Abstract
The interaction of membrane-embedded voltage-activated potassium channels (Kv) with intracellular scaffold proteins, such as the postsynaptic density 95 (PSD-95) protein, is mediated by the channel C-terminal segment. This interaction underlies Kv channel clustering at unique membrane sites and is important for the proper assembly and functioning of the synapse. In the current study, we address the molecular mechanism underlying Kv/PSD-95 interaction. We provide experimental evidence, based on hydrodynamic and spectroscopic analyses, indicating that the isolated C-terminal segment of the archetypical Shaker Kv channel (ShB-C) is a random coil, suggesting that ShB-C belongs to the recently defined class of intrinsically disordered proteins. We show that isolated ShB-C is still able to bind its scaffold protein partner and support protein clustering in vivo, indicating that unfoldedness is compatible with ShB-C activity. Pulldown experiments involving C-terminal chains differing in flexibility or length further demonstrate that intrinsic disorder in the C-terminal segment of the Shaker channel modulates its interaction with the PSD-95 protein. Our results thus suggest that the C-terminal domain of the Shaker Kv channel behaves as an entropic chain and support a “fishing rod” molecular mechanism for Kv channel binding to scaffold proteins. The importance of intrinsically disordered protein segments to the complex processes of synapse assembly, maintenance, and function is discussed.
Keywords: channel clustering, intrinsically disordered, PSD-95
Voltage-activated potassium channels (Kv) are modular membrane-spanning proteins that undergo conformational transitions between closed and open states in response to changes in membrane potential (1–3). This form of gating underlies many fundamental biological processes, in particular the generation of nerve and muscle action potentials (4). The involvement of Kv channels in shaping action potentials is primarily based on the tight interaction between the channel's voltage-sensing and pore domains (4). Effective transmission of the action potential to a target cell across the synaptic cleft requires precise channel localization and clustering (5, 6), a process mediated by the C-terminal segment of certain Kv channels (7, 8). It has been demonstrated that a conserved PDZ-binding motif present at the end of the C-terminal segment of the prototypical Shaker Kv channel is an important determinant for binding to the Drosophila postsynaptic density 95 (PSD-95) scaffold protein, Dlg, a member of the membrane-associated guanylate kinase family (5, 6, 9). This interaction is responsible for channel clustering at the fly neuromuscular junction and has been implicated in synaptic growth and plasticity (5, 6).
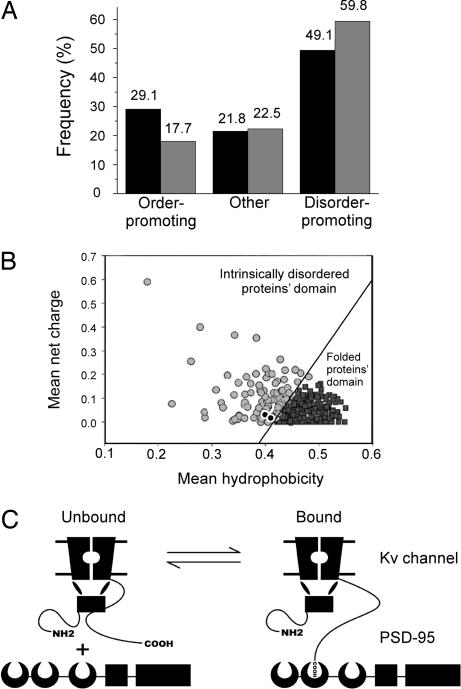
The molecular mechanism underlying the interaction of PSD-95 and Kv channels is not yet clear. Previous studies highlighted the role played by the 4-aa-long C-terminal PDZ-binding motif of the channel while ignoring the long stretch of residues to which this motif is attached. Recently, however, we have presented bioinformatics evidence to argue that intrinsic disorder at the C-terminal tail segments of Kv channels preceding the PDZ-binding motif is another important determinant for channel binding to the PSD-95 scaffold protein partner (10). This conclusion was based on the unusual sequence characteristics of the C-terminal segments of many Kv channel members. Inspection of the C-terminal sequence of the Shaker Kv channel (sw: P08510), for example, reveals an enrichment in hydrophilic amino acids, depletion of hydrophobic residues, and repetitive strings of glutamines. Analysis of amino acid usage in the Shaker C-terminal sequence reveals a depletion in order-promoting amino acids (i.e., W, C, F, I, Y, V, and L) and an enrichment in disorder-promoting residues (i.e., S, A, P, R, E, K, and Q), relative to the overall amino acid usage in the entire Drosophila melanogaster proteome (Fig. 1A). Such sequence characteristics, together with consideration of mean net charge and mean hydrophobicity values, explains why the C-terminal segments of both the Shaker and rat Kv 1.2 channels, for instance, are expected to belong to the recently defined intrinsically disordered protein family (Fig. 1B) (11). Protein segments belonging to this group bear sequence characteristics that oppose folding and are, therefore, unstructured or intrinsically disordered under physiological conditions (12–17). The lack of or ambiguity regarding structure at the C-terminal segments of the Kv 1.2 and Shaker channels, as revealed by x-ray crystallography (18) and cryo-EM (19) analyses, respectively, lends credence to this conclusion.
Fig. 1.
