Abstract
T cell receptor (TCR) recognition of peptide-MHC (pMHC) is central to the cellular immune response. A large database of TCR–pMHC structures is needed to reveal general structural principles, such as whether the repertoire of TCR/MHC docking modes is dictated by a “recognition code” between conserved elements of the TCR and MHC genes. Although ≈17 cocrystal structures of unique TCR–pMHC complexes have been determined, cocrystallization of soluble TCR and pMHC remains a major technical obstacle in the field. Here we demonstrate a strategy, based on NMR chemical shift mapping, that permits rapid and reliable analysis of the solution footprint made by a TCR when binding onto the pMHC surface. We mapped the 2C TCR binding interaction with its allogeneic ligand H–2Ld–QL9 and identified a group of NMR-shifted residues that delineated a clear surface of the MHC that we defined as the TCR footprint. We subsequently found that the docking footprint described by NMR shifts was highly accurate compared with a recently determined high-resolution crystal structure of the same complex. The same NMR footprint analysis was done on a high-affinity mutant of the TCR. The current work serves as a foundation to explore the molecular dynamics of pMHC complexes and to rapidly determine the footprints of many Ld-specific TCRs.
Keywords: chemical shift mapping, dynamics, NMR, cellular immunity, protein–protein interaction
T cells modulate the nature and extent of an immune response based primarily on specific T cell receptor (TCR) recognition of peptide in the context of a MHC molecule (1, 2). The TCR is a genetically recombined receptor, analogous to an antibody, that is composed of α and β chains. The binding site of the TCR comprises six loops called complementarity-determining regions (CDRs), with each chain contributing three loops, called CDR 1, 2, and 3. The CDR1 and CDR2 loop sequences are encoded by the variable (V) genes, but because there is no somatic mutation in TCR-V genes, these are referred to as “germ line-derived.” In contrast, the CDR3 loops, which are largely involved in antigenic peptide contact and thus specificity, are derived from recombination of V(D)J segments and vary in an almost unlimited fashion. MHC gene products present antigenic peptides to the TCR through a composite peptide-MHC (pMHC) surface composed of a highly variable element (i.e., peptide) in a groove surrounded by more conserved residues on the MHC helices (2).
The interaction between TCR and pMHC has been studied through cocrystallization of ≈17 unique complexes (1, 3). Some general principles have emerged from the current database of complexes (4). Briefly, the TCR has an orientation over the pMHC surface that generally places the germline-encoded CDR1 and CDR2 loops in contact with the conserved helical residues of the MHC, whereas the highly variable, somatically recombined CDR3 loops primarily interact with the peptide. This docking orientation, also called the “footprint,” shows a high degree of variability (±60°) in different pMHC complexes, and from analysis of the structures, there appear to be few conserved contacts that are responsible for the orientation. This dilemma has raised the question of whether a “recognition code” could exist between TCR and MHC, with the implication that there would be a limited set of footprints. On the other hand, if the orientation is largely dictated by the identity of the antigenic peptide, there could be an unlimited array of orientations (5–7). To solve this puzzle, a much larger set of structures needs to be determined so that docking orientations can be systematically correlated with V-gene usage and MHC allele.
Although x-ray crystallography provides detailed, high-resolution structures of TCR–pMHC complexes, protein expression and crystallization of these low-affinity complexes [dissociation constant (Kd) > 1 μM] present technical barriers. In the 10 years since the structures of the first TCR–pMHC complexes were first reported, only ≈17 unique structures have been determined, each of which has been the result of major resource, time, and personnel commitments over periods of years (1). In contrast, well over 100 antibody–antigen complex structures exist in the Protein Data Bank (8). Although the pace of TCR–pMHC structure determination is increasing, it remains slow. Although production of single-chain TCR in Escherichia coli can be achieved for many TCR, crystallization of the low-affinity complexes is not a certainty. This continuing technical challenge increases the value of rapid lower-resolution methods to probe docking orientations of the TCR on MHC. Given a large set of TCR that binds to a single MHC, would we find an unrestricted array of docking orientations or would there be subsets depending on the Vα and Vβ chain usage by the TCR? For this question, high-resolution structures are not necessary; instead, a method that could paint the pMHC surface with the TCR footprint would suffice. In this way, footprint mapping could serve as a filtering step to characterize a collection of TCRs specific for a given pMHC. Then, a focused effort could be made to crystallize specific TCR–pMHC complexes of interest.
