Abstract
The 2-μm yeast plasmid, a benign high-copy nuclear parasite, propagates itself with nearly the same fidelity as the chromosomes of its host. Equal plasmid segregation is absolutely dependent on the cohesin complex assembled at the plasmid partitioning locus STB. However, the mechanism of cohesin action in the context of multiple plasmid copies, resident within two separate clusters after DNA replication, is unknown. By using “single-copy” derivatives of the 2-μm plasmid, we demonstrate that recruitment of cohesin at STB during S phase indeed translates into cohesion between plasmid molecules. Through binary fluorescence tagging, we reveal that segregation of replicated plasmids occurs in a sister-to-sister fashion. Thus, cohesin serves the same fundamental purpose in plasmid and chromosome segregation.
Keywords: 2-μm circle, cohesin complex, plasmid cohesion, sister-to-sister segregation
The 2-μm plasmid, a high-copy extrachromosomal selfish DNA element resident in the yeast nucleus, propagates itself stably with the assistance of a partitioning system (1). The plasmid-coded proteins Rep1p and Rep2p and the cis-acting locus STB comprise the partitioning system, which is designed to channel central components of the chromosome segregation machinery toward plasmid segregation (2). The histone H3 variant Cse4p, which has so far been thought to be unique to centromeres (3), is present also at STB and is essential for equal plasmid segregation (4). The maturation of Cse4p-containing STB chromatin into its functional state appears to be mediated through the RSC2 chromatin remodeling complex (4–6). The yeast cohesin complex, required for one-to-one segregation of sister chromatids, is assembled at STB in a Rep1p- and Rep2p-assisted manner during early S phase, and this association lasts until anaphase (7, 8). Like chromosome–cohesin association, plasmid–cohesin association also requires the loading factors Scc2 and Scc4 (ref. 9; S. Mehta and M.J., unpublished work). However, in contrast to chromosomes, the plasmid fails to acquire cohesin when the mitotic spindle is disassembled (10). Consistent with a potential role for the RSC2 complex in remodeling STB chromatin, inactivation of the complex blocks cohesin assembly on the plasmid (5, 11). The timely recruitment of cohesin at STB and its timely disassembly are critical events in 2-μm-plasmid segregation (7).
An amplification system consisting of the Flp recombinase and its target sites (FRTs) augments the partitioning system in the high-copy persistence of the 2-μm circle. Under steady state conditions, the amplification system appears to be negatively regulated by a bipartite Rep1p–Rep2p repressor complex (12). Amplification is triggered only when a rare missegregation event leads to a copy-number drop in the plasmid. The generally accepted amplification mechanism is a carefully timed Flp recombination event that converts a pair of bidirectional replication forks into unidirectional ones by DNA inversion (13). The dual rolling-circle replication generates a plasmid concatamer that may be resolved by Flp recombination or homologous recombination.
Despite a copy number of ≈40–60 per cell, the 2-μm plasmid exists as a single tight-knit cluster of foci and segregates as a cluster (14, 15). This copy-number reduction, effectively to unity, makes an active partitioning mechanism imperative for stable propagation. Based on the role of cohesin in chromosome segregation, one might imagine that cohesin assembly pairs duplicated plasmid clusters and cohesin cleavage triggers their disjunction (7). However, there is no direct evidence for this “pairing–unpairing” mode of plasmid segregation. Furthermore, it is not clear how pairing might be effected between twin clusters, each of which contains multiple plasmid copies. The question regarding whether plasmid clusters segregate in a chromosome-tethered state, by hitchhiking on sister chromatids, or do so independently of chromosomes remains open.
The multicopy nature of the plasmid and its clustered state have so far precluded direct demonstration of plasmid cohesion and subsequent separation. Assuming that pairing by cohesin does occur, two related questions become important: (i) Are replicated plasmid molecules randomly organized into two roughly equal clusters and then paired by cohesin assembled at STB? In this scheme, the intercluster pairing may be limited to a subset of plasmid molecules within the individual clusters. (ii) Alternatively, does each plasmid molecule become paired with its sister molecule in a replication-coupled manner? Neither model requires cohesin for plasmid clustering per se, which is likely mediated through the Rep proteins in conjunction with associated host factors. Consistent with this notion, the clustered plasmid organization is not altered in G1-arrested cells even though they lack cohesin at STB (7, 15).
Using single-copy 2-μm-circle-derived reporter plasmids, we now provide concrete evidence for plasmid pairing by cohesin assembly and unpairing by cohesin disassembly. In addition, our results strongly favor a segregation mechanism in which each plasmid molecule is paired with its sister molecule. Thus, the 2-μm circle cluster is fundamentally analogous to a yeast chromosome in its mechanism of segregation.