The C-terminal tail domain of the Shaker Kv channel is predicted to be intrinsically disordered. (A) Comparison of the relative order- and disorder-promoting amino acid content across the entire Drosophila proteome (black bars) and in ShB-C (gray bars). (B) The mean net charge and mean hydrophobicity values of the Shaker and Kv 1.2 channels (black circles) lie within the intrinsically disordered protein domain of Uversky's phase-space diagram. The solid line is an empirical linear regression that defines the boundary between folded (black squares) and intrinsically disordered (gray circles) proteins (adapted with permission from ref. 11). (C) A fishing rod mechanism for Kv channel binding to scaffold proteins. A voltage-gated K+ channel interacts with the PSD-95 scaffold protein upon binding of the C-terminal PDZ-binding motif hook, tethered to an intrinsically disordered extended chain. The moon-shape, box, and rectangular shapes represent the PDZ, SH3, and guanylate kinase domains of the PSD-95 protein, respectively.
To understand the functional advantages of an intrinsically disordered C-terminal segment for the Kv channel protein, phylogenetic inference analysis of the Kv channel family was conducted (10). Such analysis provided evidence that, throughout evolution, the appearance of intrinsic disorder at the channel's C-terminal tail is associated with the presence of the PDZ-binding motif in the same part of the protein, implying that both tail region traits are important for PSD-95 scaffold protein binding (10). Based on this assertion, an intermolecular “fishing rod” mechanism for Kv channel binding to scaffold proteins was proposed (Fig. 1C) (10) in which the membrane-embedded Kv channel is anchored at unique membrane sites by binding to a scaffold protein using a C-terminal sequence that contains a flexible string (an intrinsically disordered chain) and a hook (the PDZ-binding motif) at its tip. Disorder at the channel C terminus would provide the orientational freedom needed for searching and successfully connecting to the PSD-95 scaffold protein partner.
In the present report, we have experimentally addressed predictions emerging from sequence and phylogenetic analyses to assess the feasibility of the fishing rod mechanism for Kv channel clustering proposed above. We demonstrate that the isolated C-terminal domain of the Shaker channel is indeed intrinsically disordered, that it can bind its PSD-95 scaffold protein partner, and that it can support clustering in vivo in the absence of the membrane and channel portion of the protein. Furthermore, we demonstrate that the intrinsically disordered nature of the Shaker channel C-terminal tail modulates its interaction with the PSD-95 scaffold protein, thus supporting the fishing rod mechanism of Kv channel clustering by PSD-95.
Results
The C-Terminal Domain of the Shaker K+ Channel Is Intrinsically Disordered.
Previous structural analysis of the Kv 1.2 channel by x-ray crystallography (18) showed no electron density corresponding to the C-terminal region. Difference density calculations from 2D averages and 3D reconstructions derived from electron microscopy images of full-length Shaker channels vs. a C-terminally truncated mutant (19) indicated a region that could correspond to all or part of the C-terminal segment, although the mass associated with this region could not be reliably determined. Taken together, these results are consistent with a lack of ordered structure or a flexible orientation for the C-terminal region of the channel relative to the membrane-spanning portion.
To experimentally assess the structural state of the Shaker channel C-terminal segment, ShB-C, one must first determine an appropriate N-terminal boundary for this region. Boundaries of long intrinsically disordered loops inserted into structural domains were shown to correlate with the boundaries of exons in the encoding gene (20). Although ShB-C corresponds to an extradomain segment and not an intradomain loop, we examined the Shaker channel gene structure and noted that the codon encoding Val-513 marked the start of the exon following the sixth transmembrane helix of the protein. This objective assignment of the N-terminal boundary of ShB-C proved useful, as protein constructs starting upstream of Val-513 aggregated during purification (not shown).
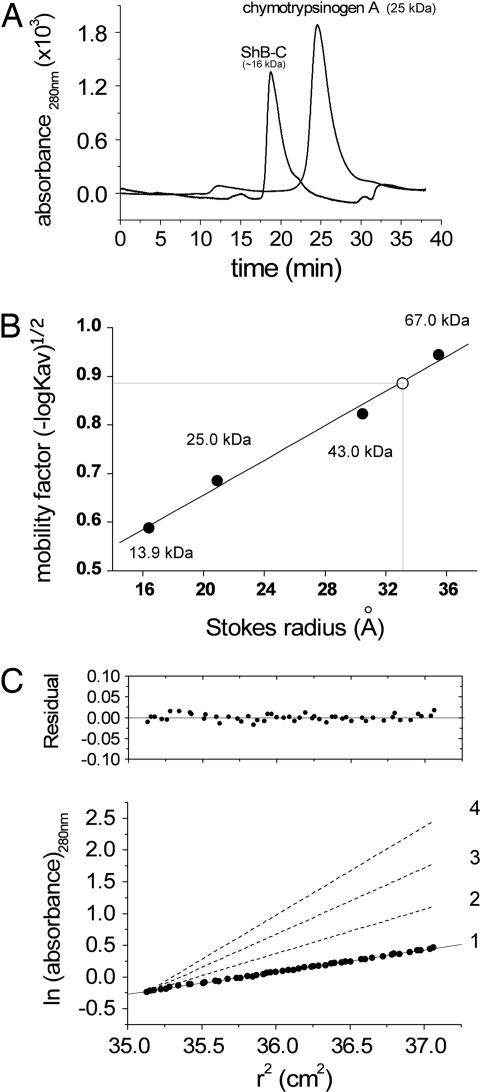
We next cloned, expressed, and purified the C-terminal domain of the Shaker channel [see supporting information (SI) Text] starting from Val-513 and examined, using hydrodynamic and spectroscopic approaches, its structural state when isolated from the rest of the channel and the membrane. ShB-C migrates in a size-exclusion column unusually fast for a 15.9-kDa protein (Fig. 2A). Compared with the 19.7-Å Stokes radius expected for a compact globular protein of this size (21), a Stokes radius of 32.9 Å was estimated for ShB-C, based on a calibration curve created using standard proteins of known molecular weight (Fig. 2B). This anomaly could be explained either by ShB-C oligomerization or by an expanded or elongated monomeric state of this domain. To discriminate between the two possibilities, analytical ultracentrifugation was performed. The data obtained were consistent with a monodispersed population of protein with molecular mass of 15.5 kDa, in good agreement with the monomer molecular mass calculated from the protein sequence (15,916 Da), confirmed by mass spectrometry. The gel filtration and analytical ultracentrifugation results can be reconciled by calculation of the expected Stokes radius for an intrinsically disordered monomeric protein of 15.9 kDa. The calculation, which yielded a value of 33.1 Å, was performed according to Uversky (21), who observed that the experimentally measured Stokes radii of known random coil proteins correlated linearly with the proteins' molecular weight [this relation is described by: logRs(coil) = −0.551 + 0.493 × logM, where Rs(coil) is the expected Stokes radius for a random coil chain, and M is the protein's molecular weight]. Thus, we conclude that ShB-C behaves as a random coil with very low compactness. Dynamic light scattering further supports this conclusion, yielding a Stokes radius of 37.2 Å (data not shown; see SI Text for further details).