Solution NMR spectroscopy is well suited for experimental characterization of the TCR–pMHC footprint in a relatively short amount of time. After assignment of the protein backbone NMR resonances, structural information can be obtained through chemical shift perturbation mapping (9). This approach is most effective when the structures of one or both of the components are known, which increases the information contained within resonance assignments. The protein that has a known structure is isotopically labeled, ligand is added, and the resonances that correspond to residues involved in intermolecular contacts are identified by changes in chemical shift. These perturbations are then mapped on the known structure to define a binding site. In this fashion, many ligands can be mapped on a single assigned and isotopically labeled protein. NMR is sensitive to interactions over a wide range of affinities. In the present application, we envision that the footprints of many TCR could be mapped rapidly onto a common pMHC molecule, whose structure was previously known. Although such footprints would not constitute high-resolution 3D structures, they would provide information about docking footprint orientation. In addition, NMR allows investigation of TCR–MHC dynamics over a range of time scales.
There are two main challenges in studying TCR–pMHC interactions by NMR. First, milligram quantities of soluble protein must be produced through E. coli expression for isotopic labeling with 15N and/or 13C. Second, the size of the proteins must be reduced to minimize spectral complexity. For the former requirement, methods have been developed that permit the production of many, but not all, TCR molecules in E. coli as either full-length heterodimers or single-chain “Fv” VαVβ heterodimers (3). In fact, the NMR structure of a free single-chain TCR (scTCR) has been determined and was tantalizingly informative about aspects of binding site dynamics, which were not further explored with experiments adding the pMHC ligand (10). Many MHC molecules, primarily class I, can also be produced in E. coli, and so in principle, TCR–pMHC NMR studies are feasible. However, with the exception of one study using a 19F-labeled peptide to study conformational peptide isomers when bound to I-Ek, there are no published NMR experiments on pMHC alone or TCR–pMHC interactions (11). The large size of the TCR–pMHC complex (≈100 kDa) has remained the major obstacle, because NMR is particularly effective up to molecular weights of 40 kDa. At higher molecular weights, slow tumbling leads to spectral broadening of the protein resonances. In addition, as the number of protein residues increases, spectral overlap hinders analysis. Relatively novel techniques such as transverse relaxation-optimized spectroscopy (TROSY) experiments and per-deuteration of protein samples have advanced the protein size limit, whereas selective labeling has decreased the spectral overlap problem (12). Here, we use TCR and pMHC molecules whose sizes have been reduced to the point that NMR can be used quickly and economically to determine footprints of multiple TCR–pMHC complexes. We then relate the information provided by this analysis to previously determined crystal structures of the TCR, pMHC, and the TCR–pMHC complex.
Results
The 2C TCR clone has the ability to bind two structurally unique class I pMHC: the self peptide, dEV8, bound to the syngeneic MHC H–2Kb and the self peptide, QL9, bound to the allogeneic MHC H–2Ld (13). The available x-ray crystal structures of the unliganded H–2Ld molecule (14), 2C TCR (15), 2C–H–2Kb–dEV8 (16), and of the recently determined 2C–H2Ld–QL9 (17) provide complementary information to NMR studies of TCR–H–2Ld–QL9 complexes. A major enabling component to our experiments is that we used yeast surface display combined with directed protein engineering to produce a functional platform or “mini” H2–Ld molecule composed of only the α1 and α2 domains of the heavy chain and peptide. This mini-MHC, which binds 2C with wild-type affinity (17, 18), has a molecular mass of 21 kDa and refolded from E. coli together with the QL9 peptide (18). The structure of this mini-MHC determined at 2.4 Å shows it to be essentially identical to that of the full-length MHC (rms deviation for carbon-α atoms is 0.81 Å) (17). We also expressed a 2C scTCR (19) by direct secretion of folded soluble TCR into the periplasmic space of E. coli with a molecular mass of 25 kDa. Therefore, the 2C–Ld complex was reduced to ≈45 kDa, which is suitable for high-field NMR investigation.
Our strategy was to produce 15N-labeled H–2Ld refolded with unlabelled QL9 peptide, assign the NMR resonances of the labeled H–2Ld, and then add unlabelled 2C scTCR to monitor the resonances in the H–2Ld spectra that shift, indicating a TCR contact position. We do not label the peptide because we are initially interested in the TCR footprint on the MHC helices, which defines the docking orientation analogous to the hands on a clock. Footprints can range from perpendicular (12 and 6 o'clock), to diagonal (≈2 and 8 o'clock). Based on previous TCR–pMHC structures, the central residues of the peptide are a pivot point and will be contacted by the centrally disposed TCR CDR3s; hence, chemical shifts of peptide residues would provide little information about the peripheral helix contacts that define the overall TCR orientation. Also, the additional peptide residues would further complicate the crowded H–2Ld NMR spectra, so their elimination simplifies our data analysis.