Results
Single-Copy Derivatives of 2-μm Circle Reporter Plasmids.
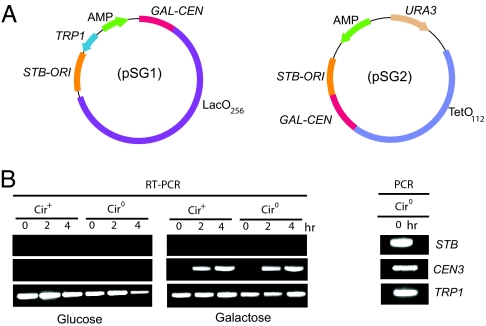
In reporter plasmids containing both STB and a centromere, the latter is dominant in copy-number control. By incorporating CEN3 into STB plasmids (Fig. 1A), we could bring down their copy number to unity or close to unity in >80% of the cell population. A CEN–STB plasmid, when tagged by fluorescence, appeared as a single focus in the nucleus, analogous to standard CEN plasmids [supporting information (SI) Fig. 7A]. The “unit-copy” status was further ascertained by Southern blot analysis (SI Fig. 7B).
Fig. 1.
CEN–STB containing single-copy derivatives of 2-μm circle reporter plasmids. (A) The plasmids pSG1 and pSG2 contain, in addition to the 2-μm circle replication origin and STB, the CEN3 sequence flanked by the GAL promoter and the CYC1 terminator; pSG1 harbors a [LacO]256 array, and pSG2 harbors a [TetO] 112 array. (B) Total RNA from [cir+] and [cir0] strains harboring pSG1 was isolated after transferring raffinose-grown log-phase cells to medium containing glucose or galactose for the indicated times. RT-PCR was performed by using primers specific to the CEN3, STB, and TRP1 sequences. A DNA sample isolated from the [cir0]/pSG1 strain before galactose induction was amplified with the same primers as those used in the RT-PCR assays to provide the reference bands.
Two CEN–STB reporter plasmids used in this study (Fig. 1A) harbored either the LacO (256 copies) or TetO (112 copies) arrays, and could be fluorescence-tagged in cells expressing GFP-LacI or RFP-TetR, respectively. The centromeres in these plasmids could be conditionally inactivated by driving transcription through them from the inducible GAL1 promoter (16). As a result, in galactose medium, their segregation was under STB control, provided the Rep1 and Rep2 proteins were supplied in trans. We confirmed that transcription through the centromere was galactose dependent (Fig. 1B). The mitotic stability of the reporter plasmids in a [cir0] host strain (lacking Rep1 and Rep2 proteins) was nearly 70% after 10 generations of nonselective growth in glucose and close to 5% after similar growth in galactose, verifying transcriptional inactivation of the centromere (results not shown).
The data from the experiments detailed in the sections to follow were effectively restricted to single-copy plasmids by excluding from the analyses those cells that contained more than two foci of a particular fluorescence type (either green or red). For example, in cohesion assays in metaphase cells, two fluorescent dots would indicate predominantly unpaired, although replicated, plasmids with perhaps an insignificant contribution from two unreplicated plasmid copies. Conversely, a single fluorescent dot would signal cohesion between replicated plasmid molecules, with negligible contamination due to replication failure.
Pairing of Replicated Plasmid Molecules: Does Cohesin Assembled at STB Mediate Plasmid Cohesion?
Conventionally, sister-chromatid cohesion is assayed in G2/M-arrested cells after microtubule depolymerization by using an antimitotic drug. The chromosomes are thus free from pulling forces exerted by the spindle. However, the test for cohesion between replicated plasmids was carried out in normally cycling cells because microtubule integrity is essential for cohesin assembly at STB (10).
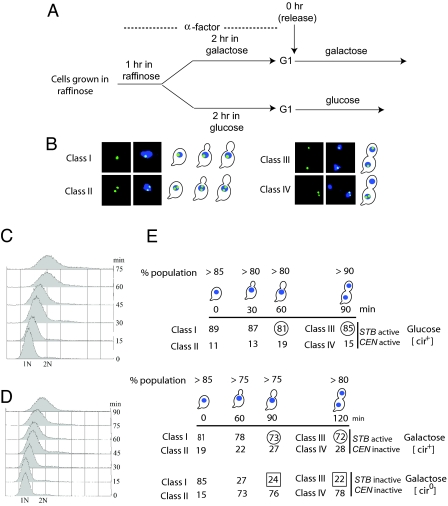
Isogenic [cir+] and [cir0] wild-type strains harboring the STB–CEN reporter were released from G1 arrest to traverse the cell cycle at 30°C (Fig. 2A) under conditions where (i) the centromere and STB were active, (ii) STB alone was active, and (iii) neither the centromere nor STB was active. Plasmid fluorescence, as well as DAPI staining patterns scored at progressive intervals in the assay, was grouped into four types (classes I–IV, Fig. 2B). Cells completed DNA replication by 60 min in glucose medium and 90 min in galactose medium, as judged by FACS analysis (Fig. 2 C and D). At these time points, the majority of cells (>75%) displayed an enlarged bud with a single DAPI staining zone near the bud neck within the mother. Among such cells, deemed to be in metaphase, ≈70–80% revealed plasmid pairing (class I; single fluorescence dot) when STB alone was active or both STB and CEN3 were active (circled numbers in column 3 of Fig. 2E). This fraction dropped to 24% when both CEN3 and STB were inactive (boxed number in column 3 of Fig. 2E).