Fig. 2.
Hydrodynamic analyses reveal an ShB-C structure with low compactness. (A) Size-exclusion chromatography elution profiles of ShB-C and chymotrypsinogen A, monitored at 280 nm. (B) Mobility [(−logKav)1/2; see Materials and Methods] vs. Stokes radius plot, used for Stokes radius determination of ShB-C, based on analytical size-exclusion chromatography of standard monomeric molecular weight markers. The open circle corresponds to ShB-C. The black circles correspond to molecular weight markers, as indicated. (C) Equilibrium sedimentation of ShB-C at 5°C and 19,000 rpm. Representative data are plotted as ln (absorbance) against the square of the radius from the axis of rotation. The slope is proportional to the molecular mass (see SI Text). Dashed lines with increasing slopes indicate calculated values for monomeric ShB-C (1), dimeric (2), trimeric (3), and tetrameric (4) forms of ShB-C. The data are consistent with a monomeric model for ShB-C, as judged by the residuals plot.
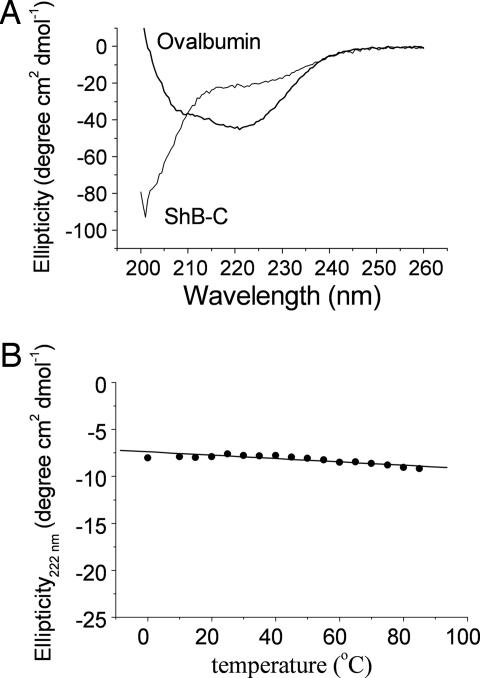
CD is a sensitive spectroscopic method for analyzing the presence or absence of secondary structural elements, in particular α-helices, within a protein. The far-UV CD spectrum for ShB-C (Fig. 3A) lacked the typical signatures of secondary structure. Neither negative peaks at 222 and 208 nm, typical for α-helices (Fig. 3A, see ovalbumin trace) nor the negative peak at ≈217 nm, typical for β-sheets, were observed in the CD spectrum of ShB-C. Instead, ShB-C exhibited a negative peak at 200 nm, indicative of a strong contribution from disordered structural elements, characteristic of a protein in a random coil conformation. Although ShB-C contains eight aromatic residues, its CD spectrum in the near-UV range (250–330 nm) was devoid of any ellipticity (not shown), indicating the absence of oriented aromatic residues typically present in the hydrophobic cores of compact globular proteins. The ellipticity of ShB-C at 222 nm did not change appreciably with increasing temperature up to 100°C and failed to reveal any cooperative transition (Fig. 3B). The slight negative slope of the signal as a function of temperature is further typical of random coil proteins. Although changes in environmental conditions can influence the distribution of conformations of intrinsically disordered proteins (21), the random coil signature of ShB-C, as revealed by CD analysis, was not altered over a wide range of pH values (3–12), salt concentrations (20–300 mM), or different concentrations of SDS or trifluoroethanol (not shown). The CD results indicate that ShB-C does not possess significant secondary structure and suggest a lack of fixed tertiary structure as well.
Fig. 3.
CD spectroscopic analysis reveals an extended conformation for ShB-C. (A) Comparison of the far-UV CD spectra of ovalbumin and ShB-C (0.5 mg/ml), obtained at room temperature (25°C). The CD spectrum of ShB-C was essentially similar at 90°C (not shown). (B) Temperature dependence of the molar ellipticity of ShB-C (0.125 mg/ml), followed at 222 nm.