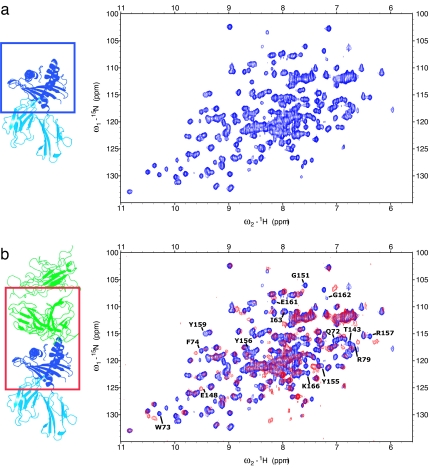
H–2Ld gave good quality 1H–15N HSQC (heteronuclear sequential quantum correlation) spectra (Fig. 1), indicating a folded protein. Backbone amide 15N and 1H resonances of H–2Ld, in addition to other NH moieties in protein side chains, were obtained from 1H15N-HSQC experiments (Fig. 1). Assignments were obtained from NOESY and scalar coupling-based 3D experiments. Uniformly, 13C–15N-labeled protein was used for scalar-coupling experiments. The most sensitive of such experiments, HNCA (amide proton and nitrogen to Cα carbon correlation), gave resonances for ≈70% of the residues, whereas less than half the expected resonances were present in CBCACONH (Cb and Cβ carbon to amide nitrogen and proton via carbonyl carbon correlation) experiments and <20 of 181 residues gave signal in HNCACB (amide proton and nitrogen to Cα and Cβ carbon correlation) or HCCH-TOCSY (′H-′3C-′3C-′H-total correlation spectroscopy) experiments. Assignments were completed with 15N-HSQC-NOESY experiments on a uniformly 15N and 50% (nominally) 2D-labeled MHC sample. NMR assignments were obtained for the two helices that form the peptide-binding cleft and for almost every residue in the protein loops. Helical residues are characterized by the presence of strong sequential HN(i)–HN(i + 1) NOEs and by the up-field shift of alpha protons in the 3.6/4.2 ppm range, as expected [Fig. 2 and supporting information (SI) Figs. 5 and 6]. Assignments of some residues in the helical region were independently verified with an MHC sample in which all Tyr residues were selectively 15N-labeled. Most of the amino acids in the β-sheet region gave weak NMR signals and could not be unambiguously assigned (Fig. 3a and SI Fig. 7). Although it is expected that signals from the supposedly rigid β-core are broader and therefore less observable, it is rather unusual for them to completely disappear in a 21-kDa protein such as the platform MHC. Although the MHC is monodisperse by gel filtration, transient aggregation at the high concentrations in the NMR tube (≈0.3 mM) is the likely reason for the loss of signal, because improved spectral quality was observed for more dilute samples. The aggregation could possibly be due to the solvent exposure of apolar residues underneath the β-sheet platform, which are normally shielded from solvent by the α3 domain and β2m in the full QL9/Ld molecule. Buffer conditions were tested and three solvent-exposed hydrophobic residues in the β-sheet were mutated to hydrophilic amino acids based on the Ld structure with no improvement in spectral quality. Because the β-sheet region is not in the part of the MHC that is expected to interact with TCR, the lack of these assignments did not affect our ability to map the TCR-binding interface.
Fig. 1.
800 MHz 1H-15N HSQC spectra collected on the platform MHC. (a) Free MHC. (b) Superposition of the free MHC (blue) and its complex with 2C TCR (red). Only the region of the MHC highlighted in the box is present in our NMR sample; the TCR and peptide are not isotopically labeled and not visible in the spectra. Some of the peaks that shift upon complex formation are highlighted.
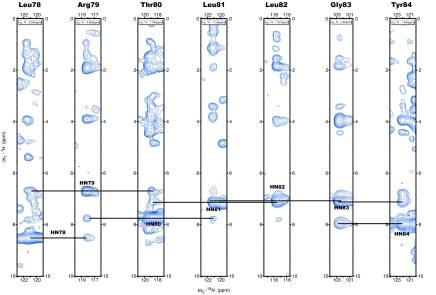
Fig. 2.
