Abstract
Purkinje cells are one of the major types of neurons that form the neural circuitry in the cerebellum essential for fine control of movement and posture. During development, Purkinje cells also are critically involved in the regulation of proliferation of progenitors of granule cells, the other major type of neurons in the cerebellum. The process that controls differentiation of Purkinje cells from their early precursors is poorly understood. Here we report that two closely related LIM-homeobox genes, Lhx1 and Lhx5, were expressed in the developing Purkinje cells soon after they exited the cell cycle and migrated out of the cerebellar ventricular zone. Double-mutant mice lacking function of both Lhx1 and Lhx5 showed a severe reduction in the number of Purkinje cells. In addition, targeted inactivation of Ldb1, which encodes a cofactor for all LIM-homeodomain proteins, resulted in a similar phenotype. Our studies thus provide evidence that these transcription regulators are essential for controlling Purkinje cell differentiation in the developing mammalian cerebellum.
Keywords: CNS, development, embryo, mouse, transcription
The neural circuitry in the cerebellum plays an essential role in controlling movement and posture. Granule cells and Purkinje cells constitute the two principal types of neurons that form this circuitry. One major challenge in understanding development of the cerebellum is to elucidate the mechanisms underlying the generation of these cells.
The generation of the granule cells has been known to involve intricate regulations by a number of signaling molecules and transcription factors. The Shh signaling molecule (1, 2) and its downstream transcription factor Gli2 (3), as well as the Notch signaling pathway (4) and other transcription factors such as Ru49/Zipro1 (5), Zic1, and Zic2 (6, 7), are important for regulation of proliferation of the granule cell progenitors. The BMP signaling molecules (8–10) and transcription factors such as Math1 (11, 12) and NeuroD (13) are critically involved in the specification or differentiation of the granule cells.
The factors that control the generation of Purkinje cells are largely unknown (for reviews, see refs. 14–17). Recent studies have revealed that Ptf1a, a basic helix–loop–helix transcription factor, is required for the specification of progenitors of all cerebellar GABAergic neurons, including Purkinje cells, in the ventricular zone of the cerebellum (18, 19). However, the factors that regulate further differentiation of Purkinje cells after they become postmitotic and migrate out of the ventricular zone have remained unclear.
The LIM-homeodomain proteins represent transcription factors that play essential roles in the differentiation of many specific types of neurons throughout the developing central nervous system (20, 21). They function in the context of protein complexes that involve a key cofactor called Ldb1 (22–24). RNAs encoding two closely related LIM-homeodomain proteins, Lhx1 and Lhx5, previously have been detected in Purkinje cells in postnatal and adult mice and rats (refs. 25 and 26 and data not shown). More recently, the expression of Lhx1 and Lhx5 proteins and of Lhx1- and Lhx5-directed Egfp reporters have been observed in developing Purkinje cells soon after they exit the cell cycle in the cerebellum (27). To begin to address the role of LIM-homeodomain proteins in cerebellar development, we examined the expression of Lhx1, Lhx5, and Ldb1 mRNAs in the developing cerebellum. We found that all are expressed in the developing Purkinje cells. Moreover, our analysis of mutant mouse embryos lacking function of these genes revealed that they are essential for Purkinje cell differentiation in the developing cerebellum.
Results
Expression of Lhx1 and Lhx5 mRNAs in the Developing Purkinje Cells.
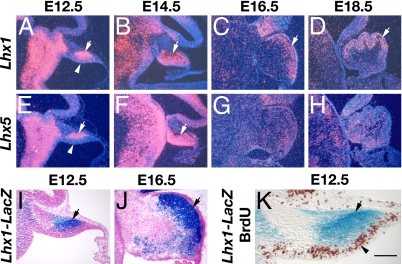
The Purkinje cells are derived from progenitors in the ventricular neuroepithelium of the developing cerebellar anlage (28). During mouse embryonic development, Purkinje cells are born between embryonic day (E)11 and E13, as their progenitors exit the cell cycle and start to migrate out of the ventricular zone (15). By in situ hybridization, we detected mRNAs encoding Lhx1 and Lhx5, two closely related LIM-homeodomain transcription factors, in cells that had just migrated out of the ventricular zone of the cerebellum at E12.5 (Fig. 1 A and E). Lhx1 and Lhx5 mRNAs continuously were present in the migrating cells that had left the ventricular zone at E14.5 (Fig. 1 B and F). By E16.5 and E18.5, we found Lhx1 mRNA in the emerging Purkinje cell layer of the cerebellum (Fig. 1 C and D), whereas the expression of Lhx5 was reduced (Fig. 1 G and H). The activity of the Lhx1 gene also was analyzed in Lhx1LacZ/+embryos, in which a part of one copy of the Lhx1 gene was replaced by a LacZ reporter (29). 5′-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) staining revealed that β-galactosidase, a product of the LacZ reporter gene, was expressed in the cerebellum (Fig. 1 I and J) in a pattern identical to that of Lhx1 mRNA shown by in situ hybridization (Fig. 1 A and C). We next performed X-gal and anti-5-bromo-2′-deoxyuridine (BrdU) double staining of sections from Lhx1LacZ/+ embryos that also were pulse-labeled by BrdU. This analysis allowed us to determine that Lhx1 was expressed only in BrdU-negative postmitotic cells in the differentiating zone but not in BrdU-positive proliferating cells in the ventricular zone of the cerebellum (Fig. 1K). The expression patterns of Lhx1 and Lhx5 mRNAs and Lhx1–LacZ reporter shown here are identical to those of Lhx1 and Lhx5 proteins and Lhx1-and Lhx5–Egfp reporters that have been described elsewhere (27).
Fig. 1.
Expression of Lhx1 and Lhx5 genes in the developing mouse cerebellum. Lhx1 (A–D) and Lhx5 (E–H) mRNAs were detected in E12.5 (A and E), E14.5 (B and F), E16.5 (C and G), and E18.5 (D and H) embryos by in situ hybridization. β-Galactosidase expressed from a LacZ reporter gene inserted into the Lhx1 gene locus was analyzed in E12.5 (I) and E16.5 (J) Lhx1LacZ/+ embryos by X-gal staining. (K) A section of the cerebellum from a BrdU-injected E12.5 Lhx1LacZ/+ embryo double labeled with X-gal staining (blue) to detect the activity of the LacZ reporter gene and with anti-BrdU staining (brown) to monitor proliferating cells. Arrows in A–F and I–K point at the region where Lhx1 or Lhx5 was expressed. Arrowheads in A, E, and K show the ventricular zone of the cerebellum. (Scale bar: A–H, 300 μm; I and J, 170 μm; K, 100 μm.)
Severe Reduction in Number of Purkinje Cells in Lhx1/Lhx5 Double Mutants.
Previous studies have revealed essential roles for Lhx1 and Lhx5 in neuronal differentiation in the developing spinal cord and hippocampus (29, 30). The expression patterns of Lhx1 and Lhx5 in the cerebellum suggest that these factors may be involved in controlling Purkinje cell differentiation. To determine the role of Lhx5 in development of the cerebellum, we performed histological analysis in Lhx5-null mice. Most of the Lhx5-null mutants die soon after birth, but a few survive to adulthood (30, 31). The cerebellum of mutant E18.5 embryos and adults was found to be indistinguishable from that of wild-type controls (data not shown) (31). Germ-line deletions of the Lhx1 result in early lethality of the mutant embryos that lack anterior head regions, including the cerebellum (29, 32). In an effort to analyze the role of Lhx1 in development of the cerebellum, we generated two different conditional Lhx1 mutant alleles, Lhx1fx1 [supporting information (SI) Fig. 6] and Lhx1fx2 (33), by inserting loxP sites into introns flanking the Lhx1 coding sequence. By crossing the Lhx1 conditional mutants with mice that express the Cre recombinase controlled by regulatory elements of either the Nestin (Lhx1fx1/fx1; Nes-Cre+) or Engrailed-1 (Lhx1fx2/fx2; En1Cre/+) gene, we were able to target Lhx1 ablation to the cerebellum by E11.5 or E12.5 (SI Figs. 6 and 7). However, histological analysis at E18.5 revealed no significant defects in the developing cerebellum of these Lhx1-deficient mutants (SI Figs. 8 and 9). The Lhx1fx1/fx1; Nes-Cre+ conditional mutants died soon after birth, possibly because of a failure in kidney function. Cre also was expressed in the developing kidney in these mutants (data not shown), which may well impair the function of Lhx1 that is essential for kidney development (32, 34, 35). The Lhx1fx2/fx2; En1Cre/+ conditional mutant was viable to adulthood and showed no abnormal phenotype.
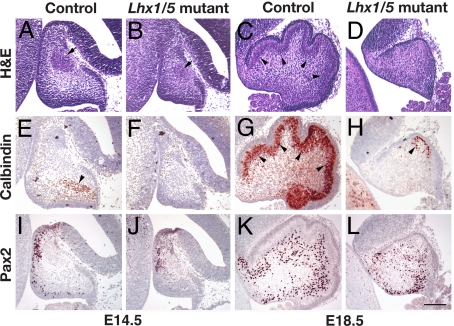
Lhx1 and Lhx5 share high sequence similarity (36) and overlapping expression patterns in the developing cerebellum (Fig. 1), suggesting that these two genes may have redundant roles in the development of the cerebellum. To examine this possibility, we generated double-mutant embryos lacking expression of both Lhx1 and Lhx5 in the developing cerebellum. At E14.5, the cerebellum of the Lhx1/Lhx5 double-mutant embryos (Lhx1fx1/LacZ, Nes-Cre+; Lhx5−/−) was small as compared to that of controls (wild type or any embryos with at least one copy of the Lhx1 or Lhx5 gene intact) (Fig. 2 A and B). At this stage, the expression of the calcium-binding protein calbindin signals the appearance of differentiated Purkinje cells in the control embryos (15) (Fig. 2E). By contrast, the staining of calbindin was absent in the cerebellum of the Lhx1/Lhx5 double-mutant embryos (Fig. 2F). At E18.5, the cerebellum in Lhx1/Lhx5 double mutants remained small as compared to that in the controls. It lacked the Purkinje cell layer and the foliation that had emerged in the controls (Fig. 2 C and D). Although a few calbindin-positive cells were present in the dorsal region, Purkinje cells were largely absent in Lhx1/Lhx5 double mutants as compared to the controls (Fig. 2 G and H). Similar defects also were observed in Lhx1/Lhx5 double mutants in which a different Lhx1 conditional mutant allele (Lhx1fx2) had been inactivated by Cre expressed under the control of En1 regulatory elements (Lhx1fx2/LacZ, En1Cre/+; Lhx5−/−) (SI Figs. 9 and 10). In those mutants, the staining of calbindin in the cerebellum essentially was negative (SI Fig. 10F). In contrast to calbindin, Pax2, a marker for cerebellar GABAergic interneurons such as the basket and stellate cells (19, 37), was detected in many cells of the Lhx1/Lhx5 double mutants as well as in the controls at E14.5 (Fig. 2 I and J) and E18.5 (Fig. 2 K and L). Thus, inactivation of Lhx1 and Lhx5 appears specifically to affect development of the Purkinje cells in the cerebellum.
Fig. 2.
Defects of the cerebellum in Lhx1/Lhx5 double mutants (Lhx1fx1/LacZ; Nes-Cre+; Lhx5−/−). The cerebellum of mutant embryos (B, D, F, H, J, and L), as compared with that of control embryos (A, C, E, G, I, and K), was analyzed by hematoxylin and eosin staining (A–D) and immunostaining of calbindin (E–H) and Pax2 (I–L) at E14.5 (A, B, E, F, I, and J) and E18.5 (C, D, G, H, K, and L). Arrows in A and B point at the cells of the developing deep cerebellar nuclei. Arrowheads in C, E, and G point out the developing Purkinje cells in the cerebellum. The arrowhead in H shows residual calbindin-positive cells present in the Lhx1/Lhx5 double mutants. (Scale bar: 100 μm.)
Defect in Purkinje Cell-Derived Cell–Cell Signaling in Lhx1/Lhx5 Double Mutants.
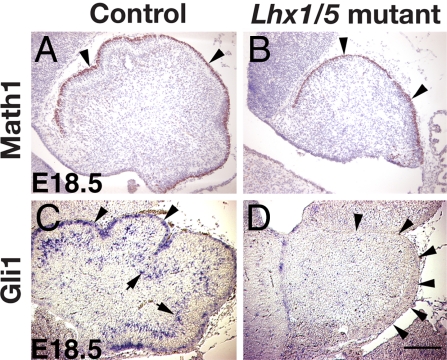
Despite the defect in Purkinje cell development, granule cells, marked by Math-1 immunostaining (11), were specified properly in Lhx1/Lhx5 double mutants (Fig. 3 A and B). Purkinje cells have been reported to interact with granule cell precursors via a Shh signaling pathway (1–3). Gli1, a Shh target gene, was detected in both Purkinje and granule cells of control embryos (3) (Fig. 3C). Consistent with the lack of Purkinje cells in Lhx1/Lhx5 double mutants, the expression of Gli1 was missing in the cerebellum of the mutants (Fig. 3D).
Fig. 3.
Purkinje cell-derived cell–cell signaling defect in the cerebellum of Lhx1/Lhx5 double mutants. (A and B) At E18.5, the granule cells in the developing cerebellum of both control (A) and Lhx1/Lhx5 double-mutant (B) embryos were labeled by immunostaining of Math1 (arrowheads). (C and D) Gli1 mRNA was detected by in situ hybridization in the Purkinje (arrows) and granule (arrowheads) cells of control embryos (C) but was largely missing in Lhx1/Lhx5 double-mutant embryos (D). Arrowheads in D point at the perimeter of the cerebellum. (Scale bar: 100 μm.)
Disappearance of Lhx1-Expressing Cells in the Cerebellum of Lhx1/Lhx5 Double Mutants.
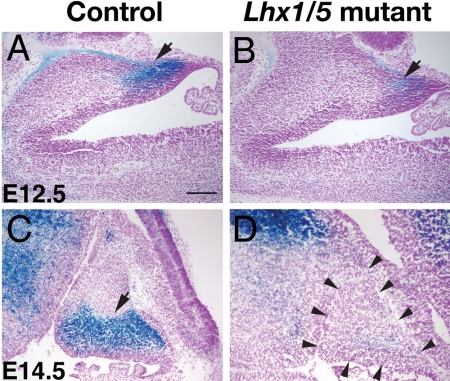
To follow the cells that normally express Lhx1 in the cerebellum of the Lhx1/Lhx5 double mutants, we analyzed the activity of β-galactosidase expressed from the LacZ reporter gene that replaced the Lhx1 coding sequence. At E12.5, β-galactosidase activity was detected in postmitotic cells that had migrated out of the ventricular zone of the developing cerebellum, but the staining was significantly reduced in Lhx1/Lhx5 double mutants compared to that of the controls (Fig. 4 A and B). At E13.5, E14.5, and E15.5, the β-galactosidase-positive cells were largely missing in the cerebellum of Lhx1/Lhx5 double mutants, whereas they were present in the controls (Fig. 4 C and D; and SI Fig. 11). Because β-galactosidase-positive cells still were detected in other brain regions of the Lhx1/Lhx5 double mutants (Fig. 4D), it appeared that the cells that normally express Lhx1 are competent for maintaining Lhx1 expression in the absence of Lhx1 and Lhx5 function. Our results suggest that these cells may start to disappear soon after they migrate out of the ventricular zone of the cerebellum in Lhx1/Lhx5 double mutants and subsequently are largely eliminated.
Fig. 4.
Gradual reduction of Purkinje cells expressing the Lhx1–LacZ reporter gene in the cerebellum of Lhx1/Lhx5 double mutants. Cells expressing the Lhx1–LacZ reporter gene in the cerebellum of control (A and C) and Lhx1/Lhx5 double-mutant (B and D) embryos were labeled by X-gal staining for β-galactosidase activity (arrows). (A and B) E12.5. (C and D) E14.5. Arrowheads in D point at the perimeter of the cerebellum. (Scale bar: 100 μm.)
Defect in Purkinje Cell Development in Ldb1 Conditional Mutants.
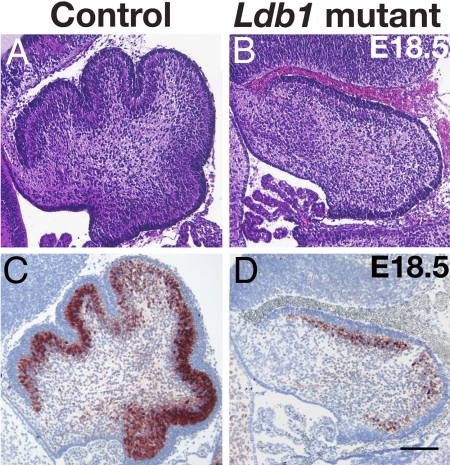
The function of LIM-homeodomain proteins is mediated by the obligatory cofactor Ldb1 (22). Ldb1 mRNA was expressed extensively in the developing cerebellum (SI Fig. 12). A germ-line deletion of Ldb1 causes very early embryonic lethality (24). In an effort to analyze the role of Ldb1 in the development of the cerebellum, we generated a conditional allele by inserting loxP sites into the Ldb1 locus flanking the Ldb1 coding sequence (SI Fig. 12) (38). To target Ldb1 deletion in postmitotic neurons in the developing cerebellum, we crossed the Ldb1 conditional mutants with the Nes-Cre+ mice. At E18.5, the cerebellum of the conditional Ldb1 mutants appeared small and lacked the foliation seen in control embryos (Fig. 5 A and B). Calbindin-positive Purkinje cells were greatly reduced in number as compared to those in the controls (Fig. 5 C and D). Thus, the deletion of Ldb1 resulted in a cerebellar phenotype that resembled that of Lhx1/Lhx5 double mutants.
Fig. 5.
Defects in the development of the cerebellum in Ldb1 conditional mutants. E18.5 conditional Ldb1 mutant (B and D) and control (A and C) embryos were analyzed by hematoxylin and eosin (A and B) and by immunostaining of calbindin (C and D). (Scale bar: 100 μm.)
Discussion
Our results show that the LIM-homeobox genes Lhx1 and Lhx5 were expressed in precursors that just exited the cell cycle and migrated out of the cerebellar ventricular zone to assume the Purkinje cell identity. When the function of both Lhx1 and Lhx5 was inactivated, the differentiation of most of these postmitotic cells was compromised, resulting in a severe reduction in number of Purkinje cells. Thus, the LIM-homeodomain factors encoded by these two genes play essential roles in the differentiation of Purkinje cells in the developing cerebellum. In addition, our study has underscored the obligatory function of the Ldb1 cofactor in this process.
A number of factors are known to be involved in Purkinje cell migration or survival. The signaling molecule reelin and its downstream transducer Dab1 are essential for the regulation of Purkinje cell migration (39–41). Survival of Purkinje cells is mediated by genes encoding nuclear factors Ror-α (42, 43) and Nna1 (44), and by neurotrophic factors such as the nerve growth factor, brain-derived neurotrophic factor, ciliary neurotrophic factor, neurotrophin-4/5, and acetylcholine (45, 46). Most recently, the transcription factor Ptf1a has been shown to be required for the specification of progenitors of all GABAergic neurons, including Purkinje cells in the ventricular zone of the developing cerebellum (18, 19). Our studies went beyond these analyses in defining factors such as Lhx1 and Lhx5 that were directly involved in Purkinje cell differentiation. This finding opens a passage toward disclosing the complex genetic program that controls the identity of this important type of neurons in the developing central nervous system.
Despite the severe reduction in number of Purkinje cells in Lhx1/Lhx5 double mutants (Lhx1fx1/LacZ, Nes-Cre+; Lhx5−/−), a few calbindin-positive cells were present in the dorsal region of the cerebellum. The presence of these cells suggests that some of the precursors might escape the conditional inactivation of function of Lhx1 and eventually undergo proper differentiation. Consistent with this idea, the calbindin-positive cells rarely were observed when we used the En1Cre allele for inactivation of Lhx1 (Lhx1fx2/LacZ, En1Cre/+; Lhx5−/−, see SI Fig. 10). In these double mutants, Cre likely was turned on earlier than it was in mutants carrying the Nes-Cre+ allele. The presence of reduced number of calbindin-positive cells in the cerebellum of the Ldb1 conditional mutants likewise may represent escapers, in this case caused by incomplete functional inactivation of Ldb1.
Although the number of Purkinje cells was severely reduced in Lhx1/Lhx5 double mutants, the cerebellar development appeared normal after inactivation of either Lhx1 or Lhx5 alone, which indicates that Lhx1 and Lhx5 are functionally redundant in controlling Purkinje cell differentiation in the cerebellum. Functional redundancy of LIM-homeodomain proteins including Lhx1 and Lhx5 also has been observed in controlling differentiation of subtypes of neurons in the developing spinal cord (47, 48).
Our study of Lhx1 and Lhx5, together with recent studies of Ptf1a (18, 19, 49), suggests that similar mechanisms of transcriptional regulation may operate in the cerebellum and the spinal cord to control the generation of specific types of neurons. In the cerebellum, Ptf1a is required for the specification of progenitors of all GABAergic neurons, including Purkinje cells (18, 19), in which Lhx1 and Lhx5 are expressed and required for their normal differentiation. Ptf1a and Lhx1/Lhx5 also are involved in the generation of subsets of GABAergic interneurons in the dorsal spinal cord (48, 49). Ptf1a and Lhx1/Lhx5 thus seem to function at sequential steps in a same genetic program that controls the generation of specific neuronal cell types in different regions of the developing central nervous system.
Materials and Methods
Animals.
A conditional mutant allele of Lhx1 (Lhx1fx1) was generated by inserting a loxP site and a loxP-flanked neomycin-resistance gene (Neo) cassette (loxP–Neo–loxP) into the first and fourth introns of the Lhx1 gene, respectively, with the use of homologous recombination in R1 mouse embryonic stem cells (SI Fig. 6A). The targeted embryonic stem cell clones were double-selected by G418 and gancyclovir and screened with Southern hybridization analysis (SI Fig. 6B). Targeted embryonic stem cells were injected into C57BL/6 (B6) blastocysts to generate chimeric mice that carry the mutant allele in the germ line. The chimeric mice were mated to wild-type B6 mice to generate heterozygous animals (Lhx1fx1/+) that subsequently were crossed to produce homozygous Lhx1 conditional mutants (Lhx1fx1/fx1). The Lhx1fx1/fx1 animals appeared healthy. They were fertile and lived a normal lifespan. The Lhx1fx1 allele was genotyped by PCR using a pair of primers (L1-PsiS, 5′-atcagaggtctgtgtggctcta-3′; L1-PsiA, 5′-tgctgagctgaagccatttcaga-3′) flanking the loxP site in the first intron of the Lhx1 gene. The PCR amplified a 230-bp band from the wild-type Lhx1 allele and a 280-bp band from the Lhx1fx1 allele, respectively.
The Ldb1 conditional mutant allele was generated by inserting a loxP site and a loxP–Neo–loxP cassette into the upstream region and the ninth intron of the Ldb1 gene, respectively (SI Fig. 12D) (38). Mice heterozygous for the Ldb1 conditional allele were crossed with EII–Cre transgenic mice (50) that express Cre transiently in the zygote to produce chimeric offspring with a selective deletion of the loxP–Neo–loxP cassette. These chimeric animals were mated to wild-type B6 mice to generate heterozygous Ldb1 conditional mutants (Ldb1fx/+) that did not contain the Neo gene. The Ldb1fx/+ mice were crossed to produce homozygous Ldb1 conditional mutants (Ldb1fx/fx). The Ldb1fx/fx mice were healthy, fertile, and lived a normal lifespan. The Ldb1fx allele was genotyped by PCR using a pair of primers (Ldb1-sense, 5′-cagcaaacggaggaaacggaagatgtcag-3′; Ldb1G, 5′-cttatgtgaccacagccatgcatgcatgtg-3′) flanking the loxP site in the upstream region of the Ldb1 gene. The PCR amplified a 350-bp band from the Ldb1 wild-type allele and a 400-bp band from the Ldb1fx allele, respectively.
The generation of other transgenic mice used in this study, which included another Lhx1 conditional mutant (Lhx1fx2/fx2) (33), the Lhx1LacZ/+ mutant (29), Lhx5 mutant (30), En1Cre/+ (51), Nes-Cre+ (52), and Rosa26stop−LacZ (53) mice, have been described previously. The breeding, maintenance, and use of animals were in compliance with guidelines approved by the animal use committees of the National Institute of Child Health and Human Development and the University of Texas M. D. Anderson Cancer Center, respectively.
Histology, in Situ Hybridization, and Immunohistochemistry.
To obtain embryos for analysis, mice were mated and checked daily for presence of vaginal plugs. Noon of the day when a plug was detected was designated as E0.5. Embryonic tissues were collected at various developmental stages and fixed in 4% paraformaldehyde in 0.1 M sodium phosphate buffer (pH 7.4) overnight at 4°C. After several washes in PBS, tissues were either dehydrated in ethanol, embedded in paraffin, and sectioned at a thickness of 5 μm or incubated in 30% sucrose in PBS, frozen in OCT compound (Sakura Finetek, Torrance, CA), and sectioned at a thickness of 14 μm. For histological analysis, paraffin sections were stained with hematoxylin and eosin (Sigma, St. Louis, MO). For in situ hybridization, either paraffin or frozen sections were hybridized with 33P-labeled or digoxigenin-labeled probes, respectively, following published procedures (54, 55). The probes used included those for Lhx1 (56), Lhx5 (36), and Gli1 (3). Immunohistochemical staining of Math1 was performed on frozen sections by using a rabbit anti-Math1 antibody (1:100; a gift from Jane Johnson, University of Texas, Dallas, TX) and the ABC elite kit (Vector Laboratory, Burlingame, CA) following procedures suggested by the manufacturer. Immunostaining of calbindin was performed on paraffin sections with a rabbit antibody (1:1,000; Chemicon, Temecula, CA) by following published procedures (57). For immunodetection of Pax2, paraffin sections were pretreated for antigen retrieval (PickCell 2100-Retriever; Electron Microscopy Sciences, Hatfield, PA) and then stained by using a rabbit antibody (1:200; Invitrogen, Carlsbad, CA) and the ABC elite kit.
X-Gal Staining for β-Galactosidase Activity.
X-gal staining of whole embryos (E12.5 or younger) was performed by following published procedures (58). After the staining, embryos were postfixed in 4% paraformaldehyde in phosphate buffer (pH 7.4), embedded in paraffin, and sectioned for further observation. Tissues from embryos older than E12.5 were fixed in 4% paraformaldehyde in phosphate buffer (0.1 M, pH 7.4) for 2–4 h at 4°C. After washes in PBS, the tissues were incubated in 30% sucrose, frozen in OCT compound, and sectioned at a thickness of 14 μm. The sections were stained in the same solution used for whole-embryo X-gal staining.
BrdU Labeling of Proliferating Cells.
For labeling of proliferating cells in developing embryos, pregnant females were injected i.p. with BrdU dissolved in saline (100 μg per g of body weight; Sigma). After an hour, the animals were killed. Embryos were dissected and first processed for X-gal staining as described above. Thereafter, they were paraffin-embedded, sectioned, and processed for detection of BrdU by a peroxidase-labeled anti-BrdU antibody (Roche, Indianapolis, IN) following published procedures (30, 57).
Northern Blot Analysis.
Total RNAs were isolated from the heads of E11.5 Lhx1 conditional mutant (Lhx1fx1/fx1; Nes-Cre+) and control embryos with TriPure RNA isolation reagent (Roche). The RNAs were separated on a gel, transferred to nylon membrane, and hybridized with a 500-bp cDNA probe derived from a part of the loxP-flanked region of the Lhx1 gene following published procedures (57). The blot also was hybridized with a probe corresponding to the GAPDH housekeeping gene to ensure equal loading and transfer of the various RNA samples.
Supplementary Material
Acknowledgments
We thank Drs. Thomas Jessell (Columbia University, New York, NY) and Jane Johnson (University of Texas, Dallas, TX) for providing Lhx1LacZ/+ mice and the antibody to Math1, respectively. We thank SingPing Huang and Donna C. Morales for technical assistance. This work was supported by funds from the Intramural Research Program of the National Institute of Child Health and Human Development (to H.W.), National Institutes of Health Grant HD30284, and the Ben F. Love Chair for Cancer Research Fund (to R.R.B.).
Abbreviation
- En
embryonic day n.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705464104/DC1.
References
- 1.Wechsler-Reya RJ, Scott MP. Neuron. 1999;22:103–114. doi: 10.1016/s0896-6273(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 2.Lewis PM, Gritli-Linde A, Smeyne R, Kottmann A, McMahon AP. Dev Biol. 2004;270:393–410. doi: 10.1016/j.ydbio.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Corrales JD, Rocco GL, Blaess S, Guo Q, Joyner AL. Development (Cambridge, UK) 2004;131:5581–5590. doi: 10.1242/dev.01438. [DOI] [PubMed] [Google Scholar]
- 4.Solecki DJ, Liu XL, Tomoda T, Fang Y, Hatten ME. Neuron. 2001;31:557–568. doi: 10.1016/s0896-6273(01)00395-6. [DOI] [PubMed] [Google Scholar]
- 5.Yang XW, Wynder C, Doughty ML, Heintz N. Nat Genet. 1999;22:327–335. doi: 10.1038/11896. [DOI] [PubMed] [Google Scholar]
- 6.Aruga J, Minowa O, Yaginuma H, Kuno J, Nagai T, Noda T, Mikoshiba K. J Neurosci. 1998;18:284–293. doi: 10.1523/JNEUROSCI.18-01-00284.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aruga J, Inoue T, Hoshino J, Mikoshiba K. J Neurosci. 2002;22:218–225. doi: 10.1523/JNEUROSCI.22-01-00218.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alder J, Lee KJ, Jessell TM, Hatten ME. Nat Neurosci. 1999;2:535–540. doi: 10.1038/9189. [DOI] [PubMed] [Google Scholar]
- 9.Rios I, Alvarez-Rodriguez R, Marti E, Pons S. Development (Cambridge, UK) 2004;131:3159–3168. doi: 10.1242/dev.01188. [DOI] [PubMed] [Google Scholar]
- 10.Qin L, Wine-Lee L, Ahn KJ, Crenshaw EB., III J Neurosci. 2006;26:1896–1905. doi: 10.1523/JNEUROSCI.3202-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ben-Arie N, Bellen HJ, Armstrong DL, McCall AE, Gordadze PR, Guo Q, Matzuk MM, Zoghbi HY. Nature. 1997;390:169–172. doi: 10.1038/36579. [DOI] [PubMed] [Google Scholar]
- 12.Gazit R, Krizhanovsky V, Ben-Arie N. Development (Cambridge, UK) 2004;131:903–913. doi: 10.1242/dev.00982. [DOI] [PubMed] [Google Scholar]
- 13.Miyata T, Maeda T, Lee JE. Genes Dev. 1999;13:1647–1652. doi: 10.1101/gad.13.13.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hatten ME, Heintz N. Annu Rev Neurosci. 1995;18:385–408. doi: 10.1146/annurev.ne.18.030195.002125. [DOI] [PubMed] [Google Scholar]
- 15.Goldowitz D, Hamre K. Trends Neurosci. 1998;21:375–382. doi: 10.1016/s0166-2236(98)01313-7. [DOI] [PubMed] [Google Scholar]
- 16.Wang VY, Zoghbi HY. Nat Rev Neurosci. 2001;2:484–491. doi: 10.1038/35081558. [DOI] [PubMed] [Google Scholar]
- 17.Chizhikov V, Millen KJ. Mol Genet Metab. 2003;80:54–65. doi: 10.1016/j.ymgme.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 18.Hoshino M, Nakamura S, Mori K, Kawauchi T, Terao M, Nishimura YV, Fukuda A, Fuse T, Matsuo N, Sone M, et al. Neuron. 2005;47:201–213. doi: 10.1016/j.neuron.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Pascual M, Abasolo I, Mingorance-Le Meur A, Martinez A, Del Rio JA, Wright CV, Real FX, Soriano E. Proc Natl Acad Sci USA. 2007;104:5193–5198. doi: 10.1073/pnas.0605699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shirasaki R, Pfaff SL. Annu Rev Neurosci. 2002;25:251–281. doi: 10.1146/annurev.neuro.25.112701.142916. [DOI] [PubMed] [Google Scholar]
- 21.Zhao Y, Malik N, Westphal H. In: Transcription Factors in the Nervous System: Development, Brain Function, and Diseases. Thiel G, editor. Weinheim, Germany: Wiley; 2006. pp. 75–94. [Google Scholar]
- 22.Agulnick AD, Taira M, Breen JJ, Tanaka T, Dawid IB, Westphal H. Nature. 1996;384:270–272. doi: 10.1038/384270a0. [DOI] [PubMed] [Google Scholar]
- 23.Thaler JP, Lee SK, Jurata LW, Gill GN, Pfaff SL. Cell. 2002;110:237–249. doi: 10.1016/s0092-8674(02)00823-1. [DOI] [PubMed] [Google Scholar]
- 24.Mukhopadhyay M, Teufel A, Yamashita T, Agulnick AD, Chen L, Downs KM, Schindler A, Grinberg A, Huang SP, Dorward D, Westphal H. Development (Cambridge, UK) 2003;130:495–505. doi: 10.1242/dev.00225. [DOI] [PubMed] [Google Scholar]
- 25.Furuyama T, Inagaki S, Iwahashi Y, Takagi H. Neurosci Lett. 1994;170:266–268. doi: 10.1016/0304-3940(94)90334-4. [DOI] [PubMed] [Google Scholar]
- 26.Hayes WP, Yangco N, Chin H, Mill JF, Pu LP, Taira M, Dawid IB, Gallo V. J Neurosci Res. 2001;63:237–251. doi: 10.1002/1097-4547(20010201)63:3<237::AID-JNR1017>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 27.Morales D, Hatten ME. J Neurosci. 2006;26:12226–12236. doi: 10.1523/JNEUROSCI.3493-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hallonet ME, Le Douarin NM. Eur J Neurosci. 1993;5:1145–1155. doi: 10.1111/j.1460-9568.1993.tb00969.x. [DOI] [PubMed] [Google Scholar]
- 29.Kania A, Johnson RL, Jessell TM. Cell. 2000;102:161–173. doi: 10.1016/s0092-8674(00)00022-2. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Y, Sheng HZ, Amini R, Grinberg A, Lee E, Huang S, Taira M, Westphal H. Science. 1999;284:1155–1158. doi: 10.1126/science.284.5417.1155. [DOI] [PubMed] [Google Scholar]
- 31.Paylor R, Zhao Y, Libbey M, Westphal H, Crawley JN. Physiol Behav. 2001;73:781–792. doi: 10.1016/s0031-9384(01)00515-7. [DOI] [PubMed] [Google Scholar]
- 32.Shawlot W, Behringer RR. Nature. 1995;374:425–430. doi: 10.1038/374425a0. [DOI] [PubMed] [Google Scholar]
- 33.Kwan KM, Behringer RR. Genesis. 2002;32:118–120. doi: 10.1002/gene.10074. [DOI] [PubMed] [Google Scholar]
- 34.Tsang TE, Shawlot W, Kinder SJ, Kobayashi A, Kwan KM, Schughart K, Kania A, Jessell TM, Behringer RR, Tam PP. Dev Biol. 2000;223:77–90. doi: 10.1006/dbio.2000.9733. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi A, Kwan KM, Carroll TJ, McMahon AP, Mendelsohn CL, Behringer RR. Development (Cambridge, UK) 2005;132:2809–2823. doi: 10.1242/dev.01858. [DOI] [PubMed] [Google Scholar]
- 36.Sheng HZ, Bertuzzi S, Chiang C, Shawlot W, Taira M, Dawid I, Westphal H. Dev Dyn. 1997;208:266–277. doi: 10.1002/(SICI)1097-0177(199702)208:2<266::AID-AJA13>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 37.Maricich SM, Herrup K. J Neurobiol. 1999;41:281–294. doi: 10.1002/(sici)1097-4695(19991105)41:2<281::aid-neu10>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 38.Suleiman H, Heudobler D, Raschta AS, Zhao Y, Zhao Q, Hertting I, Vitzthum H, Moeller MJ, Holzman LB, Rachel R, et al. Dev Biol. 2007;304:701–712. doi: 10.1016/j.ydbio.2007.01.020. [DOI] [PubMed] [Google Scholar]
- 39.D'Arcangelo G, Miao GG, Chen SC, Soares HD, Morgan JI, Curran T. Nature. 1995;374:719–723. doi: 10.1038/374719a0. [DOI] [PubMed] [Google Scholar]
- 40.Sheldon M, Rice DS, D'Arcangelo G, Yoneshima H, Nakajima K, Mikoshiba K, Howell BW, Cooper JA, Goldowitz D, Curran T. Nature. 1997;389:730–733. doi: 10.1038/39601. [DOI] [PubMed] [Google Scholar]
- 41.Howell BW, Hawkes R, Soriano P, Cooper JA. Nature. 1997;389:733–737. doi: 10.1038/39607. [DOI] [PubMed] [Google Scholar]
- 42.Dussault I, Fawcett D, Matthyssen A, Bader JA, Giguere V. Mech Dev. 1998;70:147–153. doi: 10.1016/s0925-4773(97)00187-1. [DOI] [PubMed] [Google Scholar]
- 43.Hamilton BA, Frankel WN, Kerrebrock AW, Hawkins TL, FitzHugh W, Kusumi K, Russell LB, Mueller KL, van Berkel V, Birren BW, et al. Nature. 1996;379:736–739. doi: 10.1038/379736a0. [DOI] [PubMed] [Google Scholar]
- 44.Fernandez-Gonzalez A, La Spada AR, Treadaway J, Higdon JC, Harris BS, Sidman RL, Morgan JI, Zuo J. Science. 2002;295:1904–1906. doi: 10.1126/science.1068912. [DOI] [PubMed] [Google Scholar]
- 45.Larkfors L, Lindsay RM, Alderson RF. J Neurochem. 1996;66:1362–1373. doi: 10.1046/j.1471-4159.1996.66041362.x. [DOI] [PubMed] [Google Scholar]
- 46.Mount HT, Dreyfus CF, Black IB. J Neurochem. 1994;63:2065–2073. doi: 10.1046/j.1471-4159.1994.63062065.x. [DOI] [PubMed] [Google Scholar]
- 47.Sharma K, Sheng HZ, Lettieri K, Li H, Karavanov A, Potter S, Westphal H, Pfaff SL. Cell. 1998;95:817–828. doi: 10.1016/s0092-8674(00)81704-3. [DOI] [PubMed] [Google Scholar]
- 48.Pillai A, Mansouri A, Behringer R, Westphal H, Goulding M. Development (Cambridge, UK) 2007;134:357–366. doi: 10.1242/dev.02717. [DOI] [PubMed] [Google Scholar]
- 49.Glasgow SM, Henke RM, Macdonald RJ, Wright CV, Johnson JE. Development (Cambridge, UK) 2005;132:5461–5469. doi: 10.1242/dev.02167. [DOI] [PubMed] [Google Scholar]
- 50.Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, Alt FW, Westphal H. Proc Natl Acad Sci USA. 1996;93:5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kimmel RA, Turnbull DH, Blanquet V, Wurst W, Loomis CA, Joyner AL. Genes Dev. 2000;14:1377–1389. [PMC free article] [PubMed] [Google Scholar]
- 52.Tronche F, Kellendonk C, Kretz O, Gass P, Anlag K, Orban PC, Bock R, Klein R, Schutz G. Nat Genet. 1999;23:99–103. doi: 10.1038/12703. [DOI] [PubMed] [Google Scholar]
- 53.Soriano P. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 54.Robinson GW, Wray S, Mahon KA. New Biol. 1991;3:1183–1194. [PubMed] [Google Scholar]
- 55.Schaeren-Wiemers N, Gerfin Moser A. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. (1993) [DOI] [PubMed] [Google Scholar]
- 56.Fujii T, Pichel JG, Taira M, Toyama R, Dawid IB, Westphal H. Dev Dyn. 1994;199:73–83. doi: 10.1002/aja.1001990108. [DOI] [PubMed] [Google Scholar]
- 57.Zhao Y, Morales DC, Hermesz E, Lee WK, Pfaff SL, Westphal H. Mech Dev. 2006;123:605–613. doi: 10.1016/j.mod.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 58.Sanes JR, Rubenstein JL, Nicolas JF. EMBO J. 1986;5:3133–3142. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.