Analysis of ShB-C by NMR provides unequivocal evidence for the lack of tertiary structure for this protein segment. The varied chemical environments experienced by protons of a folded protein result in a complex NMR spectrum with many nonoverlapping peaks dispersed over a wide range of chemical shifts. When, however, a protein is found to lack a fixed conformation, any given proton is under the influence of only those atoms to which it is bonded, thereby giving rise to a spectrum similar to that of noninteracting amino acids. Such a spectrum represents a summation of the NMR spectra of individual amino acids and will appear narrowly dispersed along the chemical shift axis with few resonating peaks. Accordingly, the 1H NMR spectrum of ShB-C (SI Fig. 6A) lacks the chemical shift dispersion typical of folded proteins, as exhibited by the 9-kDa snake venom toxin, α-bungarotoxin (22), shown for comparison (SI Fig. 6B). Rather, the profile of ShB-C resembles the 1H NMR spectrum of the C-terminal domain of gliotactin, a protein shown to be intrinsically disordered (SI Fig. 6C) (23). Hence, our results suggest that the C-terminal portion of the Shaker channel is an intrinsically disordered random chain.
The Isolated Shaker Channel Tail Domain Interacts with Its PSD-95 Scaffold Protein Partner to Mediate Protein Clustering in Vivo.
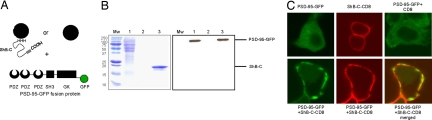
We next examined whether the intrinsically disordered isolated ShB-C, out of its membrane and channel contexts, is capable of binding its PSD-95 scaffold protein partner, and whether this interaction can mediate protein clustering in vivo. For this purpose, we transfected the Drosophila Schneider cell line with DNA encoding a PSD-95-GFP fusion protein and conducted pulldown assays using ShB-C as bait, as schematically depicted in Fig. 4A. Such an experimental design mimics the spatial constraint of membrane-integrated ShB-C, allowing us to probe the interaction of this Kv channel segment with PSD-95. The results reveal that His6-tagged ShB-C, immobilized on Ni2+ beads via its N terminus, is able to capture PSD-95 from a crude soluble protein extract of transformed Drosophila Schneider cells (Fig. 4B), leading us to conclude that ShB-C retains its function in this protocol. To assess whether the intrinsically disordered chain of ShB-C can support protein clustering in vivo, we next transfected Drosophila Schneider cells to express PSD-95-GFP and ShB-C fused to the CD8 membrane protein (6), either together or separately, and evaluated PSD-95 membrane association and ShB-C-CD8 clustering by confocal light microscopy (see SI Text). As presented in Fig. 4C, transfection with DNA encoding the PSD-95-GFP fusion construct alone resulted in homogeneous protein expression throughout the cytoplasm (Fig. 4C Upper Left). In the case of cells expressing the CD8-ShB-C chimera alone, homogeneous protein expression was observed along the plasma membrane, as reflected in the anti-CD8 antibody-stained red fluorescence pattern (Fig. 4C Upper Center). Cotransfection with constructs encoding the PSD-95-GFP chimera and the CD8 protein by itself yielded no change from the homogeneous cytoplasmic green fluorescence pattern obtained with PSD-95-GFP alone (Fig. 4C Upper Right). By contrast, cotransfection with both the PSD-95-GFP- and CD8-ShB-C-encoding constructs resulted in PSD-95 membrane association, as reflected in the green PSD-95-GFP-derived fluorescence pattern detected at the cell edge (Fig. 4C Lower Left). This time, however, the red CD8-ShB-C-derived fluorescence pattern, obtained using anti-CD8 antibodies and a suitable wavelength of detection, revealed a nonhomogeneous membrane distribution of CD8-ShB-C (Fig. 4C Lower Center). These discrete membrane domains of enhanced fluorescence intensity may reflect some degree of protein clustering. Indeed, examination of the membrane-associated yellow coloring of the merged cell images (Fig. 4C Lower Right) clearly shows overlap between the membrane association of PSD-95 and the redistribution of CD8-ShB-C in the presence of this scaffolding protein. The ability of the Shaker channel C-terminal tail to support protein clustering mediated by the PSD-95 protein was, furthermore, demonstrated at the whole organism level (6). As such, we suggest that ShB-C represents a functional module of the intact channel, and that the intrinsically unfolded state of the isolated ShB-C region is compatible with its biological function in the context of the intact protein.
Fig. 4.
Isolated ShB-C binds its PSD-95 scaffold protein partner and supports protein clustering in vivo. (A) Schematic depiction of the experimental setup used for the batch pulldown assay. Ni2+-NTA bead-bound ShB-C protein containing the PDZ-binding motif at its C terminus (gray rectangular box) served as bait for the capture of the modular PSD-95 partner protein. (B) SDS/PAGE (Left) and Western blot (Right) analyses of eluted fractions of a pulldown experiment demonstrating the molecular interaction between ShB-C and PSD-95. Left lane corresponds to protein molecular weight markers. Lane 1 corresponds to a crude extract of Drosophila S2 Schneider cells transfected to express the PSD-95-GFP fusion protein. Lanes 2 and 3 correspond to the Drosophila crude extract incubated with ShB-C-free or -bound beads, respectively. Western blot analysis of the same gel shown (Right) was performed by using anti-GFP primary antibodies (see SI Text). (C) Confocal microscopic analysis of Drosophila S2 Schneider cells expressing PSD-95-GFP and/or ShB-C fused to the CD8 membrane-targeting sequence, either separately (Upper) or together (Lower).