1H-15N planes extracted from a 15N-edited 3D NOESY experiment at 800 Mhz. Frequencies of the amide proton of L78 to T84 are shown. Strong sequential amide–amide interactions characteristic of helical conformation are observed in the spectrum and are annotated in the figure.
Fig. 3.
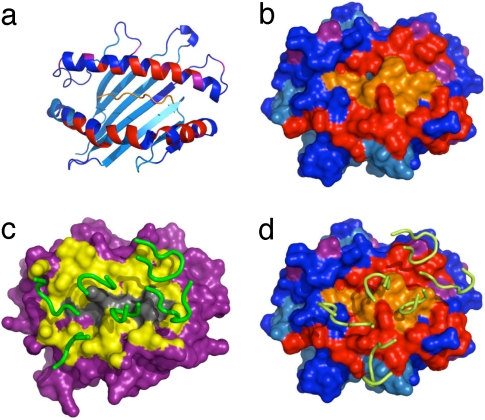
Sequence alignment of Ld and Kb, and identification of Ld residues that shift upon TCR binding. (a) Amino acid sequence alignment of the α1 and α2 domains of class I MHCs Kb (Top), Ld (Middle), and stabilized mutant Ld–m31 (Bottom). Residues that differ between Kb and Ld are highlighted in orange. Residues that differ between Ld (wild-type) and Ld–m31 are highlighted in light blue. Previous studies (24) have identified residues in the helices of Ld that are important for TCR binding to Ld (denoted by a green box below the amino acid). Residue positions that showed changes in chemical shifts of their NMR spectra upon 2C TCR binding are red and all assigned residues are yellow. (b) Ld α1 and α2 helices are shown in blue, with Ld residues that showed chemical shifts upon TCR complex formation shown in red. The majority of residues that are influenced by TCR binding line the peptide-binding groove. In contrast, residues that do not shift upon complex formation (blue) tend to point away from the groove.
Each peak in the 1H-15N HSQC corresponds to a specific protein residue and its chemical shifts are exquisitely sensitive to the local chemical and structural environment. The sensitivity is such that 15N HSQC spectra are often referred to as protein fingerprints because they provide a unique representation of a molecule. Upon complex formation, the chemical environment of residues close to the interface will change and the corresponding peaks in the 15N HSQC spectrum will shift. Consequently, assigning each peak to its specific protein residue allows identification of the residues involved in intermolecular interactions. To map the TCR–pMHC interface, NMR data were collected on samples of 15N-labeled platform MHC and unlabelled 2C TCR mixed in equimolar amount. The TCR was left unlabelled so that its signal would not be present in NMR spectra, thus simplifying data analysis, while maintaining its effect on the resonances of the platform MHC. The free and bound 15N HSQC of the platform MHC were compared (Fig. 1) and residues were denoted as a significant chemical shift difference if, upon complex formation, their position changed by >0.02 ppm in the proton dimension or 0.2 ppm in the nitrogen dimension (SI Fig. 6). The same strategy can be applied to 13C HSQC experiments, which provide information on protein side chains instead of protein backbone amides. We did not follow chemical shift changes in 13C HSQCs' spectra because such spectra are usually less informative than 15N HSQCs and because 13C-labeling is roughly 10-fold more expensive than 15N-labeling, making it less economical for rapid mapping of many TCRs. Likewise, the peptide was not labeled in the NMR experiments.
Significant chemical shift changes were observed in 28 assigned residues upon complex formation with the TCR (Figs. 1 and 3 and SI Fig. 6). Differences between free and bound states reflect a change in the local chemical environment, which can result from either direct interaction between the MHC and the TCR or local structural rearrangement; the NMR data itself cannot differentiate between these two situations. Nonetheless, MHC helix α1 residues 69–77 and MHC helix α2 residues 154–159 are the pivotal regions of the NMR determined footprint, positioned along a diagonal across the peptide-binding cleft (Figs. 3 and 4 a and b). MHC residues I63 and V66, which face the peptide, also shift, whereas the residues found on the opposite side of the peptide-binding groove do not shift, providing a way to localize the TCR (Fig. 3b). Residues pointing away from the peptide-binding site or the TCR tend not to undergo chemical shift changes (Fig. 3b), supporting the interpretation that chemical shift changes arise directly from interactions with the TCR. It is clear that residues in the α1 and α2 helices are either influenced directly by TCR interactions, or they are involved as peptide contact residues that line the groove and are affected indirectly by the TCR (Fig. 3b). As most of the side chains of shifted residues shown in Fig. 3b are likely involved in contacts with the peptide, the NMR results provide evidence that the dynamics of the entire TCR–pMHC complex could be influenced through indirect interactions that may not be apparent from crystallographic approaches. Although residues W147 and Q149 are marked as not being affected by binding, their overlap with other residues in the NMR spectra did not allow us to determine their status in the complex.
Fig. 4.
Residues that shift upon complex formation mapped on crystal structure. Residues with ambiguous or missing assignments are shown in cyan, residues with significant shift are shown in red, and residues with a small shift are shown in purple; the unlabelled peptide, for which we have no information, is shown in orange. (a) Cartoon representation of the H–2Ld–peptide structure with NMR shifts colored. (b) Surface representation of H–2Ld–peptide with NMR shifts colored. (c) Surface representation of the x-ray structure of the 2C–H–2Kb–dEV8 complex (16); the CDR loops are shown in green and residues within 6 Å of the MHC–TCR interface expected to shift in a mapping experiment are in yellow. (d) Surface representation of the x-ray structure of the 2C–H–2Ld–QL9 complex (17). The 2C CDR loops are shown in yellow, and residues are colored to indicate shift mapping result as in b.
Although resonances from the region of helix α2 between E154 and E163 were difficult to assign because of the weak NMR signal, preparation of an MHC sample with selectively 15N-labeled tyrosines greatly simplified the spectra and allowed identification of Y155 and Y156, as well as confirmation of neighboring residue assignments. Their involvement in the intermolecular interface was confirmed by adding unlabelled TCR to the sample. Residues Y159–L160 could not be unambiguously assigned, but the assignments that have been made in the region E154–E163 give sufficient information to define an overall footprint (Fig. 4). When comparing these results to a 2.4-Å crystal structure of the 2C–H–2Kb–dEV8 structure (Fig. 4c), residues E154–E163 were proximal to the TCR, indicating that the results are consistent with, but not identical to, the structure of another related TCR–pMHC complex. The observed shifts fit well with the 2.35-Å crystal structure of 2C–H–2Ld–QL9, which is rotated toward the α2 helix (Fig. 4d).
Residues I142–K146 and E166–H169 in helix α2 show significant chemical shift changes upon complex formation, but are at the edge of the interface, not directly in contact with the TCR. The shift might be caused by a rearrangement of the TCR loops or simply by local, minor changes in the MHC, because NMR shifts simply reflect a change in the chemical environment.
Comparison of Binding of Two TCR Mutants with Different Affinity.
To test this method with another related H2–Ld-specific TCR, we chose to look at a high-affinity mutant of 2C, called m6. Yeast display was used to isolate the m6 high-affinity mutant by varying the sequence of the CDR3α of the 2C TCR (20). We subsequently conducted chemical-shift mapping experiments on wild-type 2C TCR with an affinity of 2 μM for Ld (described above) and high-affinity 2C TCR variant m6 with an affinity of 8 nM (CDR3α sequence 98-SGFASAL-104 in wild-type was changed to 98-SHQGRYL-104 in m6) (18). The advantage of analyzing the 2C TCR m6 was an improvement in the robustness of the NMR chemical shift spectral quality because of the higher affinity of the TCR, which results in a longer residence time of the TCR–pMHC complex. The intermediate exchange regime of the low-affinity complex moved toward a more favorable slow exchange in the high-affinity complex. When binding affinity is low, the protein can alternate between free and bound states, which results in the broadening of the NMR signal; in a complex with higher affinity, the bound form is stabilized and the resulting signal is sharper. In addition, we could then compare the binding of two different TCRs to the same pMHC, allowing us to test and validate the method of footprint mapping by chemical shift.
All residues that shifted upon complex formation with 2C TCR did so in the 2C TCR high-affinity mutant (m6), producing a nearly identical footprint of MHC residues that shift. The only exception was MHC residue L168, which shifts in the high-affinity mutant but not in the wild-type. Although the footprint of residues that shifted was maintained, the extent that they shifted differed between the two complexes. Increased shifts are particularly found in the 154–163 region of helix α2, part of which could not be unambiguously assigned. The peaks for these residues show some of the larger chemical shift differences between 2C and m6, and display a significantly weaker signal in the 2C versus the higher-affinity m6 complex. