Abstract
Functional neuroimaging and electrophysiological studies have documented a dynamic baseline of intrinsic (not stimulus- or task-evoked) brain activity during resting wakefulness. This baseline is characterized by slow (<0.1 Hz) fluctuations of functional imaging signals that are topographically organized in discrete brain networks, and by much faster (1–80 Hz) electrical oscillations. To investigate the relationship between hemodynamic and electrical oscillations, we have adopted a completely data-driven approach that combines information from simultaneous electroencephalography (EEG) and functional magnetic resonance imaging (fMRI). Using independent component analysis on the fMRI data, we identified six widely distributed resting state networks. The blood oxygenation level-dependent signal fluctuations associated with each network were correlated with the EEG power variations of delta, theta, alpha, beta, and gamma rhythms. Each functional network was characterized by a specific electrophysiological signature that involved the combination of different brain rhythms. Moreover, the joint EEG/fMRI analysis afforded a finer physiological fractionation of brain networks in the resting human brain. This result supports for the first time in humans the coalescence of several brain rhythms within large-scale brain networks as suggested by biophysical studies.
Keywords: brain rhythms, multimodal imaging, resting fluctuations
A fundamental issue in neuroscience is understanding how large neuronal assemblies cooperate in the brain, and what mechanisms underlie this cooperation, that is the basis for all sensory, cognitive, and motor activities. Traditional physiological models of brain function emphasize the importance of spike rate as a medium for encoding and transferring signals in the brain, and often delineate electrophysiological spontaneous activity as internal noise (1, 2). More recent models propose that spontaneous activity may also play an important functional role by providing important endogenous or top-down constraints to sensory-, cognitive-, or motor-driven activity or temporal windows of opportunity for long-range communication (3–5).
Functional neuroimaging studies have provided evidence for a baseline of neuronal ongoing activity, from which transient changes induced by specific perceptual and cognitive tasks, generally named activations, arise (6, 7). Interestingly, spontaneous activity, as measured with blood oxygen level-dependent (BOLD) functional MRI (fMRI) in the resting awake or anesthetized brain, is organized in multiple highly specific functional anatomical networks (resting state networks, RSNs) (8, 9). These RSNs fluctuate at frequencies between 0.01 and 0.1 Hz, and strongly overlap with sensory–motor, visual, auditory, attention, language, and default networks that are commonly modulated during active behavioral tasks (10–20).
A critical step toward understanding the functional role of spontaneous activity is to clarify the neurophysiological basis of these RSNs, and more generally the neuronal dynamics underlying intrinsic activity (21, 22). An important clue that low-frequency BOLD coherence and neuronal activity may be related was the discovery that low-frequency fluctuations of the band-limited power of local field potential, recorded from a grid of cortical electrodes in the awake monkey, fluctuates approximately at the same frequency as the BOLD signal (23). Moreover, in humans, several studies have reported significant correlations between alpha (24–29) and beta (27) power in the EEG, and simultaneously recorded BOLD signal fluctuations within specific brain networks.
From a theoretical standpoint, however, the assumption that a single cerebral rhythm is associated with a specific cerebral functional network is not likely. In fact, detailed biophysical studies reveal that even single neurons exhibit complex dynamics, including the capacity to oscillate at multiple frequencies (3, 30). Moreover, brain regions do not generally display pure oscillations. Conversely, a combination of delta, theta, alpha, beta, and gamma rhythms, generally ascribed to network operations in cortico-thalamic systems, has been reported (31, 32). Furthermore, although adjacent frequency bands within the same neuronal network are typically associated with different brain states and compete with each other, several rhythms can coexist in the same area or interact among different structures (4, 31).
These theoretical considerations strongly suggest that large-scale functional-anatomical networks, including RSNs, should show electrophysiological oscillations in multiple frequency bands, and that different frequency bands should be coupled to mediate brain operations. To address this issue, we developed a completely data-driven general approach for investigating the relationship between neuronal oscillatory processes in different EEG frequency bands and coherent fMRI fluctuations. We determined independent spatiotemporal patterns in the BOLD signal that correspond to six specific cortico-thalamic RSNs (8, 9); next, we examined their correlations with EEG power fluctuations in delta, theta, alpha, beta, and gamma bands. Each brain network was associated with a specific combination of EEG rhythms, a neurophysiological signature that constitutes a baseline for evaluating changes in oscillatory signals during active behavior.
Results
For further details, see supporting information (SI) Figs. 4–12 and SI Tables 1–3.
The complete procedure developed for associating EEG rhythmic activity with coherent fMRI fluctuations is illustrated in SI Fig. 4. Spectral analysis of the EEG was carried out after the removal of artifacts induced by the fMRI scanning (33); SI Figs. 5 and 6 and SI Table 1 show that the procedure produces artifact-free EEG signals. Five reference waveforms representing the time course of EEG power in the delta, theta, alpha, beta, and gamma bands were reconstructed. Analysis of these waveforms revealed positively correlated power fluctuation across different frequency bands, with correlation coefficients ranging between 0.11 and 0.47 (SI Table 2). This positive correlation suggests that slow and fast rhythms are not independent, and may underlie patterns of cooperation on a variety of spatial and temporal scales (34). Hence, analysis of simultaneous EEG/fMRI data requires methods that consider the whole frequency spectrum rather than single frequency bands (24–29).