Fig. 2.
Plamsid cohesion mediated through STB or CEN during metaphase in cycling cells. (A) The experimental regimen for cell cycle arrest and release is schematically outlined. (B) The four representative classes of cells scored in the assay are displayed. Classes I and II are preanaphase cells showing one and two fluorescent plasmid foci, respectively. Classes III and IV are postanaphase cells containing 1:1 and 2:0 patterns of plasmid segregation, respectively. (C and D) DNA replication was followed by FACS analysis. (E) The cell populations were assayed at the indicated times after release from G1 arrest and binned into classes I–IV.
At 90 min and 120 min into the cell cycle in glucose and galactose, respectively, the population comprised almost entirely (80–90%) of late anaphase or telophase cells. Among these, >80% showed equal plasmid segregation (class III) when CEN and STB were active, and >70% showed equal plasmid segregation when STB alone was active (circled numbers in column 4 of Fig. 2E). Lack of an active CEN or STB resulted in a precipitous fall in equal segregation to ≈20% (boxed number in column 4 of Fig. 2E).
When the Rep–STB system is functional, replicated plasmid molecules remain paired during metaphase and become unpaired during anaphase. The timing of these events suggests that cohesin assembly and disassembly are responsible for pairing and unpairing, respectively.
The assays carried out in [cir+] cells examine a single-copy fluorescent reporter plasmid resident within a cluster of native 2-μm circles that remain invisible in the analysis. By contrast, the assays in [cir0] cells report on a true single-copy plasmid. The observed differences in plasmid cohesion between [cir0] and [cir+] strains were unrelated to copy number. The results from the [cir0] strain were similar to those from a strain harboring a multicopy 2-μm-circle derivative lacking the REP1 gene (data not shown).
We have verified that inactivating STB in the GAL–CEN and STB-containing reporter plasmid is functionally equivalent to deleting it. The cohesion and segregation patterns of a GAL–CEN plasmid (lacking STB) in either a [cir+] or a [cir0] strain were similar to those of the GAL–CEN plus STB reporter resident in a [cir0] strain (inactive STB). The percentage of metaphase cells showing plasmid cohesion was roughly 75% in glucose for both reporters (active CEN) (data not shown); this value dropped to nearly 20% in galactose (inactive CEN) (Fig. 2E).
Tanaka et al. (17) noted that duplicated copies of a fluorescence-tagged minichromosome harboring a 130-bp minimal centromere were precociously separated in more than half of the metaphase cells due to spindle force. We observed this phenotype in ≈20% of the metaphase cells when the centromere was functional. Perhaps the larger (320 bp) CEN fragment housed by our reporter plasmids, the simultaneous presence of STB and CEN in them, or some feature of the GAL-promoter-controlled CEN could have partially mitigated the spindle force.
Cohesin Is the Agent of Plasmid Pairing, and Plasmid Separation Follows Cohesin Disassembly.
To critically verify the notion that pairing of replicated plasmid copies is cohesin mediated, we exploited previously established features of plasmid–cohesin association. Cohesin recruitment at STB is absolutely dependent on Rep1 and Rep2 proteins (7, 11). The STB chromatin harbors the histone H3 variant Cse4p (4), and inactivation of Cse4p by a temperature-sensitive mutation blocks STB–cohesin association. Will disrupting cohesin assembly at STB or directly inactivating cohesin result in lack of cohesion between replicated plasmids?
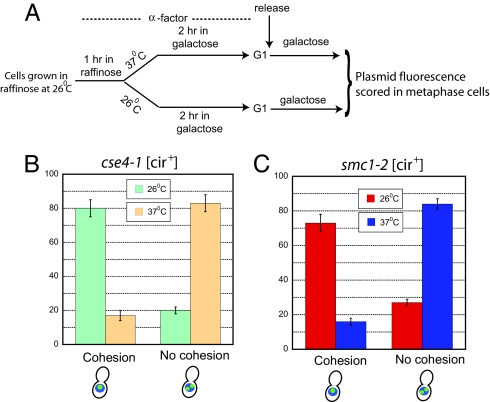
We know that withdrawal of Rep proteins (in the [cir0] background) results in lack of plasmid cohesion in metaphase cells (Fig. 2E). We also assayed plasmid cohesion in a [cir+] host strain expressing the cse4-1 allele (18). This mutant allele, like cse4-107 tested previously (4), fails to support cohesin assembly at STB at 37°C (S.H. and M.J., unpublished data). Cells released from G1 arrest into galactose medium after centromere inactivation on the reporter plasmid (Fig. 3A) completed DNA replication by 100 min and 75 min at 26°C and 37°C, respectively (inferred by FACS analysis; data not shown). The plasmid fluorescence pattern was scored at these time points in “metaphase” cells as described for the assays shown in Fig. 2. The single-dot pattern (cohesion) was predominant over the two-dot pattern (no cohesion) at 26°C, whereas the reverse was true at 37°C (Fig. 3B). Thus, inactivation of Cse4p, which blocks STB–cohesin association, also prevents plasmid cohesion. Consistent with the notion that plasmid cohesion is a central step in equal plasmid segregation, at 37°C the cse4-1 mutation caused a much larger proportion of 2:0 segregation (≈70%) relative to 1:1 segregation (SI Fig. 8). At 26°C, the principal mode of segregation was 1:1 (≈80%).
Fig. 3.
Plasmid cohesion in cse4-1 [cir+] and smc1-2 [cir+] cells at permissive and nonpermissive temperatures. (A) The protocol for assaying plasmid cohesion is schematically indicated. (B and C) The fractions of metaphase cells displaying one fluorescent dot (cohesion) or two fluorescent dots (lack of cohesion) were estimated in the cse4-1 and smc1-2 strains at 26°C and 37°C.
For testing the effect of disabling a cohesin component on plasmid pairing, we performed the cohesion assay in the smc1-2 [cir+] strain. Cells arrested in G1 in galactose medium were released into the same medium at 26°C and 37°C, and plasmid fluorescence was assayed after 100 min and 75 min, respectively, in metaphase cells. FACS analysis confirmed that ≈90% of cells in the assayed populations harbored 2N DNA content (data not shown). At 26°C, one fluorescent dot outnumbered two dots, ≈3:1; at 37°C, the strong bias was in the other direction, ≈4:1 (Fig. 3C).
We conclude that cohesin assembled at the Cse4p-containing STB chromatin with assistance of the Rep proteins is responsible for holding together duplicated 2-μm plasmids. In the absence of functional cohesin, plasmid pairing is interrupted. During a normal cell cycle, the paired plasmids separate when cohesin is inactivated during anaphase. Experiments analogous to those reported by Mehta et al. (7) showed that assembly of a noncleavable version of cohesin at STB blocks the separation of the duplicated single-copy reporter plasmid (S.K.G. and M.J., unpublished data). Failure to establish cohesion or disassemble cohesin at the appropriate times in the cell cycle leads to high frequency missegregation of the plasmid. Thus, the fundamental mechanism used for sister-chromatid pairing and segregation also operates on the 2-μm circle even though the native plasmid segregates as a pair of sister clusters, each containing multiple copies of the plasmid.
Cohesin–STB Association and Establishment of Cohesion: Dependence on the Mitotic Spindle.
In several respects, the 2-μm plasmid is similar to chromosomes in the recruitment of cohesin and the establishment of cohesion. The loading factors Scc2p and Scc4p, which are responsible for deposition of cohesin at centromeres and chromosome arm sites, are essential for the assembly of cohesin at STB (S. Mehta and M.J., unpublished data). ChIP analysis revealed that Scc2p associates with STB in a Rep1p–Rep2p-dependent manner (SI Fig. 9). Upon inactivating Ctf7p, which is required for establishing chromosomal cohesion (19), plasmid cohesion is disrupted (SI Fig. 10) without cohesin being excluded from STB (S. Mehta and M.J., unpublished data). In contrast to chromosomes, the plasmid depends on spindle integrity for enlisting cohesin at STB (10). Plasmid molecules replicate in the absence of spindle and can acquire cohesin when the spindle is allowed to reassemble after replication. However, this postreplication assembly of cohesin at STB is not functional in segregation. These earlier observations, coupled with current results, suggest that spindle disassembly results in failure of plasmid cohesion, and subsequent spindle polymerization does not reinstate cohesion. We wished to verify these possibilities.
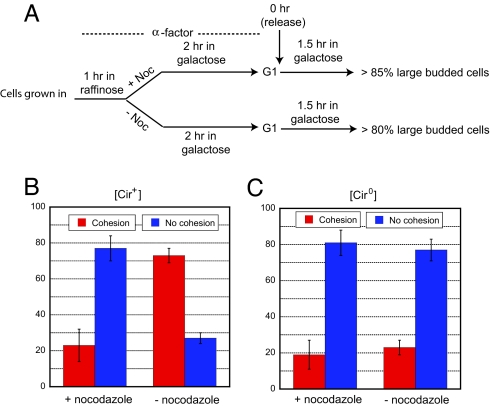
In a nocodazole-treated and G2/M-arrested [cir+] population (Fig. 4A), >70% of the cells revealed lack of plasmid cohesion (Fig. 4B). This fraction was comparable to that observed in metaphase cells in an isogenic [cir0] population released from G1 arrest in nocodazole-free medium (Fig. 4C). In the [cir0] background lacking the partitioning system, nocodazole treatment did not further increase the frequency of unpaired plasmid dots (Fig. 4C).
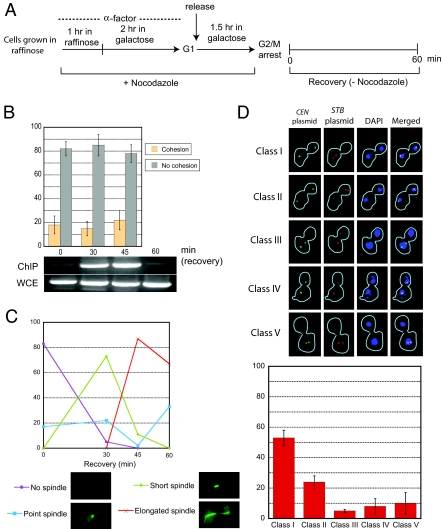
Fig. 4.
Spindle integrity in the establishment of plasmid cohesion. (A) The effect of spindle disassembly on plasmid cohesion was followed according to the outlined experimental procedure. (B and C) The frequencies of one fluorescent plasmid dot (cohesion) or two fluorescent plasmid dots (lack of cohesion) were estimated in metaphase cells during the cell cycle (− nocodazole) or in cells arrested at G2/M (+ nocodazole).
In cells released from nocodazole arrest (Fig. 5A), lack of cohesion persisted at 30 min and 45 min into the recovery, even though cohesin was associated with STB at these time points (Fig. 5B). At 30 min, nearly all of the cells contained a point spindle or a short spindle; by 45 min, spindle extension and nuclear elongation (as inferred from microtubule and DAPI staining) were more pervasive (Fig. 5C and data not shown). The cohesion results for the 45 min time point refer only to the fraction of preanaphase cells containing a single DAPI mass in the mother cell compartment. At 60 min, nearly all of the cells had traversed anaphase, with a small fraction having completed cytokinesis. As expected, cohesin was absent from STB at this time point. In cells displaying clearly segregated chromosomes, as indicated by DAPI staining, an STB plasmid (red) showed a high incidence of missegregation (2:0; cell types I, II, and V in Fig. 5D). By contrast, a coexisting CEN plasmid (green) segregated equally (1:1) in most of the cells (except cell types III and V in Fig. 5D).
Fig. 5.
Lack of plasmid cohesion in cells released from nocodazole arrest into nocodazole-free medium. (A) The experimental scheme for nocodazole-mediated arrest of cells at G2/M and their subsequent release to resume the cell cycle in the absence of the drug is diagrammed schematically. (B) Plasmid cohesion was assayed at the indicated time points during recovery. In parallel, the association of the cohesin component Mcd1p (expressed from the native promoter as an HA-tagged version) with STB was followed by ChIP. (C) The restoration of the mitotic spindle was monitored in recovering cells. The spindle was visualized in fixed cells by indirect immunofluorescence by using primary antibodies against Tub1p followed by fluorescein-conjugated secondary antibodies. (D) The segregation patterns of a CEN plasmid (green) and a cohabiting CEN–STB plasmid (red) under Rep-STB control were assayed in postanaphase cells 75 min after release from nocodazole arrest.
The lack of a functional spindle at the time of replication aborts plasmid cohesion and causes high rates of plasmid missegregation. Because chromosomal cohesion is spindle-independent, once the spindle is reorganized, paired sister chromatids and CEN plasmids go on to segregate equally.
Segregation of Plasmid Occurs in a Sister-to-Sister Fashion.
How are replicated plasmid molecules, which comprise a population of identical individuals, chosen for pairing by cohesin? Two distinct models can be envisaged (Fig. 6 A and B). In a global and approximate counting model, roughly equal pools of plasmid molecules are organized into two clusters at completion of DNA replication. The two clusters become bridged when cohesin assembled at STB pairs plasmid molecules across them. In this scheme, not every STB need participate in the bridging event (Fig. 6B Left). In an alternative precise counting model, each plasmid molecule is paired with its sister during the process of replication, and the two sisters then occupy separate clusters (Fig. 6B Right). In other words, the sisterhood between the duplicated clusters as a whole applies to every individual molecule within them. One feature common to both models is that the cohesin bridge defines the boundary between the two clusters. In their cohesed metaphase context, the clusters are referred to as “two” in a prescient sense, anticipating their binary split and segregation upon cohesin cleavage. Does 2-μm-circle segregation conform to one of the two mechanisms diagrammed in Fig. 6 A or B?
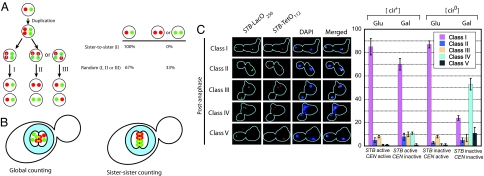
Fig. 6.
A sister-to-sister binary counting mechanism for the segregation of the 2-μm plasmid. (A) The possible modes of cohesin-mediated plasmid counting and segregation are schematically diagrammed. Replication of the single-copy reporter plasmids (one tagged by green fluorescence and the other by red fluorescence) generates two green and two red plasmid molecules. A strict sister-to-sister counting mode will pair red with red and green with green (I). A random counting mechanism may follow the pairing scheme I or mediate red-to-green pairings as indicated by II and III. Segregation according to I will not generate daughter nuclei containing two red or two green plasmids. However, equal probabilities of I, II, and III segregation modes will result in 33% frequency of such nuclei. (B) The distribution and organization of duplicated plasmid molecules according to the global counting mechanism (Left) and the sister-to-sister counting mechanism (Right) are schematically represented. The cohesin bridge (yellow) that pairs plasmid molecules (red and green) defines the boundary between the sister clusters that separate from each other upon cohesin disassembly. (C) Plasmid segregation was assayed in postanaphase cells under conditions where the plasmid-borne CEN and STB were both active, only one of the two was active, and neither one was active. The observed patterns were divided into classes I–V. Class I, with each prescient daughter cell containing one red and one green plasmid, is the only class predicted by the sister-to-sister segregation mode. Note that, even with an active centromere, class I was <100%. Class V, with would-be daughter cells containing two red or two green plasmids, is excluded by sister-to-sister segregation; however, it is a significant entity (33%) according to the random pairing mode. Note the rarity of class V in the presence of an active CEN or STB. Classes II–IV signify 3:1 and 4:0 missegregation events.
We introduced two variants of the single-copy STB-reporter plasmids, pSG1 and pSG2 (see Fig. 1A), into isogenic [cir+] and [cir0] strains expressing both GFP-LacI and RFP-TetR. Each plasmid could be distinguished by its fluorescence tag, green for pSG1 and red for pSG2. After releasing G1-arrested cells into a synchronous cell cycle, plasmid cohesion and plasmid segregation were assayed. As expected, cohesion was observed in most metaphase cells when either CEN or STB was active or when both were active (SI Fig. 11). There was a clear lack of cohesion when neither was active. We then examined the red (R)-to-green (G) segregation profiles in postanaphase cells with clearly separated chromosomes (Fig. 6B). The 1:1 green/1:1 red (RG:RG) (class I) pattern was dominant when at least one of the two, CEN or STB, was functional. In sharp contrast, the incidence of 2:0 green/0:2 red (RR:GG) (class V) was quite low. The 3:1 and 4:0 unequal segregation patterns (RRG:G, GGR:R, and GGRR:0) (classes II–IV) were also highly infrequent. CEN (or the CEN–STB combination) was slightly superior to STB in effecting the faithful R-to-R and G-to-G segregation, ≈80–85% versus 70%. In the [cir0] host strain, with neither CEN nor STB being active, the segregation became largely unequal, with a strong tendency for both green and red to stay together in the mother cell (class IV). Of particular significance is the result that the class V (RR:GG) pattern, which should be a sizable fraction (33%) according to the random counting model (Fig. 6A), was almost nonexistent during STB-mediated segregation. It was as low as that encountered during CEN-assisted segregation.
The data from the population assays presented above were consistent with time-lapse analysis of red-green segregation carried out on individual cells [see SI Fig. 12 and time-lapse movies (SI Movies 1–3)]. The STB-mediated sister-to-sister segregation observed in the [cir+] strain was not conditioned by the reporter plasmids resident within a larger cluster of native 2-μm circles. Nearly identical results were obtained when the two-color segregation was assayed in a [cir0] strain expressing Rep1 and Rep2 proteins from the bidirectional GAL1–GAL10 promoter (SI Fig. 13).
In summary, the mechanisms for CEN-based and Rep–STB-based faithful propagation of chromosome and plasmid, respectively, share the common logic of cohesin-mediated sister-to-sister segregation.
Discussion
The stable persistence of the 2-μm plasmid in yeast derives from its ability to appropriate components of the chromosome segregation machinery and channel them for its own partitioning (1, 4, 7, 20–22). The plasmid partitioning proteins Rep1 and Rep2 mediate the incorporation of the histone H3 variant Cse4p into chromatin at the STB locus (4). This special nucleosome tag, regarded until recently to be unique to centromeres, is critical for assembling the plasmid partitioning complex. The maturation of STB chromatin into its functionally competent state, apparently mediated by the RSC2 remodeling complex (5, 6, 11), permits the recruitment of the cohesin complex as a key step in equal plasmid segregation. The mitotic spindle strongly influences the plasmid partitioning pathway by promoting the association of host factors, including cohesin, at STB. The mechanism by which cohesin promotes equal segregation of multiple plasmid copies arranged into two clusters has been conceptually and technically difficult to address. The work described here provides a rational solution to this problem.
The unit-copy CEN–STB reporter plasmids display similar temporal patterns of cohesion and separation during the cell cycle, regardless of whether they segregate using the Rep–STB or the CEN pathways. Plasmid pairing under a functional Rep–STB system is mediated through the cohesin complex. Inactivating cohesin directly or blocking cohesin assembly at STB at the appropriate time in the cell cycle abrogates plasmid pairing and leads to missegregation. Plasmid–cohesin association requires the deposition factors Scc2p and Scc4p, and the former has been detected at STB by ChIP. Cohesin assembly at STB per se does not lead to plasmid cohesion, as revealed by inactivation of Ctf7p or the postreplication enlistment of cohesin in cells recovering from nocodazole treatment. Thus, the 2-μm plasmid is generally similar to chromosomes in recruitment of cohesin and establishment of cohesion (23). However, it differs conspicuously from chromosomes in using its Rep proteins and depending on spindle integrity for funneling cohesin for its selfish needs.
Despite the multicopy and clustered status of the 2-μm circle, cohesin-mediated pairing of plasmid molecules occurs in a sister-to-sister fashion. This mechanism was suggested by the equal segregation of a fluorescently tagged single-copy reporter plasmid within a cluster of untagged native plasmids and further confirmed by the one-to-one segregation of two reporter plasmids harboring distinct fluorescence tags. The timing of plasmid cohesion during the cell cycle and the coupling between replication and cohesion are consistent with this mechanism. This pairing rule implicates a high-order organization within the plasmid cluster and suggests how that order might be engendered in the sister cluster after DNA replication. A plasmid cluster templates its sister cluster through cohesin-assisted pairing of sister molecules. Current evidence suggests that adhesion of plasmid molecules within a cluster is likely mediated by the Rep proteins, together with host factors other than cohesin (7). The function of cohesin is to bridge the two “would-be” clusters that split asunder in anaphase.
Strictly, our interpretation of sister-to-sister segregation applies to a single-plasmid focus rather than a single-plasmid molecule. Incorporating CEN into a reporter plasmid reduces its copy number to nearly unity (not necessarily unity). However, given the similarities established here between cohesin-mediated CEN and STB segregation mechanisms, sister-to-sister segregation of the 2-μm circle at the single-plasmid level is a reasonable extrapolation.
The aggregation of the plasmid molecules in association with the partitioning proteins and host factors is not unlike the clustering of centromeres in association with the kinetochore complex. Both these multiDNA/multiprotein assemblies appear to remain congressed throughout the cell cycle. Yet, in the face of these organizational constraints, the plasmid molecules and centromeres replicate efficiently, acquire cohesin, and pair faithfully with their respective sisters. The size of the plasmid entity in segregation (60 × 6 = 360 kbp), roughly one and a half times the size of the smallest yeast chromosome (230 kbp), further accentuates the analogy between sister plasmid clusters and sister chromatids in partitioning. What mediates the opposite movement of the sister clusters during segregation remains an open challenge. Plausible alternatives to be entertained include hitchhiking on sister chromatids, spindle-assisted segregation independent of chromosomes, and cosegregation with a nuclear entity other than chromosomes that evenly partitions between daughter cells.
Materials and Methods
Strains and Plasmids.
The yeast strains and plasmids used in this study are listed in SI Table 1. The strains used for ChIP harbored an integrated copy of either the MCD1 or SCC2 gene tagged with the HA epitope (ref. 7 and this study).
The reporter plasmids pSG1 and pSG2 were constructed as described in SI Appendix.
RT-PCR.
Total cellular RNA was isolated from cultures grown either in glucose or galactose at the indicated time points by using RNeasy kit (Qiagen, Valencia, CA). RT-PCR was performed by using One Step RT-PCR kit (Qiagen) with ≈2 μg of RNA as template per reaction.
Antibodies.
Antibodies used in this study are described in ref. 4.
Other Protocols.
Synchronized cell populations were obtained by α factor-induced arrest in G1 phase, followed by release as described in ref. 15. Nocodazole arrest and subsequent release are detailed in refs. 10 and 15. Fluorescence microscopy of reporter plasmids, indirect immunofluorescence assays for cytoskeletal structures, and ChIP followed protocols standardized in prior studies (4, 7, 15).
Supplementary Material
Acknowledgments
We thank C. Chan (University of Texas), D. Koshland and V. Guacci (Carnegie Institution, Baltimore, MD), K. Nasmyth (University of Oxford, Oxford, U.K.), and R. Rothstein (Columbia University, New York, NY) for yeast strains and plasmids; and Chien-Hui Ma for excellent technical assistance. This work was supported by National Institutes of Health Grant GM64363. Partial support was provided by the Robert F. Welch Foundation Award F-1274.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0702996104/DC1.
References
- 1.Ghosh SK, Hajra S, Paek A, Jayaram M. Annu Rev Biochem. 2006;75:211–241. doi: 10.1146/annurev.biochem.75.101304.124037. [DOI] [PubMed] [Google Scholar]
- 2.Jayaram M, Mehta S, Uzri D, Velmurugan S. Plasmid. 2004;51:162–178. doi: 10.1016/j.plasmid.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 4.Hajra S, Ghosh SK, Jayaram M. J Cell Biol. 2006;174:779–790. doi: 10.1083/jcb.200603042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang J, Hsu JM, Laurent BC. Mol Cell. 2004;13:739–750. doi: 10.1016/s1097-2765(04)00103-0. [DOI] [PubMed] [Google Scholar]
- 6.Wong MC, Scott-Drew SR, Hayes MJ, Howard PJ, Murray JA. Mol Cell Biol. 2002;22:4218–4229. doi: 10.1128/MCB.22.12.4218-4229.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mehta S, Yang XM, Chan CS, Dobson MJ, Jayaram M, Velmurugan S. J Cell Biol. 2002;158:625–637. doi: 10.1083/jcb.200204136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nasmyth K. Annu Rev Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- 9.Ciosk R, Shirayama M, Shevchenko A, Tanaka T, Toth A, Shevchenko A, Nasmyth K. Mol Cell. 2000;5:243–254. doi: 10.1016/s1097-2765(00)80420-7. [DOI] [PubMed] [Google Scholar]
- 10.Mehta S, Yang XM, Jayaram M, Velmurugan S. Mol Cell Biol. 2005;25:4283–4298. doi: 10.1128/MCB.25.10.4283-4298.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang XM, Mehta S, Uzri D, Jayaram M, Velmurugan S. Mol Cell Biol. 2004;24:5290–5303. doi: 10.1128/MCB.24.12.5290-5303.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Som T, Armstrong KA, Volkert FC, Broach JR. Cell. 1988;52:27–37. doi: 10.1016/0092-8674(88)90528-4. [DOI] [PubMed] [Google Scholar]
- 13.Futcher AB. J Theor Biol. 1986;119:197–204. doi: 10.1016/s0022-5193(86)80074-1. [DOI] [PubMed] [Google Scholar]
- 14.Scott-Drew S, Murray JA. J Cell Sci. 1998;111:1779–1789. doi: 10.1242/jcs.111.13.1779. [DOI] [PubMed] [Google Scholar]
- 15.Velmurugan S, Yang XM, Chan CS, Dobson M, Jayaram M. J Cell Biol. 2000;149:553–566. doi: 10.1083/jcb.149.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill A, Bloom K. Mol Cell Biol. 1987;7:2397–2405. doi: 10.1128/mcb.7.7.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanaka T, Cosma MP, Wirth K, Nasmyth K. Cell. 1999;98:847–858. doi: 10.1016/s0092-8674(00)81518-4. [DOI] [PubMed] [Google Scholar]
- 18.Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- 19.Skibbens RV, Corson LB, Koshland D, Hieter P. Genes Dev. 1999;13:307–319. doi: 10.1101/gad.13.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Velmurugan S, Mehta S, Jayaram M. Curr Top Dev Biol. 2003;56:1–24. doi: 10.1016/s0070-2153(03)01005-6. [DOI] [PubMed] [Google Scholar]
- 21.Malik HS. J Cell Biol. 2006;174:747–749. doi: 10.1083/jcb.200608107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yeh E, Bloom K. EMBO Rep. 2006;7:985–987. doi: 10.1038/sj.embor.7400793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uhlmann F, Nasmyth K. Curr Biol. 1998;8:1095–1101. doi: 10.1016/s0960-9822(98)70463-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.