Intrinsic Disorder at the Shaker Channel Tail Domain Regulates Its Interaction with PSD-95.
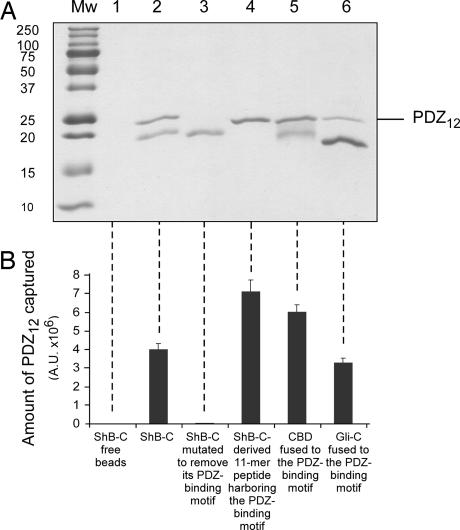
To what extent does the intrinsically disordered nature of ShB-C regulate its interaction with the PSD-95 scaffold protein? To address this question, we exploited the pulldown experimental setup described above to qualitatively evaluate the interaction between the PSD-95 scaffold protein and various versions of ShB-C exhibiting different flexibilities or chain lengths. To achieve fast readout in our pulldown assays, we challenged the anchored stationary ShB-C protein or its mutants with a mobile phase containing not full-length PSD-95 but rather its purified tandem PDZ-1 and -2 domains (PDZ12), previously shown to directly interact with the Kv channel C-terminal tail (24). The use of this experimental design to assess the interaction of the Kv channel C-terminal tail with PSD-95, despite presenting a simplified view of the process, is, nonetheless, informative. Capture of the purified PDZ12 protein (21.5 kDa) by bead-fixed ShB-C can be evaluated by SDS/PAGE analysis with high sensitivity, as reflected in Fig. 5A (lane 2), where the ability of full-length ShB-C protein to capture significant amounts of PDZ12 is shown. By contrast, as expected from the results of previous studies (5–7), no interaction occurred when an ShB-C mutant protein lacking its C-terminal PDZ-binding motif was used as bait (compare lanes 2 and 3). A significantly higher amount of PDZ12 protein was trapped using an ShB-C mutant protein lacking the entire intrinsically disordered chain except for the last 11 C-terminal residues, which include the PDZ-binding motif (compare lanes 2 and 4). To next test whether flexibility of the Shaker C-terminal tail affected its interaction with PSD-95, we performed a pulldown experiment in which the entire intrinsically disordered sequence of ShB-C, apart from the last nine amino acids containing the PDZ-binding motif, was replaced by a globular cellulose-binding domain (CBD) (see SI Text). The folded 17-kDa CBD moiety fused to the Shaker channel PDZ-binding motif captured PDZ12 to a greater extent than did the similarly sized but intrinsically unstructured full-length ShB-C (compare lanes 2 and 5). When, however, the intrinsically disordered C-terminal domain of gliotactin (Gli-C), a nonrelated cell adhesion protein (23) identical in length but envisaged by all disorder predictors to be more flexible, was fused to the Shaker channel PDZ-binding motif, capture of PDZ12 was achieved to a much lower extent (lane 6) (refer to SI Text for calculation of disorder tendencies of ShB-C and Gli-C). Differences in the amount of captured PDZ12 by each construct (Fig. 5B), relative to ShB-C, were all found to be statistically significant (P < 0.0001). These observed differences cannot be attributed to changes in long-range electrostatic interactions, because the amount of PDZ12 captured by the various ShB-C-modified chains was not affected by changes in ionic strength over a wide range of salt concentrations (50–300 mM; data not shown). Our binding experiments thus demonstrate that, relative to ShB-C, shorter or less flexible protein chains have higher affinity for the PDZ domains of the PSD-95 protein than do longer or more flexible chains. Assuming that the intrinsically disordered chain of ShB-C does not fold upon binding to PDZ12 (see SI Appendix), these observations can be explained by considering the contribution of entropy to binding. Long chains, sampling more conformational states, lose more entropy than do shorter or rigid structures upon association, because the configurational space of the longer chains is restricted to a greater degree. Taken together, our findings imply that, in addition to its PDZ-binding motif, the intrinsically disordered nature of the Shaker channel C-terminal tail preceding this motif also acts as an important determinant in mediating the binding of the channel to its PSD-95 scaffold protein partner.
Fig. 5.
The intrinsically disordered nature of ShB-C modulates its interaction with the PSD-95 scaffold protein. (A) SDS/PAGE analysis of eluted fractions of pulldown experiments using different ShB-C-derived chains as bait for the PDZ12 protein (see Materials and Methods). Lanes 1–5 correspond to pulldown experiments using (i) beads alone or beads containing the following chains as bait: (ii) ShB-C, (iii) the ShB-C protein mutated in its PDZ-binding motif, (iv) an ShB-C mutant lacking the entire C-terminal intrinsically disordered segment apart from the 11 last amino acids, (v) an ShB-C mutant protein in which the intrinsically disordered segment but not the PDZ-binding motif was replaced by the folded cellulose-binding domain, or (vi) an ShB-C mutant protein in which the complete intrinsically disordered segment but not the PDZ-binding motif was replaced by the intrinsically disordered C-terminal domain of gliotactin (Gli-C). (B) Densitometric analysis of the results presented in A. Each reported value represents an average of eight independent measurements. Differences in the amount of captured PDZ12 in each category, relative to ShB-C, were all found to be statistically significant, as judged by two-sided Student's t test. Because multiple comparisons of ShB-C to the other protein chains are involved, we followed Bonferroni's correction and used a more stringent criteria to reject the null hypothesis that the two compared groups are identical, based on a P value <1%.