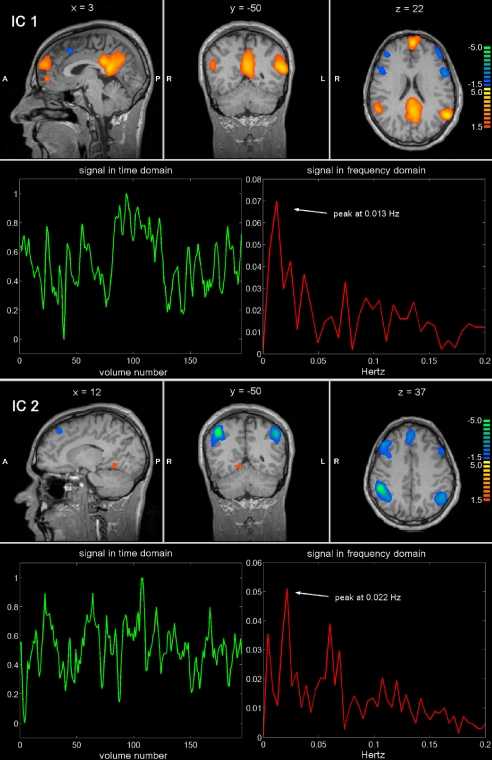
Resting state patterns were identified from the BOLD signals by using independent component analysis (ICA), a technique that extracts maximally independent patterns of brain activity (or independent components, ICs). Each IC consists of a temporal waveform and an associated spatial map; the latter is expressed in terms of z-scores that reflect the degree to which a given voxel time-course correlates with the specific IC temporal waveform (35). Fig. 1 provides an example of two independent components in one subject. The first component involves a characteristic lateral parietal, medial parietal, and frontal pattern, with a peak in the frequency domain at 0.013 Hz, consistent with the so-called default-mode network (15–19); the other consists of a bilateral fronto-parietal pattern, including intraparietal sulcus and frontal eye field, with a frequency peak at 0.022 Hz, that corresponds to the dorsal attention network (14, 20). In general, the waveforms of these spontaneous activity patterns showed slow fluctuations, with frequency content in the range 0.01–0.1 Hz as observed in previous works (10, 36).
Fig. 1.
Example of two ICs separated by ICA from a single-subject fMRI data set. Sagittal, coronal and axial functional maps, are shown, along with the related BOLD signal in time and frequency domains. Brain areas are respectively colored in yellow-orange or in azure-blue in case of positive and negative correlation with the IC waveform.
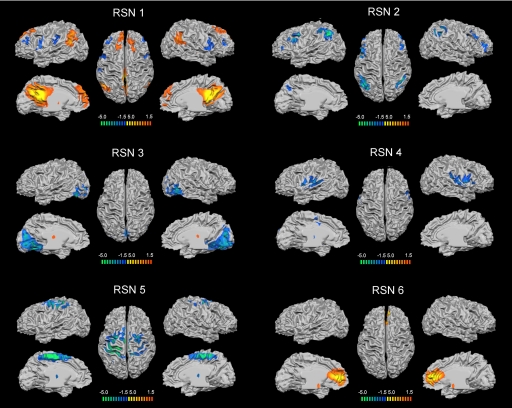
Across subjects, we identified six RSNs (Fig. 2):
RSN 1: a network corresponding to most regions of the default-mode network putatively associated with internal processing (15–19). This network involves bilateral inferior parietal lobule (mainly angular gyrus), posterior cingulated/precuneus, bilateral superior frontal gyrus and medial frontal gyrus.
RSN 2: a network corresponding to the dorsal attention network mediating goal-directed stimulus-response selection (14). This network includes bilaterally the intraparietal sulcus, cortex at the intersection of precentral and superior frontal sulcus near/at the human frontal eye field, ventral precentral, and middle frontal gyrus.
RSN 3: a posterior network involving the retinotopic occipital cortex and the temporal-occipital regions including human MT, dedicated to visual processing (13).
RSN 4: a network primarily including the bilateral superior temporal cortex, corresponding to the auditory-phonological system (12).
RSN 5: a network including the precentral, postcentral, and medial frontal gyri, the primary sensory-motor cortices, and the supplementary motor area (11).
RSN 6: a network including the medial-ventral prefrontal cortex, the pregenual anterior cingulate, the hypothalamus, and the cerebellum, putatively related to self-referential mental activity (37).
Fig. 2.
Cortical representation of the six RSNs. For each RSN. (Left) Lateral and medial views of left hemisphere. (Center) Dorsal view. (Right) Lateral and medial views of right hemisphere.
There was also significant evidence of thalamo-cortical connectivity for RSNs 1, 3, 4, and 5 (SI Fig. 7), showing the participation of the thalamus in the modulation of resting cerebral fluctuations (8, 38). Furthermore, the detection of hippocampal activity in RSN 1 (SI Table 3) further supported the involvement of the default-mode system in memory processes (39).
The fMRI total variance explained by the RSNs was 44 ± 6%. Their replicability was examined with a multidimensional scaling visualization plot kindly provided by Esposito and Goebel (40) (SI Fig. 8). This analysis showed that the homogeneity of RSNs 2–4 was reduced with respect to RSNs 1, 5, and 6, because some maps referring to RSNs 2–4, although they were correctly clustered on the basis of intra- and intercluster similarities, presented partially overlapping activations. Their consistency was demonstrated by replicating the same ICA decomposition separately for the BOLD signals recorded in the first two, and in the last two minutes. These blocks of time can be considered largely independent in the domain of interest as the temporal autocorrelation values fell from one to zero at 95% confidence interval in <18 s for every RSN. We found significant correlations (all RSNs, P < 0.001) between the corresponding spatial maps: 0.59 for RSN 1, 0.77 for RSN 2, 0.64 for RSN 3, 0.54 for RSN 4, 0.40 for RSN 5, and 0.68 for RSN 6 (SI Fig. 9).
Next, we analyzed the electrophysiological correlates of each network. The BOLD signal time-course corresponding to each independent component was correlated with the EEG reference waveforms of the various frequency bands (SI Fig. 4). These maps contain regions with a positive (warm colors, yellow-orange) or a negative (cool colors, azure-blue) correlation of the BOLD signal with the power fluctuations in the various EEG bands (Figs. 1 and 2). It is assumed that a positive correlation of electrical rhythmic activity with BOLD signals underlies neuronal oscillatory synchronization whereas a negative correlation underlies neuronal desynchronization (23, 41).
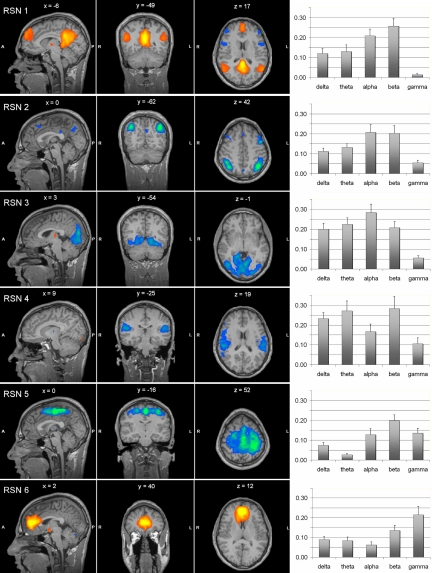
Fig. 3 shows the spatial map for each RSN, and the bar plots of the correlation between the BOLD signal time course and the power fluctuation in each frequency band. In general correlation levels were not high, ranging between 0.03 and 0.28; among them, only correlations >0.20 were significant (P < 0.05, with Bonferroni correction). More than one rhythm was associated with the same network, confirming that neurons oscillating at different frequencies may contribute to the same functional system. RSNs 1 (default) and 2 (dorsal attention) had stronger relationship with alpha and beta rhythms, albeit in opposite directions, with RSN1 showing positive whereas RSN2 showing negative correlation with alpha and beta rhythms; RSNs 3 (visual) with all rhythms with the exclusion of the gamma rhythm; RSN 4 (auditory) with delta, theta, and beta rhythms; RSN 5 (somato-motor) with beta rhythm; RSN 6 (self-referential) with gamma rhythm.
Fig. 3.
Association between EEG rhythms and fMRI RSNs. (Left) Sagittal, coronal, and axial spatial maps of the six RSNs. (Right) Bar plots of the average correlations between the brain oscillatory activity in the delta, theta, alpha, beta, and gamma bands, and the RSN time courses.
To test the consistency of this association, we performed the same split-half analysis on the BOLD signal-EEG power correlation. No statistical difference, assessed by a two-sample Kolmogov–Smirnov test (α = 0.05), was found between the correlations calculated separately for data collected in the first two, and in the last two minutes (SI Fig. 10). Again, these two blocks of time can be considered largely independent given that the EEG power fluctuations were calculated from data at high temporal resolution. Hence the relationship between BOLD signal RSNs and EEG power was both robust and reliable.
Finally, we performed inferential statistical testing for evaluating whether these multiband patterns of EEG power correlation were specific and discriminated among different RSNs. To test the specificity of the association between BOLD RSNs and EEG distribution of power in different bands, we performed a nonparametric one-way ANOVA (Kruskal–Wallis test) on the correlation values, with the RSNs as groups. This analysis showed for each EEG band that the profile of correlation was statistically different across RSNs with P < 0.05 (SI Fig. 11). Next, to study the source of this variation, we carried out a linear discriminant analysis (LDA), a statistical technique able to classify objects into mutually exclusive and exhaustive groups based on a set of measurable object's features. By means of LDA, we verified that the RSNs could be separated on the basis of their specific EEG power profile at α = 0.05 (SI Fig. 12).
Discussion
Spontaneous brain activity in the awake resting state is organized in a finite set of spatiotemporal patterns that we have characterized by using a completely data-driven approach, linking EEG brain rhythms and low-frequency coherent fluctuations of the BOLD signal.
Methodological Considerations.
We have investigated the EEG signal variations over a large frequency range (1–50 Hz), using an optimized method for the reconstruction of the EEG data with negligible contamination of the disturbances induced by simultaneous fMRI scanning (33). The obtained EEG reference waveforms represent rhythm power fluctuations in different frequency bands, and have been previously associated to BOLD fluctuations over very long time scales (23); in this study, we found that these waveforms are positively cross-correlated, suggesting that neuronal signals are dynamically coupled in different frequency bands (31, 32). This coupling could be expected on the basis of a previous work by Bruns et al. (34), who demonstrated with a similar method the presence of long-range cooperation of neuronal assemblies oscillating at different frequencies. These findings indicate the importance of analyzing multiple frequency bands simultaneously, rather than limiting the analysis to one or two EEG bands.
With regard to the fMRI BOLD data analysis, we found six replicable functional brain networks that simultaneously fluctuate at slightly different BOLD signal frequencies (<0.1 Hz). These networks are similar to those obtained by other research groups using ICA (8, 9), and more generally correspond to the main functional networks reported in the literature (10–20). Interestingly, each of these networks could be generally associated with more than one kind of electric oscillation, confirming that the brain does not generally display pure rhythms within distinct frequency bands, mostly generated in restricted neuronal circuits, but a coalescence of rhythms. Not surprisingly, our spatial maps are very similar to the maps obtained with the previous EEG/fMRI studies on alpha-power correlation. In particular, RSNs 3 and 4 have been found by Goldmann et al. (24) and Moosmann et al. (23), RSN 3 has been found by Feige et al. (28), and Laufs et al. have been able to associate a map similar to that of RSN 2 with the alpha rhythm and a map similar to that of RSN 1 with the beta rhythm in the frequency range 17–23 Hz (26, 27).
One important limitation of this approach is that the EEG analysis is limited in the frequency domain (<50 Hz) because of the attenuation of the electric field through the skull, and that behaviorally important neuronal activity also occurs at much higher frequencies (42). Another important limit is that our analysis is based on the correlation between BOLD signal and EEG power time courses, independent of their amplitude.
Neurobiological Considerations.
An important question is whether these networks reflect only anatomical connectivity or yet uncovered physiological mechanisms. Although their topographical organization strongly suggest a relationship to anatomical connectivity (8), our combined BOLD-EEG results show that anatomical connectivity alone cannot account for the obtained findings. The best example is the separation of the dorsal attention network (RSN 2) from the rest of the visual system (RSN 3). Visual areas within the intraparietal sulcus (14, 43) are in fact bidirectionally connected with many other visual areas, especially in the middle temporal region (44, 45), yet these two networks can be separated on the basis of BOLD signal fluctuations and EEG power fluctuations, with the dorsal network more weighted toward alpha and beta rhythms, and the visual cortex more weighted toward delta and theta rhythms. This separation breaks down during visual processing when these areas show common task variability (48).
Our current hypothesis is that these resting state networks represent a finite set of spatiotemporal basis function from which task-networks are then dynamically assembled and modulated during different behavioral states. One form of modulation would be the combination of different networks to mediate behavior, which is apparent in this study. Interestingly, in prior work, RSNs 1 (default) and 6 (self-referential) have been jointly considered part of the default-mode network based on task-activation (15, 47) and regions-of-interest based functional connectivity studies (36). Here, we clearly show that these two sets of regions represent different functional entities as they separate along both independent temporal components of the BOLD signal, and different EEG power spectra. These two networks are the only ones showing a positive correlation with EEG power, which is consistent with their activation in the resting state (6, 47).
Furthermore, the EEG power spectra associated with the ventro-medial prefrontal cortex (RSN 6) is strongly weighted toward gamma power, whereas the rest of the default network (RSN 1) is more strongly associated with alpha and beta power. This difference in EEG power predicts that ventro-medial prefrontal cortex should show stronger BOLD modulation in the resting state than the rest of the default network, as BOLD modulations have been more closely linked to changes in gamma power (23). This prediction is met given the ventro-medial prefrontal cortex shows a tonic deactivation during goal-directed behavior, which persists as long as subjects are engaged in an active behavior; whereas other areas of the default network (e.g., lateral and medial parietal cortex) show a much more time-locked deactivation to specific events (48).
Another form of cooperation during active behavior is the modulation in the frequency domain of networks that might be competing for resources. An interesting example is the relationship between the dorsal attention network and the default network that are modulated in opposite directions during rest and active behavior (6, 15). Although the dorsal attention network is positively modulated, the default network shows negative modulations of the BOLD signal during active tasks as compared with rest. In this study, we found that default and attention networks show a very similar correlation with EEG power in all frequency bands, especially alpha and beta as shown by prior works (26, 27). An increase in alpha and beta power at rest correlates positively with activity in the default and self-referential networks, and negatively with activity in the dorsal attention networks. Conversely, a fall in alpha and beta power during active attention-demanding behavior, or EEG desynchronization, corresponds to a relative increase in dorsal attention activity. However, our results indicate that these two networks are coupled in terms of EEG power, not BOLD functional connectivity.
It is certainly possible that these slow coupled fluctuations of EEG power and BOLD signal do not reflect a dynamic baseline of inter-areal temporal interaction from which task-evoked patterns arise, but a more basic neurophysiological mechanism unrelated to functional neuronal communication. Recent work has isolated ultra-slow (<1 Hz) fluctuations of the EEG signal in the same range as the BOLD signal fluctuations, which are positively correlated with faster EEG oscillations, but are unlikely to mediate neuronal communication (49). Another possibility is that slow BOLD signal fluctuations are related to the so-called up–down neuronal states, slow (≈1 Hz) intrinsic fluctuations of the membrane potential identified in anesthetized animals, and recently shown to fluctuate synchronously between two connected areas (50).
In summary, we have combined EEG and fMRI to study the dynamic baseline functional architecture of the human brain. Our results extend previous studies on the resting-state condition showing that the brain is never really at rest: instead of stabilizing at an uniform level, brain activity fluctuates within definite spatiotemporal patterns. This spontaneous ongoing oscillatory activity, whose existence has been noted for as long as electrical recordings have been made, depends on the dynamic interplay between distinct functional networks, each characterized by a specific electrophysiological signature.
Materials and Methods
Study Protocol.
Fifteen healthy male subjects (mean age 24, range 20–29 years) participated in the study, which was approved by the Ethics Committee of Chieti University. After giving written informed consent, subjects underwent a brief neurological examination that did not reveal any medical issues. We simultaneously recorded EEG and fMRI on all subjects over 4 min. They were instructed to simply lie with their eyes closed inside the scanner and not fall asleep; no visual or auditory stimuli were presented at any time during functional scanning.
EEG Acquisition.
A 32-channel MR-compatible BrainAmp system (Brainproducts, Munich, Germany) was used for EEG recordings, along with the BrainCap electrode cap (Falk Minow Services, Herrsching-Breitbrunn, Germany). All of the electrodes, which were placed on the scalp according to the international 10–20 system, were ring-type sintered nonmagnetic Ag/AgCl electrodes. An additional channel was dedicated to the electrocardiogram (ECG); two other channels were positioned over the subject earlobes, and their average was used as reference. The impedance of each electrode was maintained lower than 5 kΩ using electrode paste. Data were collected with a sampling rate of 5 kHz; band-pass filtering from 0.016 to 250 Hz was applied, along with 50-Hz notch filtering.
FMRI Acquisition.
Functional images were acquired with a Siemens Magnetom Vision scanner at 1.5 T by means of T2*-weighted echo planar imaging free induction decay sequences with the following parameters: echo time (TE), 60 ms; matrix size, 64 × 64; field of view (FOV), 256 mm; in-plane voxel size, 4 × 4 mm; flip angle, 90°; slice thickness, 10 mm; and no gap. Functional volumes consisted of nine bicommissural slices, acquired with a volume repeat time (TR) of 1,200 ms and a scan time gap of 280 ms. A total of 200 volumes were acquired for each subject, and the first 5 volumes were discarded to ensure steady-state longitudinal magnetization. Subsequently, a high-resolution structural volume was acquired via a 3D MPRAGE sequence (sagittal; matrix, 256 × 256; FOV, 256 mm; slice thickness, 1 mm; no gap; in-plane voxel size, 1 mm × 1 mm; flip angle, 12°; TR, 9.7 ms; TE, 4 ms) to provide the anatomical reference for the functional scan.
Artifact Removal from EEG Data.
Brain Vision Analyzer software (Brainproducts) was used for off-line correction of imaging artifact. This software implements the adaptive artifact subtraction (AAS) method, in which the imaging artifact waveforms are segmented, averaged, and iteratively subtracted from the EEG signals (51). Subsequently, data were exported to ASCII format and processed by using a self-developed software implemented in MATLAB (Mathworks, Sherborn, MA). They were down-sampled to 1 kHz and digitally filtered in the band 1–50 Hz using a Chebyshev II-type filter with 40 dB attenuation and zero-phase distortion. After visually checking them for the exclusion of movement artifacts and noisy channels from the data, a method based on temporal ICA was used for the rejection of the ballistocardiographic (BCG) artifact and the residual imaging artifact from the filtered EEG recordings. The details and a complete validation of the method have been previously published (33). In this study, the artifact removal effectiveness was evaluated by measuring the residual BCG artifact amplitude (peak-to-peak) in the EEG data, as well as the correlation of the EEG signals with the ECG trace and the imaging artifact estimate.
Rhythm Reference Waveform Reconstruction.
To determine the spectral characteristics of the artifact-free EEG data, the power spectrum was calculated by using a fast Fourier transform. The spectrum was averaged in epochs of 1,200 ms, corresponding to the volume TR, and over all EEG channels. The resulting spectrogram was divided into five subbands, corresponding to delta (1–4 Hz), theta (4–8 Hz), alpha (8–13 Hz), beta (13–30 Hz), and gamma (30–50 Hz) rhythms. The power spectral density was calculated for the five frequency bands, thus obtaining a single power time series for each brain rhythm. Interband correlations were calculated from these power time series, thus measuring the level of coherence across different bands (34). Next, they were convolved with a canonical hemodynamic response function, generated by using a gamma function (delay time, 2 s; rise time, 4 s; fall time, 6 s; undershoot, 0.2; restore time, 2 s). The five signals were normalized by subtracting the minimum value and dividing by the difference of maximum and minimum values. Finally, each rhythm reference waveform consisted of 195 samples, representing the estimate of a specific neural oscillatory activity.
Separation of BOLD Spatiotemporal Patterns.
Brain Voyager QX software (Brain Innovation, Maastricht, The Netherlands) was used for image data preparation and processing. Functional image time series were first corrected for the differences in slice acquisition times, detrended, realigned with T1 volumes, and warped into the standard Talairach anatomical space. For each data set, spatial ICA (sICA) was used for the decomposition of fMRI time-series into brain activity patterns starting from the spatial covariance of the measured signals (35). After data reduction by means of principal component analysis (PCA), 30 ICs (with waveforms and spatial maps) were estimated for each subjects using the deflation approach of the FastICA algorithm (52). For each IC, the waveform corresponds to the time course of a specific pattern of brain activity, and the intensity with which this activity is found across the voxels is expressed by the associated spatial map. To display voxels contributing most strongly to a particular IC, the intensity values in each map were scaled to z scores (37). Voxels with absolute z scores >1.5 were considered to be IC active voxels. Negative z scores indicate voxels the BOLD signals of which are modulated opposite to the IC waveform.
To extend the ICA analysis from single-subject to multisubject studies, the ICs estimated from each subject were clustered with the self-organizing group ICA (sogICA) method, according to their mutual similarities (40). Once the ICs belonging to a cluster have been retrieved, the IC maps were averaged and the resulting map was assumed as the representative of the cluster. The results of the automated clustering procedure were visually checked, and the differences between single-subject and group maps were assessed: only the clusters that were composed by reproducible maps across subjects were considered. Consequently, a reduced number of spatio-temporally distinct patterns of low-frequency coherencies were extracted, and finally associated with RSNs within the brain. To assess the homogeneity of the RSN spatial maps across subjects, the intra- and intercluster similarities were visualized in a two-dimensional space using multidimensional scaling (40).
Association of EEG Rhythms and fMRI Maps.
The correspondence between neuronal rhythms and RSNs was analyzed, estimating the similarity between the EEG reference waveforms and the time-courses of the RSNs. This was statistically assessed by using the parametric two-tailed Pearson's test. For each subject, the correlation coefficient was computed to express the amount of common variation between the single reference waveform and the single IC waveform. The correlation coefficients related to the ICs of a specific cluster were directly averaged, to obtain a group-level correlation between the brain rhythm and the RSN. Therefore, the Bonferroni correction method was adopted for addressing multiple comparisons and correctly evaluating the statistical significance of the resulting correlations.
Finally, inferential statistical testing was performed for evaluating whether the correlation patterns could be associated with electrophysiological signatures of resting state networks. To this purpose, the Kruskal–Wallis test was used to test whether the correlation patterns for each single EEG band were statistically different across RSNs. Finally, linear discriminant analysis was performed on correlation values, testing whether the RSNs could be separated on the basis of their specific EEG power profile.
Supplementary Material
Acknowledgments
We thank Francesca Petrucci, Antonio Ferretti, and Mark McAvoy for scientific discussion, Fabrizio Esposito and Rainer Goebel for methodological support, and Abraham Z. Snyder and Gordon Shulman for constructive suggestions about the manuscript. This work was supported by Italian Ministry for University and Research Grant PRIN 2005027850_001. M.C. was supported by a European Union Marie Curie Chair (MEXC-CT-2004-006783 IBSEN), which also covered some of the research costs, and by National Institute of Mental Health Grant R01MH71920-06 and National Institutes of Health Grant NS48013.
Abbreviations
- BOLD
blood oxygen level-dependent
- fMRI
functional MRI
- RSN
resting state network
- IC
independent component
- ICA
IC analysis.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The EEG and fMRI data have been deposited with the fMRI Data Center, www.fmridc.org (accession no. 2-2007-122B5).
This article contains supporting information online at www.pnas.org/cgi/content/full/0700668104/DC1.
References
- 1.Hubel DH, Wiesel TN. J Physiol. 1962;160:106–154. doi: 10.1113/jphysiol.1962.sp006837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shadlen MN, Newsome WT. J Neurosci. 1998;18:3870–3896. doi: 10.1523/JNEUROSCI.18-10-03870.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Llinas RR. Science. 1988;242:1654–1664. doi: 10.1126/science.3059497. [DOI] [PubMed] [Google Scholar]
- 4.Varela F, Lachaux JP, Rodriguez E, Martinerie J. Nat Rev Neurosci. 2001;2:229–239. doi: 10.1038/35067550. [DOI] [PubMed] [Google Scholar]
- 5.Engel AK, Fries P, Singer W. Nat Rev Neurosci. 2001;2:704–716. doi: 10.1038/35094565. [DOI] [PubMed] [Google Scholar]
- 6.Gusnard DA, Raichle ME. Nat Rev Neurosci. 2001;2:685–694. doi: 10.1038/35094500. [DOI] [PubMed] [Google Scholar]
- 7.Raichle ME, Gusnard DA. Proc Natl Acad Sci USA. 2002;99:10237–10239. doi: 10.1073/pnas.172399499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Luca M, Beckmann CF, De Stefano N, Matthews PM, Smith SM. NeuroImage. 2006;29:1359–1367. doi: 10.1016/j.neuroimage.2005.08.035. [DOI] [PubMed] [Google Scholar]
- 9.Damoiseaux JS, Rombouts SA, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF. Proc Natl Acad Sci USA. 2006;103:13848–13853. doi: 10.1073/pnas.0601417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cordes D, Haughton VM, Arfanakis K, Carew JD, Turski PA, Moritz CH, Quigley MA, Meyerand ME. Am J Neuroradiol. 2001;22:1326–1333. [PMC free article] [PubMed] [Google Scholar]
- 11.Biswal BB, Yetkin FZ, Haughton VM, Hyde JS. Magn Reson Med. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- 12.Biswal BB, Van Kylen J, Hyde JS. NMR Biomed. 1997;10:165–170. doi: 10.1002/(sici)1099-1492(199706/08)10:4/5<165::aid-nbm454>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 13.Lowe MJ, Mock BJ, Sorenson JA. NeuroImage. 1998;7:119–132. doi: 10.1006/nimg.1997.0315. [DOI] [PubMed] [Google Scholar]
- 14.Corbetta M, Shulman GL. Nat Rev Neurosci. 2002;3:201–215. doi: 10.1038/nrn755. [DOI] [PubMed] [Google Scholar]
- 15.Shulman GL, Fiez JA, Corbetta M, Buckner RL, Miezin FM, Raichle ME, Petersen SE. J Cognit Neurosci. 1997;9:648–663. doi: 10.1162/jocn.1997.9.5.648. [DOI] [PubMed] [Google Scholar]
- 16.Binder JR, Frost JA, Hammeke TA, Bellgowan PS, Rao SM, Cox RW. J Cognit Neurosci. 1999;11:80–95. doi: 10.1162/089892999563265. [DOI] [PubMed] [Google Scholar]
- 17.Mazoyer B, Zago L, Mellet E, Bricogne S, Etard O, Houde O, Crivello F, Joliot M, Petit L, Tzourio-Mazoyer N. Brain Res Bull. 2001;54:287–298. doi: 10.1016/s0361-9230(00)00437-8. [DOI] [PubMed] [Google Scholar]
- 18.Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL. Proc Natl Acad Sci USA. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greicius MD, Krasnow B, Reiss AL, Menon V. Proc Natl Acad Sci USA. 2003;100:253–258. doi: 10.1073/pnas.0135058100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fox MD, Corbetta M, Snyder AZ, Vincent JL, Raichle ME. Proc Natl Acad Sci USA. 2006;103:10046–10051. doi: 10.1073/pnas.0604187103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsodyks M, Kenet T, Grinvald A, Arieli A. Science. 1999;286:1943–1946. doi: 10.1126/science.286.5446.1943. [DOI] [PubMed] [Google Scholar]
- 22.Arieli A, Sterkin A, Grinvald A, Aertsen A. Science. 1996;273:1868–1871. doi: 10.1126/science.273.5283.1868. [DOI] [PubMed] [Google Scholar]
- 23.Leopold DA, Murayama Y, Logothetis NK. Cereb Cortex. 2003;13:422–433. doi: 10.1093/cercor/13.4.422. [DOI] [PubMed] [Google Scholar]
- 24.Goldman RI, Stern JM, Engel J, Jr, Cohen MS. NeuroReport. 2002;13:2487–2492. doi: 10.1097/01.wnr.0000047685.08940.d0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moosmann M, Ritter P, Krastel I, Brink A, Thees S, Blankenburg F, Taskin B, Obrig H, Villringer A. NeuroImage. 2003;20:145–158. doi: 10.1016/s1053-8119(03)00344-6. [DOI] [PubMed] [Google Scholar]
- 26.Laufs H, Kleinschmidt A, Beyerle A, Eger E, Salek-Haddadi A, Preibisch C, Krakow K. NeuroImage. 2003;19:1463–1476. doi: 10.1016/s1053-8119(03)00286-6. [DOI] [PubMed] [Google Scholar]
- 27.Laufs H, Krakow K, Sterzer P, Eger E, Beyerle A, Salek-Haddadi A, Kleinschmidt A. Proc Natl Acad Sci USA. 2003;100:11053–11058. doi: 10.1073/pnas.1831638100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feige B, Scheffler K, Esposito F, Di Salle F, Hennig J, Seifritz E. J Neurophysiol. 2005;93:2864–2872. doi: 10.1152/jn.00721.2004. [DOI] [PubMed] [Google Scholar]
- 29.Laufs H, Holt JL, Elfont R, Krams M, Paul JS, Krakow K, Kleinschmidt A. NeuroImage. 2006;31:1408–1418. doi: 10.1016/j.neuroimage.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Buzsaki G, Draguhn A. Science. 2004;304:1926–1929. doi: 10.1126/science.1099745. [DOI] [PubMed] [Google Scholar]
- 31.Steriade M. J Neurophysiol. 2001;86:1–39. doi: 10.1152/jn.2001.86.1.1. [DOI] [PubMed] [Google Scholar]
- 32.Steriade M. Neuroscience. 2006;137:1087–1106. doi: 10.1016/j.neuroscience.2005.10.029. [DOI] [PubMed] [Google Scholar]
- 33.Mantini D, Perrucci MG, Cugini S, Ferretti A, Romani GL, Del Gratta C. NeuroImage. 2007;34:598–607. doi: 10.1016/j.neuroimage.2006.09.037. [DOI] [PubMed] [Google Scholar]
- 34.Bruns A, Eckhorn R, Jokeit H, Ebner A. NeuroReport. 2000;11:1509–1114. [PubMed] [Google Scholar]
- 35.McKeown MJ, Makeig S, Brown GG, Jung TP, Kindermann SS, Bell AJ, Sejnowski TJ. Hum Brain Mapp. 1998;6:160–188. doi: 10.1002/(SICI)1097-0193(1998)6:3<160::AID-HBM5>3.0.CO;2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME. Proc Natl Acad Sci USA. 2005;102:9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.D'Argembeau A, Collette F, Van der Linden M, Laureys S, Del Fiore G, Degueldre C, Luxen A, Salmon E. NeuroImage. 2005;25:616–624. doi: 10.1016/j.neuroimage.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 38.Johansen-Berg H, Behrens TE, Sillery E, Ciccarelli O, Thompson AJ, Smith SM, Matthews PM. Cereb Cortex. 2005;15:31–39. doi: 10.1093/cercor/bhh105. [DOI] [PubMed] [Google Scholar]
- 39.Vincent JL, Snyder AZ, Fox MD, Shannon BJ, Andrews JR, Raichle ME, Buckner RL. J Neurophysiol. 2006;96:3517–3531. doi: 10.1152/jn.00048.2006. [DOI] [PubMed] [Google Scholar]
- 40.Esposito F, Scarabino T, Hyvarinen A, Himberg J, Formisano E, Comani S, Tedeschi G, Goebel R, Seifritz E, Di Salle F. NeuroImage. 2005;25:193–205. doi: 10.1016/j.neuroimage.2004.10.042. [DOI] [PubMed] [Google Scholar]
- 41.Logothetis NK, Pauls J, Augath M, Trinath T, Oeltermann A. Nature. 2001;412:150–157. doi: 10.1038/35084005. [DOI] [PubMed] [Google Scholar]
- 42.Siegel M, Donner TH, Oostenveld R, Fries P, Engel AK. Cereb Cortex. 2006;17:732–741. doi: 10.1093/cercor/bhk025. [DOI] [PubMed] [Google Scholar]
- 43.Sereno MI, Pitzalis S, Martinez A. Science. 2001;294:1350–1354. doi: 10.1126/science.1063695. [DOI] [PubMed] [Google Scholar]
- 44.Ungerleider LG, Desimone R. J Comp Neurol. 1986;248:190–222. doi: 10.1002/cne.902480204. [DOI] [PubMed] [Google Scholar]
- 45.Maunsell JH, Van Essen DC. J Comp Neurol. 1987;266:535–555. doi: 10.1002/cne.902660407. [DOI] [PubMed] [Google Scholar]
- 46.Hasson U, Nir Y, Levy I, Fuhrmann G, Malach R. Science. 2004;303:1634–1640. doi: 10.1126/science.1089506. [DOI] [PubMed] [Google Scholar]
- 47.Gusnard DA, Akbudak E, Shulman GL, Raichle ME. Proc Natl Acad Sci USA. 2001;98:4259–4264. doi: 10.1073/pnas.071043098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dosenbach NU, Visscher KM, Palmer ED, Miezin FM, Wenger KK, Kang HC, Burgund ED, Grimes AL, Schlaggar BL, Petersen SE. Neuron. 2006;50:799–812. doi: 10.1016/j.neuron.2006.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vanhatalo S, Palva JM, Holmes MD, Miller JW, Voipio J, Kaila K. Proc Natl Acad Sci USA. 2004;101:5053–5057. doi: 10.1073/pnas.0305375101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hahn TT, Sakmann B, Mehta MR. Nat Neurosci. 2006;9:1359–1361. doi: 10.1038/nn1788. [DOI] [PubMed] [Google Scholar]
- 51.Allen PJ, Josephs O, Turner R. NeuroImage. 2000;12:230–239. doi: 10.1006/nimg.2000.0599. [DOI] [PubMed] [Google Scholar]
- 52.Hyvärinen A. IEEE T Neural Network. 1999;10:626–634. doi: 10.1109/72.761722. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.