because of the introduction of a novel class of antitumor agents with potent/unusual clinical activities, comprised of monofunctional DNA alkylators derived from natural sources. This group includes ecteinascidin-743 (ET-743, Yondelis, trabectedin), irofulven, hedamycin, and the acronycine derivative S23906. Mechanistic studies have revealed several differences in the way these compounds react with DNA-metabolizing enzymes as well as with the repair machinery (1–11), which could, at least in part, explain their different activity spectra. A better understanding of the factors controlling the induction and removal of DNA damage induced by these compounds should not only facilitate their clinical application, but may also contribute to our general understanding of the structural and biological features governing adduct processing in mammalian cells.
ET-743 has been subject to particular scrutiny linked to its source, clinical activity, and molecular mechanisms. ET-743 is a tetrahydroisoquinoline alkaloid isolated from the Caribbean sea squirt Ecteinascidia turbinata (12), pointing to marine products as a potential new source of molecules with original chemical structures and activities. ET-743 shows a broad activity spectrum toward tumor cell lines at pM and low nM concentrations (6, 13) and has clinical activity toward ovarian cancer as well as soft tissue and bone sarcomas in heavily pretreated patients (14–16). In particular, ET-743 treatment is associated with a striking, long-term response in a subset of patients with otherwise chemoresistant soft tissue sarcomas (reviewed in ref. 17).
ET-743 binds to the minor groove of DNA with preference for GC-rich triplets and subsequently forms covalent adducts with the N2-position of guanine through its carbinolamine moiety (18). As a result, the minor groove is opened up and bended toward the major groove (19). The direction of bending is a novel feature among DNA minor groove-interactive agents, making ET-743 unique.
A particularly interesting aspect of the ET adducts is their interaction with cellular DNA repair pathways. ET lesions are not recognized by the global genome protein XPC but are recognized by transcription-coupled nucleotide excision repair (NER) as well as by proteins involved in the common NER pathway (1–3). Mammalian cells deficient in NER proteins such as the XPD and XPB helicases, and the XPG and ERCC1/XPF endonucleases showed 2- to 8-fold increased resistance to ET-743 in comparison with repair-proficient WT cells (1–3). Increased resistance to ET-743 was also reported for yeast mutants lacking the APN1 endonuclease, which particularly is involved in base excision repair (20). The unusual effect of these repair proteins on the cytotoxicity of ET-743 has been explained by the stabilization of repair complexes by the ET adducts (1–4, 21). Mismatch repair status had no detectable influence on the sensitivity to ET-743 (2, 6) whereas loss of the DNA-dependent kinase (DNA-PK) was reported to sensitize cells to ET-743 (2). Conflicting data have been reported for proteins involved in homologous recombination (HR) repair, because loss of repair function was associated with increased resistance to ET-743 in Saccharomyces cerevisiae (20) but increased sensitivity in Schizosaccharomyces pombe (21).
Although it is widely believed that the cytotoxic activity of ET-743 is based on its interaction with the NER machinery (reviewed in ref. 17), the high residual activity of the drug toward NER-deficient cells as well as the rather modest resistance levels (2- to 8-fold) suggest that this cannot be the only mechanism of action of ET-743. We speculated that the ET adducts would be unusually stable because of the absence of DNA repair and thus able to interfere with the replication machinery whenever the treated cells entered S phase, possibly triggering the formation of secondary DNA lesions with increased toxicity.
We here report that exposure to pharmacologically relevant concentrations of ET-743 is associated with the induction of replication-dependent double-strand breaks (DSBs). Cells deficient for HR repair showed pronounced sensitivity to the drug due to the continued presence of unrepaired DSBs that gave rise to chromosomal abnormalities. Our findings characterize the processing of the monofunctional ET adducts into cytotoxic lesions and identify replication and HR repair as key modulators of the cellular activity of this unique compound.
Results
ET-743 Exposure Is Associated with Potent Inhibition of DNA Synthesis.
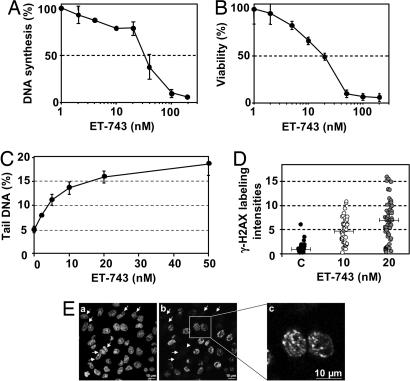
To establish whether unrepaired ET adducts affect DNA synthesis, the influence of ET-743 on the incorporation of radiolabeled thymidine was determined. The results indicate that DNA synthesis is reduced by 50% after 1-h exposure to 30 nM ET-743 (Fig. 1A). Next, cells were exposed to ET-743 for 1 h followed by postincubation in drug-free media for four cell doubling times, and the cellular viability was determined by the MTT assay (Fig. 1B). The results show that the cellular viability is reduced by 50% after exposure to 20 nM ET-743. Therefore, inhibition of DNA synthesis and loss of cellular viability take place within the same dose range.
Fig. 1.
Influence of ET-743 on DNA synthesis, cellular survival, and DSB formation. (A) HeLa cells were incubated with the indicated concentrations of ET-743 for 1 h, and the influence on DNA synthesis was measured by incorporation of radiolabeled thymidine. (B) HeLa cells were exposed to ET-743 for 1 h followed by postincubation in drug-free media for four doubling times and the MTT viability assay. (C) HeLa cells were incubated with the indicated concentrations of ET-743 for 1 h, and the formation of DSBs was determined by the neutral comet assay. The points indicate the average amounts of DNA in the comet tail. Error bars represent standard errors and are indicated when they exceed the symbol size. (D) Cells were incubated with the indicated concentrations of ET-743 for 1 h, and the formation of γ-H2AX foci was revealed by immunostaining. Total fluorescent intensities were determined by MetaMorph image analysis. Each point represents the γ-H2AX staining in an individual cell. Bars represent the average fluorescence intensities. (E) Confocal fluorescence microscopy of nuclear DNA (a) and γ-H2AX (b and c). Cells were incubated with 10 nM ET-743 for 1 h, fixed, and labeled with a γ-H2AX-directed antibody. DNA was counterstained with Topro-3. Arrows indicate cells with no detectable γ-H2AX signal.
ET-743 Exposure Is Associated with a Dose-Dependent Formation of DNA DSBs.
Single-cell electrophoresis under neutral conditions (the neutral comet assay), which almost uniquely detects DSBs (22, 23), was used to determine whether ET-743 exposure is accompanied by formation of DNA strand breaks. The results (Fig. 1C) show that 1-h exposure of HeLa cells to ET-743 was associated with a dose-dependent increase in DSBs, which could be detected at doses as low as 2 nM. The rapid formation of DSBs indicates that they are a direct result of drug action and not due to apoptotic DNA fragmentation. In agreement, exposure to 10 nM ET-743 for up to 24 h was not accompanied by any detectable increase in the fraction of apoptotic cells as determined by annexin V labeling (data not shown).
ET-743 Exposure Is Accompanied by γ-H2AX Formation.
The formation of phosphorylated histone H2AX (γ-H2AX) is considered as a sensitive surrogate marker for the formation of DSBs. Cells were exposed to ET-743 for 1 h followed by immunostaining with a γ-H2AX-directed antibody. The results show that ET-743 exposure is accompanied by a dose-dependent increase in γ-H2AX formation [Fig. 1D and supporting information (SI) Fig. 5]. Interestingly, focalized γ-H2AX staining was observed only in a subset of the cell population, indicating a possible role of the cell cycle (Fig. 1E, compare a and b; arrows indicate cells without γ-H2AX staining). ET-associated induction of γ-H2AX was observed in all cell lines examined, including HeLa cells, Chinese hamster cells, and CEM human leukemia cells (data not shown).
Influence of DNA and RNA Synthesis.
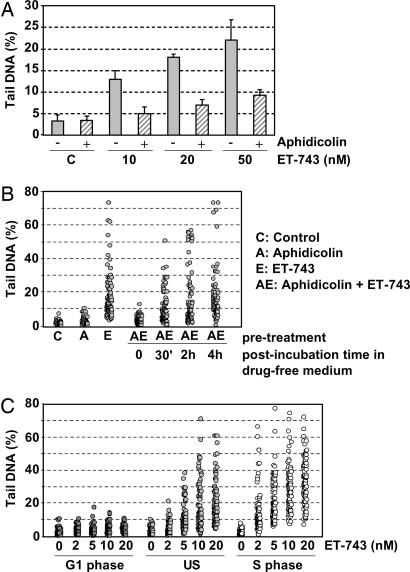
To determine whether the ET-induced DSBs were linked to macromolecular synthesis, HeLa cells were first incubated with different doses of ET-743 in the absence or presence of aphidicolin, a DNA synthesis inhibitor. The results show that aphidicolin substantially reduced the induction of DSBs even at doses as high as 50 nM ET-743 (Fig. 2A). In contrast, 5,6-dichloro-1-β-d-ribofuranosylbenzimi-dazole, a selective inhibitor of RNA polymerase II, had no detectable influence on the formation of DSBs (data not shown).
Fig. 2.
The formation of secondary DNA damage after ET-743 exposure. (A) HeLa cells were incubated with the indicated concentrations of ET-743 in the absence (filled bars) or presence (hatched bars) of aphidicolin (5 μM). The values indicate the average amounts of DNA in the comet tail from at least two experiments. Error bars represent standard errors. (B) HeLa cells were incubated with 10 nM ET-743 for 1 h in the absence or presence of aphidicolin followed by postincubation in drug-free media for the indicated times. Each point represents the amount of tail DNA in an individual cell from a typical experiment. (C) G1-synchronized, unsynchronized (US), or S-synchronized HeLa cells were exposed to the indicated concentrations of ET-743 followed by comet analysis. Each point represents the amount of tail DNA in an individual cell from a typical experiment.
To further characterize the role of DNA synthesis, HeLa cells were incubated with ET-743 in the presence of aphidicolin for 1 h followed by postincubation in drug-free media for 4 h (Fig. 2B). The results show that reinitiation of DNA synthesis during the postincubation period, as determined by incorporation of radiolabeled thymidine (data not shown), was accompanied by a gradual formation of DSBs. By 4 h after incubation, the DNA synthesis had recovered totally and the levels of DSBs had reached approximately the same levels as observed after 1 h in the presence of ET-743 alone. These results clearly indicate that the formation of ET-induced DSBs strongly, if not entirely, depends on active DNA synthesis. The results also suggest that even brief exposure to ET-743 results in the formation of long-lasting ET adducts, which are difficult to repair and can be converted into DSBs later.
ET-743-Mediated DNA Damage in Synchronized Cells.
To further confirm the role of DNA synthesis in the formation of DSBs, nonproliferating lymphocytes (that are in the G0 phase of the cell cycle) were isolated from the peripheral venous blood of healthy donors and exposed to ET-743. The results showed no formation of DNA strand breaks, even at doses as high as 100 nM ET-743 as determined by the highly sensitive alkaline comet assay, in marked contrast to what was observed for cycling CEM lymphoblastic leukemia cells (SI Fig. 6).
Next, the influence of ET-743 toward unsynchronized, G1-synchronized, or S-synchronized HeLa cells was compared. The average levels of DNA damage in ET-exposed G1 cells were comparable to those of untreated control cells (Fig. 2C). In clear contrast, ET exposure of cells synchronized in the S phase of the cell cycle was accompanied by significantly higher levels of DNA damage compared with unsynchronized cells (P ≤ 0.05 as determined by Student's t test).
Cytotoxicity of ET-743 Toward Cells Deficient in Recombination Repair.
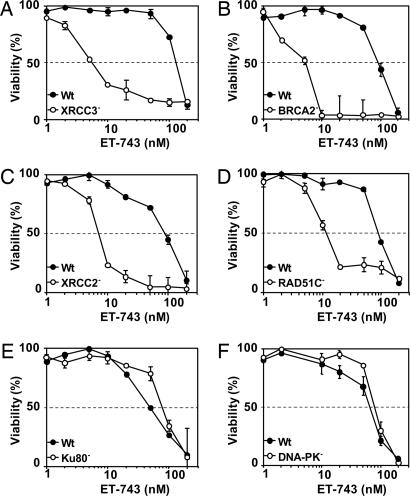
DSBs can be repaired by two major pathways, HR, which is active during S and G2, and nonhomologous end joining, which is preferentially active during the G0/G1 phase of the cell cycle. To determine the importance of the two repair pathways, the cytotoxicity of ET-743 toward cell lines deficient for HR (RAD51C, XRCC2, XRCC3, and BRCA2) or nonhomologous end-joining (Ku80 and DNA-PK) and their respective repair-proficient parental cell lines was established. The results show that cells deficient for XRCC3, BRCA2, RAD51C, and XRCC2 were 23-, 18-, 8-, and 8-fold more sensitive to ET-743, respectively, compared with the corresponding repair-proficient parental cell lines (Fig. 3 A–D). In contrast, deficiencies in Ku80 and DNA-PK had minor influence on the sensitivity to ET-743 (Fig. 3 E and F).
Fig. 3.
Cytotoxic effects of ET-743 toward recombination-deficient cells. Recombination-proficient and -deficient cells were exposed to ET-743 for 1 h followed by postincubation in drug-free media for four generation times, and the viability was determined by the MTT assay. (A) Filled circles, AA8 parental cells; open circles, irs1SF (XRCC3−). (B) Filled circles, V79 parental cells; open circles, V-C8 (BRCA2/XRCC11/FANCD1−). (C) Filled circles, V79B parental cells; open circles, CL-V4B (RAD51C−). (D) Filled circles, V79 parental cells; open circles, irs1 (XRCC2−). (E) Filled circles, V79B parental cells; open circles, XR-V15B (Ku80/XRCC5−). (F) Filled circles, M059J Fus1 (DNA-PK-proficient); open circles, M059J Fus9 (DNA-PK-deficient). Each point represents the average of at least two independent experiments, each done in duplicate. Error bars represent standard errors and are indicated when they exceed the symbol size.
Cell Cycle Progression in XRCC3-Proficient and -Deficient Cells.
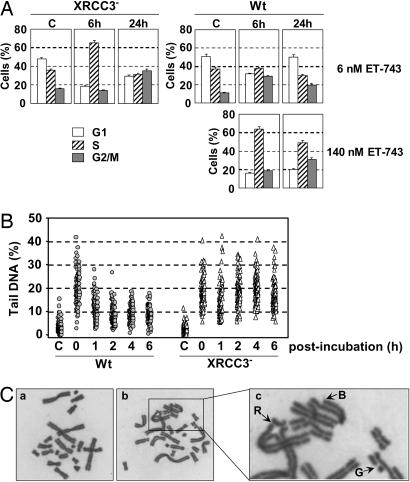
To characterize the influence of ET-743 on HR-deficient cells, WT AA8 and XRCC3-deficient irs1SF cells were treated with different doses of ET-743 for 1 h followed by postincubation in drug-free media for the indicated times (Fig. 4A). The results show that exposure of irs1SF cells to 6 nM ET-743 (corresponding to the IC50 dose determined in Fig. 3A) was accompanied by a dramatic enrichment of cells in the S phase of the cell cycle, which increased from 36% in untreated controls to 65% 6 h after incubation, suggesting rapid activation of the inter-S phase checkpoint. By 24 h after incubation, the most notable feature was a strong accumulation of cells in G2/M representing 35% of the cells compared with 13% for the untreated control cells. In contrast, exposure of parental AA8 cells to the same dose of ET-743 was not associated with any detectable effect on S-phase progression during the first 6 h (Fig. 4A and data not shown) whereas the fraction of cells in the G2/M phase of the cell cycle was enriched by 6 h and, to a lesser degree, by 24 h. Interestingly, exposure of AA8 cells to 140 nM ET-743 (corresponding to the IC50 dose) was associated with notable changes in both S phase and G2 phase progression, which closely resembled the changes observed at isotoxic doses for irs1SF. These results show that, although ET-743 induced comparable cell cycle effects in HR-proficient and -deficient cells at isotoxic concentrations, induction of a marked S-phase arrest at low drug concentrations was restricted to the XRCC-3-deficient cells.
Fig. 4.
Influence of ET-743 on HR-proficient and -deficient cells. (A) Cell cycle distribution in AA8 (WT) and irs1SF (XRCC3−) cells exposed to the indicated concentrations of ET-743 for 1 h followed by postincubation in drug-free media for the indicated times. The IC50 value for irs1SF was 6 nM, and the isotoxic dose was 140 nM for AA8. (B) Repair of ET-induced DSBs in WT and XRCC3-deficient cells. Parental AA8 and XRCC3-deficient irs1SF cells were exposed to 100 nM ET-743 for 1 h followed by postincubation in drug-free media for the indicated times and determination of DNA strand breaks by the neutral comet assay. Each point represents the amount of tail DNA in an individual cell from a typical experiment. (C) Karyotypes of V79 cells. (a) Typical metaphase in untreated V79 cells. (b and c) V79 cells treated with 100 nM ET-743 for 1 h followed by 24 h of postincubation in drug-free media. The arrows indicate chromatid gaps (G), breaks (B), or radials (R).
Formation and Removal of DSBs in XRCC3-Proficient and -Deficient Cells.
Next, WT and XRCC3-deficient cells were exposed to 100 nM ET-743 for 1 h followed by postincubation in drug-free media and comet analysis. The results (Fig. 4B) show that 1 h of exposure to ET-743 was accompanied by the induction of comparable levels of DNA strand breaks in parental and XRCC3-deficient cells, with the average levels of DNA in the comet tail being 20% for the WT cells compared with 19% for the XRCC3-deficient cells. However, striking differences between the two cell lines were revealed during the postincubation period. The DSBs were effectively repaired in the parental cells, where the average levels of DNA in the comet were reduced from 20% to 11% after 1 h and to 8% 6 h after incubation. In comparison, no repair was observed for the XRCC3-deficient cells, where the average levels of DNA in the comet remained ≈17% even 6 h after incubation. Therefore, the pronounced sensitivity of XRCC3-deficient cells to ET-743 (Fig. 3A) was associated with the persistence of unrepaired DSBs.
Induction of Chromosome Damage in BRCA2-Deficient Cells.
Unrepaired DSBs may lead to chromosomal abnormalities. To determine the influence of HR repair on the karyotype of ET-treated cells, BRCA2-proficient and -deficient cells were exposed to different doses of ET-743 for 1 h followed by postincubation in drug-free media for 24 h and karyotype analysis (Table 1). BRCA2-deficient cells were preferred for these studies because they, in contrast to their XRCC3-deficient counterparts, are readily amendable to karyotype analysis. ET exposure of BRCA2-deficient cells was accompanied by a dose-dependent increase in the number of cells showing chromosome aberrations with the most common aberrations being chromatid breaks, gaps, and fragments. At 10 nM ET-743, all metaphases examined showed at least three chromosome aberrations (Table 1).
Table 1.
Analysis of chromosome aberrations in repair-proficient V79 parental cells and BRCA2-deficient V-C8 cells after 1 h of exposure to ET-743 followed by 24 h of postincubation in drug-free media
| ET-743, nM | WT |
BRCA2− |
||||
|---|---|---|---|---|---|---|
| Undamaged chromosomes | Damaged chromosomes | End-to-end association or radials | Undamaged chromosomes | Damaged chromosomes | End-to-end association or radials | |
| 0 | 100 | 0 | 0 | 97 | 3 | 0 |
| 1 | 86 | 14 | 1 | |||
| 2 | 58 | 42 | 1 | |||
| 5 | 10 | 90 | 2 | |||
| 10 | 99 | 1 | 0 | 0 | 100 | 3 |
| 50 | 50 | 50 | 27 | |||
| 100 | 43 | 57 | 24 | |||
One hundred metaphases per treatment condition were evaluated.
In contrast to the BRCA2-deficient cells, exposure of repair-proficient parental cells to 10 nM ET-743 was associated with chromosome abnormalities in only 1% of the cells (Table 1), in agreement with the ability of HR-proficient cells to repair the ET-induced DSBs (Fig. 4B). At higher doses, ≈50% of the observed metaphases showed chromosome aberrations (Fig. 4C) of the same type as observed for the BRCA2-deficient cells. However, even at elevated doses, only some metaphases showed chromosome aberrations, in striking contrast to the highly damaged chromosomes observed for BRCA2-deficient cells at equitoxic doses (Table 1).
Discussion
In this article we establish a mechanistic link between the initial ET adducts and the subsequent downstream events leading to cell death and identify replication and HR as important modulators of ET action.
ET-743 forms monofunctional adducts with the exocyclic amino group of guanine in the minor groove of DNA (18). The results presented here indicate that the ET adducts are poorly repaired and are able to induce secondary DNA damage even a long time after the initial drug exposure. Unrepaired adducts led to formation of DSBs as determined by single-cell electrophoresis under neutral conditions and by γ-H2AX induction. Interestingly, γ-H2AX staining was completely absent in some cells, suggesting a role for the cell cycle in the formation of the ET-induced DSBs. In agreement, no DNA strand breaks were observed in resting lymphocytes or in cells synchronized in G1, whereas cells synchronized in S showed increased levels of DSBs. These findings suggest a strict requirement for DNA synthesis in the formation of the ET-induced DSBs.
The formation of secondary DNA lesions in term of DSBs is not restricted to ET-743 but has previously been described for a number of other DNA-interacting agents including the DNA cross-linking agent mitomycin C (24) and the monofunctional DNA alkylator S23906 (11) and in NER-deficient cells after UV radiation (25) and is believed to occur when the advancing DNA replication fork runs into the DNA adducts. ET-induced DSBs were accompanied by the formation of focalized γ-H2AX, which is believed to promote the assembly of HR repair complexes around the DSB (26, 27). Mammalian cells deficient in HR showed pronounced sensitivity to ET-743 whereas nonhomologous end joining repair had minor influence on the sensitivity to ET-743. Although the relation between DNA synthesis and HR is well established (for recent review, see ref. 28), their respective links to DSB formation is more ambiguous, and several possibilities have been suggested. First, we might have a sequential model where the DSBs are created by the replication fork (or its collapse) thereby triggering HR. In this case we would expect little, if any, differences in the initial levels of DSBs between HR-proficient or -deficient cells. Second, HR might be needed to prevent replication fork collapse and to promote replication restart. In this case we would expect more initial levels of DSBs in HR-deficient cells. Third, the DSBs may be reaction intermediates formed during the recombination process that had become activated by the stalled replication forks. In this case we would expect more initial levels of DSBs in HR-proficient cells. Our experimental results provide clear evidence in favor of the first model. Strikingly, although HR status had little, if any, influence on the initial levels of DSBs after short exposure to ET-743, marked differences were revealed during the postincubation period, where no detectable repair was observed in the HR-deficient cells. The prolonged presence of unrepaired DSBs was followed by the appearance of mitotic chromosomes with chromatid aberrations in terms of gaps and breaks, in agreement with the S-phase-dependent induction of DSBs.
Previous studies showed that the ET adducts were recognized by transcription-coupled NER, a process that is triggered by arrest of the elongating RNA polymerase II by the DNA adducts. Surprisingly, 5,6-dichloro-1-β-d-ribofuranosylbenzimi-dazole, a selective RNA polymerase II inhibitor, showed no detectable influence on the formation of ET-induced DSBs, suggesting that RNA synthesis does not contribute to DSB formation. Intriguingly, similar findings have previously been reported for mitomycin C, another DNA minor groove binder (29) associated with the formation of replication-dependent DSBs (24).
HR appears to be at least as important as NER in terms of modulating the response to ET, positively or negatively. Findings with mammalian repair mutants indicate that loss of NER function was associated with 2- to 8-fold increased resistance (1–3), whereas loss of proteins needed for HR repair was accompanied by 8- to 23-fold increased sensitivity (this study). The substantial residual activity of ET-743 toward NER-deficient cells suggests that the effects of NER and HR repair are, at least in part, independent events. In agreement, we observed a dose-dependent formation of DSBs in XPG-CS-deficient cells, which are deficient in both transcription-coupled NER and the common NER pathway (SI Fig. 7). In comparison, the levels of ET-induced DSBs were only slightly higher in repair-proficient WT and XPC cells, indicating that the putative DNA/ET/NER complexes are unlikely to be a major contributor to the formation of DSBs.
The conversion of the ET adducts into DSBs may, at least in part, be explained by the influence of the adducts on the properties of the local duplex DNA. Biochemical evidence as well as molecular modeling studies suggest that the ET adducts strongly stabilize the helical structure of duplex DNA, which is likely to slow down DNA separation during macromolecular synthesis (30, 31). In agreement, ET adducts were shown to strongly hamper DNA unwinding by the RecBCD helicase (9). These findings suggest that ET-743 may have functional similarities to cross-linking agents like mitomycin C, an established inducer of replication-dependent DSBs (24).
The important influence of different DNA repair pathways on the cytotoxic activity of ET-743 indicates that DNA repair functionality may be useful to assist in selection of the patients most likely to benefit from treatment with ET-743. This notion is supported by a preliminary clinical study with 92 sarcoma patients.‖ This study reports that ET-treated patients could be divided into three subgroups: one group responding with long-lasting progression-free survival, one group with an intermediate response, and one group of patients who did not benefit from the treatment. Importantly, long-term progression-free survival was associated with high expression of mRNA coding for the NER protein ERCC1 and low expression levels of the recombination proteins BRCA1 and BRCA2. In marked contrast, high expression of BRCA1/BRCA2 and low expression of ERCC1 were associated with lack of clinical response to ET-743 in every one of the 14 patients in this group.
Taken together, we have identified the formation of replication-dependent DSBs as a crucial component of the cytotoxic activity of ET-743 and provided a model explaining the involvement of replication and HR repair in the processing of ET-induced DNA lesions. Our data provide a rational to use expression levels of selected repair factors for clinical response prediction and underline the interplay between macromolecular synthesis and repair pathways in DNA lesion processing.
Experimental Procedures
Chemicals and Cells.
ET-743 was generously provided by the National Cancer Institute (Frederick, MD), and thymidine, aphidicolin, and nocodazole were purchased from Sigma (Saint-Quentin Fallavier, France). 5,6-Dichloro-1-β-d-ribofuranosylbenzimi-dazole was obtained from Calbiochem (La Jolla, CA).
HeLa cervical carcinoma cells and CEM lymphoblastic leukemia cells have been described previously (6). Recombination-deficient cells lines and their corresponding repair-proficient parental cells were generously provided by Malgorzata Zdzienicka (Leiden University Medical Center, Leiden, The Netherlands). The cells include irs1, defective in XRCC2 (32); irs1SF, defective in XRCC3 (33, 34); CL-V4B, defective in RAD51C (35); V-C8, defective in BRCA2/XRCC11/FANCD1 (36); and XR-V 15B, defective in Ku80/XRCC5 (37, 38).
DNA-PK-deficient Fus9 human M059J glioblastoma cells and DNA-PK-proficient Fus1 cells (39) were kindly provided by Bernard Salles (Centre National de la Recherche Scientifique/University Paul Sabatier, Toulouse, France). All cell lines were tested regularly for Mycoplasma contamination by PCR analysis.
Cell Synchronization.
To synchronize HeLa cells in G1, cells were treated with nocodazole (75 ng/ml) for 18 h followed by mitotic shake-off (70–90% mitotic cells) and postincubation in drug-free media for 3 h. A double thymidine block was used to synchronize cells at the G1/S border. Cells were incubated with 2 mM thymidine for 16 h followed by 8 h of recovery and a second 16-h thymidine exposure followed by 3 h postincubation in drug-free media to permit the cells to enter S (>80% S-phase cells).
For “biological” synchronization, nonproliferating lymphocytes (that are in the G0 phase of the cell cycle) were isolated from the peripheral venous blood of healthy donors by Ficoll centrifugation according to standard procedures (40).
Cell Cycle Analysis and Inhibition of DNA Synthesis.
Cell cycle analysis was determined by flow cytometry analysis, and inhibition of DNA synthesis was determined by incorporation of radioactive thymidine as described previously (7, 41). Values are given as the mean of at least two independent experiments, each done in duplicate.
Viability Assays.
The cytotoxic activity of ET-743 was measured by using the MTT colorimetric assay as previously described (6). All values are averages of at least three independent experiments, each done in duplicate.
For annexin V labeling, ET-treated cells were rinsed and stained with annexin V–FITC (Molecular Probes Invitrogen, Cergy-Pontoise, France) for 15 min at room temperature in the dark followed immediately by flow cytometry on an Epics XL/MCL flow cytometer (Beckman Coulter, Roissy, France).
Single-Cell Electrophoresis.
Cells for comet analysis were exposed to the indicated drug concentrations at 37°C in the dark and analyzed immediately according to previously published procedures (21, 23, 42). Image analysis was performed by using Komet 5.5 software (Kinetic Imaging, Nottingham, U.K.). At least 100 cells were analyzed per sample. Values represent the average of at least two independent experiments.
Immunofluorescence and Microscopy.
HeLa, AA8, or Irs1SF cells were plated onto coverslips and grown overnight before treatment with ET-743. CEM cells were treated with ET-743 and fixed onto coverslips by cytospin (4 min at 265 × g, Cytofuge 2; Statspin, Norwood, MA). After treatment, cells were fixed in freshly prepared 4% paraformaldehyde for 20 min and permeabilized with PBS/Triton X-100 (0.5%) for 20 min. Coverslips were saturated in presence of PBS+ (PBS containing 1% BSA and 0.2% gelatin; Sigma), and antigens were revealed by immunolabeling using primary mouse antibodies directed against γ-H2AX (1:100 dilution; catalog no. 05-636; Upstate Biotechnology). The secondary antibody used in this study was Cy3-conjugated donkey anti-mouse IgG antibody (1:100 dilution; catalog no. 715-165-151; Jackson ImmunoResearch, Bar Harbor, ME). All antibodies were diluted in PBS+. Topro-3 was added to label DNA (1:5,000 dilution, T3605; Molecular Probes Invitrogen), and images were collected by using either a TCS SP confocal microscope (Leica) or a Retiga 1300 camera (QImaging, Burnaby, Canada) attached to a Nikon TS 100 microscope. Fluorescence intensities were measured by using MetaMorph software (Universal Imaging, Downingtown, PA). Background over noncellular regions was subtracted.
Karyotype Analysis.
V79 parental cells and V-C8 mutant cells (BRCA2/XRCC11/FANCD1−) were exposed for 1 h to the indicated doses of ET-743. Cells were washed with PBS and postincubated in drug-free medium for 24 h, and chromosome spreads were prepared as described (22). One hundred metaphases per treatment condition were evaluated.
Supplementary Material
Acknowledgments
We warmly thank Drs. Malgorzata Zdzienicka and Bernard Salles for generously providing us with the recombination-deficient cells and Dr. Christine Perot for help with the karyotype analysis. We also thank Dr. Jose Jimeno for helpful discussions. This work was supported by Fondation Pour la Recherche Médicale, Association Pour la Recherche sur le Cancer (Villejuif, France) Grant 3795, and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior/Comité Français Pour l'évaluation de la Coopération Universitaire avec le Brésil Grant 583/07. D.G.S. was supported by Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Brasil.
Abbreviations
- DSB
double-strand break
- HR
homologous recombination
- NER
nucleotide excision repair
- DNA-PK
DNA-dependent kinase.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609877104/DC1.
Schöffski, P., Casali, P. G., Taron, M., Van Oosterom, A. T., Judson, I. R., Grosso, F., Blay, J.-Y., Maki, R. G., Tercero, J. C., Jimeno, J. M., Rosell, R. (2006) 42nd Annual Meeting of the American Society of Clinical Oncology, June 2–6, 2006, Atlanta, GA, abstr. 9522.
References
- 1.Takebayashi Y, Pourquier P, Zimonjic DB, Nakayama K, Emmert S, Ueda T, Urasaki Y, Kanzaki A, Akiyama SI, Popescu N, et al. Nat Med. 2001;7:961–966. doi: 10.1038/91008. [DOI] [PubMed] [Google Scholar]
- 2.Damia G, Silvestri S, Carrassa L, Filiberti L, Faircloth GT, Liberi , Foiani M, D'Incalci M. Int J Cancer. 2001;92:583–588. doi: 10.1002/ijc.1221. [DOI] [PubMed] [Google Scholar]
- 3.Erba E, Bergamaschi D, Bassano L, Damia G, Ronzoni S, Faircloth GT, D'Incalci M. Eur J Cancer. 2001;37:97–105. doi: 10.1016/s0959-8049(00)00357-9. [DOI] [PubMed] [Google Scholar]
- 4.Zewail-Foote M, Li VS, Kohn H, Bearss D, Guzman M, Hurley LH. Chem Biol. 2001;8:1033–1049. doi: 10.1016/s1074-5521(01)00071-0. [DOI] [PubMed] [Google Scholar]
- 5.Jaspers NG, Raams A, Kelner MJ, Ng JM, Yamashita YM, Takeda S, McMorris TC, Hoeijmakers JH. DNA Repair. 2002;1:1027–1038. doi: 10.1016/s1568-7864(02)00166-0. [DOI] [PubMed] [Google Scholar]
- 6.Poindessous V, Koeppel F, Raymond E, Comisso M, Waters SJ, Larsen AK. Clin Cancer Res. 2003;9:2817–2825. [PubMed] [Google Scholar]
- 7.Koeppel F, Poindessous V, Lazar V, Raymond E, Sarasin A, Larsen AK. Clin Cancer Res. 2004;10:5604–5613. doi: 10.1158/1078-0432.CCR-04-0442. [DOI] [PubMed] [Google Scholar]
- 8.Wu J, Ling X, Pan D, Apontes P, Song L, Liang P, Altieri DC, Beerman T, Li F. J Biol Chem. 2005;280:9745–9751. doi: 10.1074/jbc.M409350200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dziegielewska B, Beerman TA, Bianco PR. J Mol Biol. 2006;361:898–919. doi: 10.1016/j.jmb.2006.06.068. [DOI] [PubMed] [Google Scholar]
- 10.Léonce S, Perez V, Lambel S, Peyroulan D, Tillequin F, Michel S, Koch M, Pfeiffer B, Atassi G, Hickman JA, Pierré A. Mol Pharmacol. 2001;60:1383–1391. doi: 10.1124/mol.60.6.1383. [DOI] [PubMed] [Google Scholar]
- 11.Léonce S, Kraus-Berthier L, Golsteyn RM, David-Cordonnier MH, Tardy C, Lansiaux A, Poindessous V, Larsen AK, Pierré A. Cancer Res. 2006;66:7203–7210. doi: 10.1158/0008-5472.CAN-05-3946. [DOI] [PubMed] [Google Scholar]
- 12.Sakai R, Rinehart KL, Guan Y, Wang AH. Proc Natl Acad Sci USA. 1992;89:11456–11460. doi: 10.1073/pnas.89.23.11456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez EJ, Corey EJ, Owa T. Chem Biol. 2001;8:1151–1160. doi: 10.1016/s1074-5521(01)00082-5. [DOI] [PubMed] [Google Scholar]
- 14.Yovine A, Riofrio M, Blay J-Y, Brain E, Alexandre J, Kahatt C, Taamma A, Jimeno J, Martin C, Salhi Y, et al. J Clin Oncol. 2004;22:890–899. doi: 10.1200/JCO.2004.05.210. [DOI] [PubMed] [Google Scholar]
- 15.Le Cesne A, Blay J-Y, Judson I, Van Oosterom A, Verweij J, Radford J, Lorigan P, Rodenhuis S, Ray-Coquard I, Bonvalot S, et al. J Clin Oncol. 2005;23:576–584. doi: 10.1200/JCO.2005.01.180. [DOI] [PubMed] [Google Scholar]
- 16.Sessa C, De Braud F, Perotti A, Bauer J, Curigliano G, Noberasco C, Zanaboni F, Gianni L, Marsoni S, Jimeno J, et al. J Clin Oncol. 2005;23:1867–1874. doi: 10.1200/JCO.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 17.Fayette J, Coquard IR, Alberti L, Ranchère D, Boyle H, Blay J-Y. Oncologist. 2005;10:827–832. doi: 10.1634/theoncologist.10-10-827. [DOI] [PubMed] [Google Scholar]
- 18.Pommier Y, Kohlhagen G, Bailly C, Waring M, Mazumder A, Kohn KW. Biochemistry. 1996;35:13303–13309. doi: 10.1021/bi960306b. [DOI] [PubMed] [Google Scholar]
- 19.Zewail-Foote M, Hurley LH. J Med Chem. 1999;42:2493–2497. doi: 10.1021/jm990241l. [DOI] [PubMed] [Google Scholar]
- 20.Soares DG, Poletto NP, Bonatto D, Salvador M, Schwartsmann G, Henriques JA. Biochem Pharmacol. 2005;70:59–69. doi: 10.1016/j.bcp.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 21.Herrero AB, Martin-Castellanos C, Marco E, Gago F, Moreno S. Cancer Res. 2006;66:8155–8162. doi: 10.1158/0008-5472.CAN-06-0179. [DOI] [PubMed] [Google Scholar]
- 22.Wojewodzka M, Buraczewska I, Kruszewski M. Mutat Res. 2002;518:9–20. doi: 10.1016/s1383-5718(02)00070-0. [DOI] [PubMed] [Google Scholar]
- 23.Olive PL, Banáth JP. Nat Protocols. 2006;1:23–29. doi: 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- 24.Niedernhofer LJ, Odijk H, Budzowska M, van Drunen E, Maas A, Theil AF, de Wit J, Jaspers NG, Beverloo HB, Hoeijmakers JH, Kanaar R. Mol Cell Biol. 2004;24:5776–5787. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunkern TR, Kaina B. Mol Biol Cell. 2002;13:348–361. doi: 10.1091/mbc.01-05-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie A, Puget N, Shim I, Odate S, Jarzyna I, Bassing CH, Alt FW, Scully R. Mol Cell. 2004;16:1017–1025. doi: 10.1016/j.molcel.2004.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thiriet C, Hayes JJ. Mol Cell. 2005;18:617–622. doi: 10.1016/j.molcel.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 28.Eppink B, Wyman C, Kanaar R. Exp Cell Res. 2006;312:2660–2665. doi: 10.1016/j.yexcr.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 29.Sastry M, Fiala R, Lipman R, Tomasz M, Patel DJ. J Mol Biol. 1995;247:338–359. doi: 10.1006/jmbi.1994.0143. [DOI] [PubMed] [Google Scholar]
- 30.David-Cordonnier MH, Laine W, Lansiaux A, Rosu F, Colson P, de Pauw E, Michel S, Tillequin F, Koch M, Hickman JA, et al. Mol Cancer Ther. 2005;4:71–80. [PubMed] [Google Scholar]
- 31.Marco E, Gago F. Mol Pharmacol. 2005;68:1559–1567. doi: 10.1124/mol.105.015685. [DOI] [PubMed] [Google Scholar]
- 32.Keohane A, Godden J, Stratford IJ, Adams GE. Br J Cancer. 1990;61:722–726. doi: 10.1038/bjc.1990.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fuller LF, Painter RB. Mutat Res. 1988;193:109–121. doi: 10.1016/0167-8817(88)90041-7. [DOI] [PubMed] [Google Scholar]
- 34.Tebbs RS, Zhao Y, Tucker JD, Scheerer JB, Siciliano MJ, Hwang M, Liu N, Legerski RJ, Thompson LH. Proc Natl Acad Sci USA. 1995;92:6354–6358. doi: 10.1073/pnas.92.14.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Godthelp BC, Wiegant WW, van Duijn-Goedhart A, Scharer OD, van Buul PP, Kanaar R, Zdzienicka MZ. Nucleic Acids Res. 2002;30:2172–2182. doi: 10.1093/nar/30.10.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kraakman-van der Zwet M, Overkamp WJ, van Lange RE, Essers J, van Duijn-Goedhart A, Wiggers I, Swaminathan S, van Buul PP, Errami A, Tan RT, et al. Mol Cell Biol. 2002;22:669–679. doi: 10.1128/MCB.22.2.669-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zdzienicka MZ, Tran Q, van der Schans GP, Simons JW. Mutat Res. 1988;194:239–249. doi: 10.1016/0167-8817(88)90025-9. [DOI] [PubMed] [Google Scholar]
- 38.Errami A, Smider V, Rathmell WK, He DM, Hendrickson EA, Zdzienicka MZ, Chu G. Mol Cell Biol. 1996;16:1519–1526. doi: 10.1128/mcb.16.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoppe BS, Jensen RB, Kirchgessner CU. Radiat Res. 2000;153:125–130. doi: 10.1667/0033-7587(2000)153[0125:cotrmc]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 40.Boyum A. Lymphology. 1977;10:71–76. [PubMed] [Google Scholar]
- 41.Skladanowski A, Côme M-G, Sabisz M, Escargueil AE, Larsen AK. Mol Pharmacol. 2005;68:625–634. doi: 10.1124/mol.105.013995. [DOI] [PubMed] [Google Scholar]
- 42.De Meo M, Laget M, Castegnaro M, Dumenil G. Mutat Res. 1991;260:295–306. doi: 10.1016/0165-1218(91)90038-n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.