Discussion
Synaptic transmission in neuronal synapses requires the proper organization of ion channels, neurotransmitter receptors, cell adhesion proteins, cytoskeleton proteins, and signaling molecules at the synaptic junction (25). Essential for synapse organization are multidomain scaffold proteins, like the PSD-95 protein, that interact with several different proteins, thereby serving as nuclei for assembly of those macromolecular complexes underlying synaptic architecture (26–30). Through their PDZ domains, scaffold proteins anchor and cluster ion channels at specific subcellular locations by interacting with one or more consensus PDZ-binding sequence motifs located at the intracellular C-terminal tails of the channels (28–30). Insight into the molecular mechanism underlying the interaction of ion channels with scaffold proteins may come from consideration of the specific interaction between the Kv channel and PSD-95. Based on bioinformatics sequence and phylogenetic inference analyses of the Kv channel family, we recently suggested a fishing rod mechanism to describe Kv channel binding to scaffold proteins (10). In this model, the voltage-gated K+ channel interacts with PDZ domain(s) of the PSD-95 protein through binding of the channel C-terminal PDZ-binding motif “hook,” tethered to the rest of the channel protein by an extended chain.
For the fishing rod mechanism to hold true, two criteria must be met. One must first demonstrate that the C-terminal domain of the Shaker channel is indeed intrinsically disordered. Second, it must be shown that the nature of the intrinsically disordered chain of the Kv channel affects its interaction with the PSD-95 scaffold protein. The results presented in the current report provide evidence that the C-terminal tail of the Shaker Kv channel is indeed intrinsically disordered. The hydrodynamic properties of a polypeptide corresponding to the C-terminal segment of the channel, as inferred from size exclusion chromatography, dynamic light scattering, and analytical ultracentrifugation, showed ShB-C to behave as a monomeric random coil protein (Fig. 2). Far-UV CD and NMR spectroscopic analyses further indicated that ShB-C lacks secondary and tertiary structure (Fig. 3 and SI Fig. 6). Moreover, we have shown that the isolated intrinsically disordered ShB-C protein, out of its native channel and membrane contexts, remains functional, maintaining its ability to capture its Dlg scaffold protein partner from the soluble pool of Drosophila Schneider cell proteins and supporting protein clustering in vivo in the same cell line (Fig. 4). A lack of intrinsic structure may, therefore, be inherent to the C-terminal tail of the Shaker Kv channel in its native context, contributing to the function of the PDZ-binding motif (the possibility that ShB-C may acquire structure upon interacting with other channel segments or auxiliary subunits is discussed in SI Appendix).
Our results further provide evidence that the intrinsically disordered nature of the C-terminal chain modulates the interaction of the Kv channel with the PSD-95 scaffold protein, the second requirement for proof that the fishing rod model accurately describes the mechanism of Kv channel binding to scaffold proteins. Assuming a simple one-step binding reaction of the random coil chain of ShB-C to PSD-95 and considering the entropic contribution of the binding reaction, shorter or less flexible (i.e., stiffer) chains would be expected to have higher affinity for the PSD-95 partner than would a more flexible chain. The results of our pulldown experiments (Fig. 5) suggest this to be the case. Shaker Kv channel C-terminal tails in which most of the intrinsically disordered segment, but not the PDZ-binding motif, was eliminated or replaced by the globular folded cellulose-binding domain captured PDZ12 to a much greater extent than did the full length ShB-C protein. In addition, when fused to gliotactin, an intrinsically disordered chain identical in length yet more flexible than ShB-C, the Shaker PDZ-binding motif bound less PDZ12 than did ShB-C. In line with these results, systematic shortening of the ShB-C chain further revealed a monotonic increase in the amount of PDZ12 captured (not shown). Thus, properties associated with polymer chain chemistry, in particular chain length and conformational entropy, modulate the interaction strength of the channel C-terminal tail with the PSD-95 protein. Combined with limited proteolysis analysis showing that the random chain of ShB-C acquires no structure upon binding to PDZ12 (SI Text), it appears that ShB-C behaves as an entropic chain, providing the orientational freedom necessary to search for and interact with the PSD-95 scaffold protein partner.
As such, our results support the fishing rod model for Kv channel–scaffold protein interaction. From a structural perspective, this model is analogous to the “ball-and-chain” model for channel inactivation (31). In the latter, an intrinsically disordered chain, harboring at its end the “ball” peptide sequence, serves as an entropic clock that modulates the kinetics of channel inactivation by an intramolecular binding reaction between the chain-linked ball peptide and its receptor site in the pore domain of the channel (32). In the proposed intermolecular fishing rod mechanism, the length of the intrinsically disordered C-terminal entropic chain of the channel was set during evolution so as to achieve a relatively short interaction time between both proteins, giving rise to affinity in the moderate (micromolar) to low (millimolar) range (E.M., unpublished results) (24, 30). Such affinities are typical for dynamic processes, such as postsynaptic signaling and plasticity, where transient binding and unbinding reactions play an important role in the system dynamics (33).
Finally, one can ask whether insight into the mode of interaction of other ion channels with their cognate PDZ domain-containing scaffold proteins can be inferred from our bioinformatics and experimental results addressing the Kv channel–PSD-95 interaction. We find, using sequence analysis, that other PDZ-binding motif-containing ion channels previously reported to interact with the PDZ domains of scaffold proteins (34–39), including voltage-gated sodium channels, NMDA receptors, glutamate receptor subunits, transient receptor potential channels, inward-rectifier potassium channels, and aquaporins, are all predicted to bear intrinsically disordered, unfolded C-terminal domains (not shown). Accordingly, we propose intrinsically disordered protein segments to play a general role in mediating channel clustering by scaffold proteins.
The results presented in the current study provide direct functional evidence for the involvement of intrinsically disordered protein segments in processes related to synapse assembly, maintenance, and function. Only two other synaptic proteins, i.e., NACP, a protein involved in synapse formation in the brain and implicated in Alzheimer's disease (40), and the C-terminal segment of gliotactin (23), a protein belonging to the neural cell adhesion protein family involved in the formation of glutamatergic synapses during development, have been reported to be intrinsically disordered. In neither case, however, was a link between the intrinsically disordered character of the protein and its role in synapse formation established. Considering the important roles intrinsically disordered proteins play in cell signaling (41, 42), the involvement of such protein domains in neuronal systems merits further investigation.
Materials and Methods
For descriptions of molecular biology, protein purification, analytical ultracentrifugation, CD and 1H-NMR spectroscopies, disorder prediction, cell culture and transformation, confocal microscopy, and limited proteolysis procedures, refer to SI Text. Other procedures are described below.
Analytical Size-Exclusion Chromatography.
For Stokes radius determination, gel filtration of ShB-C and standard molecular weight marker proteins was performed on an analytic size-exclusion column (TSK gel G3000SW column; Tosohass, Tokyo, Japan), preequilibrated in buffer A (20 mM imidazole/0.3 M NaCl/50 mM Tris·HCl, pH 8) and run at a flow rate of 0.5 ml/min at room temperature on an AKTA FPLC system (Amersham–Amersham Pharmacia, Uppsala, Sweden). The elution volumes of the standard and ShB-C proteins (Ve) were converted into mobility-factor parameters, Kav, using the following equation: Kav = (Ve − Vo)/(Vt − Vo), where Vo is the column exclusion volume (5.3 ml), and Vt is the total column bed volume. The Stokes radius (RST) of ShB-C was estimated using a linear calibration plot of RST vs. (−log Kav)1/2 (23), obtained with the standard globular bovine proteins pancreatic ribonuclease A, chymotrypsinogen A, ovalbumin, and serum albumin. The theoretical Stokes radii of native (RSTN) or fully unfolded (RSTRC) proteins were determined as described (21).
Pulldown Assays.
To qualitatively detect interactions between the PSD-95 and ShB-C proteins, batch-mode pulldown experiments were performed. Briefly, Ni2+-NTA beads (100 μl; Qiagen, Chatsworth, CA) were washed three times in buffer A and then incubated with His6-tagged ShB-C (50 μM) for 15 min at room temperature with gentle rocking. After loading of the ShB-C stationary phase, the beads were washed five times with buffer A to remove unbound ShB-C and then challenged for 5 min with a mobile-phase solution containing either crude soluble protein extracts of PSD-95-GFP-expressing Drosophila SR2 Schneider cells or 50 μM of pure PDZ12 protein (both prepared in buffer A). After binding, the beads were washed five times with buffer A followed by elution using buffer A containing 500 mM imidazole. Captured proteins were detected by using standard SDS/PAGE (Fig. 5) or Western blot (Fig. 4B) analyses. The amounts of PDZ12 protein captured by either ShB-C or mutants thereof were quantified densitometrically.
Supplementary Material
Acknowledgments
We thank Dr. H. Shumueli for technical assistance and Dr. U. Isacoff (University of California, Berkeley) for kindly providing the ShB-C-CD8 clone. This research was supported by Israel Science Foundation Grant 323/04 (to O.Y). O.Y is the incumbent of the Belle and Murray Nathan Career Development Chair in Neurobiology.
Abbreviation
- Kv
voltage-activated potassium channels.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0704059104/DC1.
References
- 1.Sigworth FJ. Q Rev Biophys. 1994;27:1–40. doi: 10.1017/s0033583500002894. [DOI] [PubMed] [Google Scholar]
- 2.Yellen G. Q Rev Biophys. 1998;31:239–295. doi: 10.1017/s0033583598003448. [DOI] [PubMed] [Google Scholar]
- 3.Bezanilla F. Physiol Rev. 2000;80:555–592. doi: 10.1152/physrev.2000.80.2.555. [DOI] [PubMed] [Google Scholar]
- 4.Hille B. Ion Channels of Excitable Membranes. Sunderland, MA: Sinauer; 2001. [Google Scholar]
- 5.Tejedor FJ, Bokhari A, Rogero O, Gorczyca M, Zhang J, Kim E, Sheng M, Budnik V. J Neurosci. 1997;17:152–159. doi: 10.1523/JNEUROSCI.17-01-00152.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zito K, Fetter RD, Goodman CS, Isacoff EY. Neuron. 1997;19:1007–1016. doi: 10.1016/s0896-6273(00)80393-1. [DOI] [PubMed] [Google Scholar]
- 7.Kim E, Niethammer M, Rothschild A, Jan YN, Sheng M. Nature. 1995;378:85–88. doi: 10.1038/378085a0. [DOI] [PubMed] [Google Scholar]
- 8.Tiffany AM, Manganas LN, Kim E, Hsueh YP, Sheng M, Trimmer JS. J Cell Biol. 2000;148:147–158. doi: 10.1083/jcb.148.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruiz-Canada C, Koh YH, Budnik V, Tejedor FJ. J Neurochem. 2002;82:1490–1501. doi: 10.1046/j.1471-4159.2002.01092.x. [DOI] [PubMed] [Google Scholar]
- 10.Magidovich E, Fleishman SJ, Yifrach O. Bioinformatics. 2006;22:1546–1550. doi: 10.1093/bioinformatics/btl137. [DOI] [PubMed] [Google Scholar]
- 11.Uversky VN, Gillespie JR, Fink AL. Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 12.Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Biophys J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fink AL. Curr Opin Struct Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 15.Tompa P. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 16.Uversky VN. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dyson HJ, Wright PE. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 18.Long SB, Campbell EB, Mackinnon R. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 19.Sokolova O, Accardi A, Gutierrez D, Lau A, Rigney M, Grigorieff N. Proc Natl Acad Sci USA. 2003;100:12607–12612. doi: 10.1073/pnas.2235650100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukuchi S, Homma K, Minezaki Y, Nishikawa K. J Mol Biol. 2006;355:845–857. doi: 10.1016/j.jmb.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 21.Uversky VN. Eur J Biochem. 2002;269:2–12. doi: 10.1046/j.0014-2956.2001.02649.x. [DOI] [PubMed] [Google Scholar]
- 22.Scherf T, Kasher R, Balass M, Fridkin M, Fuchs S, Katchalski-Katzir E. Proc Natl Acad Sci USA. 2001;98:6629–6634. doi: 10.1073/pnas.111164298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeev-Ben-Mordehai T, Rydberg EH, Solomon A, Toker L, Auld VJ, Silman I, Botti S, Sussman JL. Proteins. 2003;53:758–767. doi: 10.1002/prot.10471. [DOI] [PubMed] [Google Scholar]
- 24.Long JF, Tochio H, Wang P, Fan JS, Sala C, Niethammer M, Sheng M, Zhang M. J Mol Biol. 2003;327:203–214. doi: 10.1016/s0022-2836(03)00113-x. [DOI] [PubMed] [Google Scholar]
- 25.Kennedy MB. Science. 2000;290:750–754. doi: 10.1126/science.290.5492.750. [DOI] [PubMed] [Google Scholar]
- 26.Baron MK, Boeckers TM, Vaida B, Faham S, Gingery M, Sawaya MR, Salyer D, Gundelfinger ED, Bowie JU. Science. 2006;311:531–535. doi: 10.1126/science.1118995. [DOI] [PubMed] [Google Scholar]
- 27.Garner CC, Nash J, Huganir RL. Trends Cell Biol. 2000;10:274–280. doi: 10.1016/s0962-8924(00)01783-9. [DOI] [PubMed] [Google Scholar]
- 28.Sheng M, Sala C. Annu Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- 29.Kim E, Sheng M. Nat Rev Neurosci. 2004;5:771–781. doi: 10.1038/nrn1517. [DOI] [PubMed] [Google Scholar]
- 30.Harris BZ, Lim WA. J Cell Sci. 2001;114:3219–3231. doi: 10.1242/jcs.114.18.3219. [DOI] [PubMed] [Google Scholar]
- 31.Hoshi T, Zagotta WN, Aldrich RW. Science. 1990;250:533–538. doi: 10.1126/science.2122519. [DOI] [PubMed] [Google Scholar]
- 32.Zhou M, Morais-Cabral JH, Mann S, MacKinnon R. Nature. 2001;411:657–661. doi: 10.1038/35079500. [DOI] [PubMed] [Google Scholar]
- 33.Sheng M, Kim MJ. Science. 2002;298:776–780. doi: 10.1126/science.1075333. [DOI] [PubMed] [Google Scholar]
- 34.Kornau HC, Schenker LT, Kennedy MB, Seeburg PH. Science. 1995;269:1737–1740. doi: 10.1126/science.7569905. [DOI] [PubMed] [Google Scholar]
- 35.Tang Y, Tang J, Chen Z, Trost C, Flockerzi V, Li M, Ramesh V, Zhu MX. J Biol Chem. 2000;275:37559–37564. doi: 10.1074/jbc.M006635200. [DOI] [PubMed] [Google Scholar]
- 36.Leonoudakis D, Mailliard W, Wingerd K, Clegg D, Vandenberg C. J Cell Sci. 2001;114:987–998. doi: 10.1242/jcs.114.5.987. [DOI] [PubMed] [Google Scholar]
- 37.Adams ME, Mueller HA, Froehner SC. J Cell Biol. 2001;155:113–122. doi: 10.1083/jcb.200106158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gee SH, Madhavan R, Levinson SR, Caldwell JH, Sealock R, Froehner SC. J Neurosci. 1998;18:128–137. doi: 10.1523/JNEUROSCI.18-01-00128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia J, Zhang X, Staudinger J, Huganir RL. Neuron. 1999;22:179–187. doi: 10.1016/s0896-6273(00)80689-3. [DOI] [PubMed] [Google Scholar]
- 40.Weinreb PH, Zhen W, Poon AW, Conway KA, Lansbury PT., Jr Biochemistry. 1996;35:13709–13715. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- 41.Uversky VN, Oldfield CJ, Dunker AK. J Mol Recognit. 2005;18:343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- 42.Dyson HJ, Wright PE. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.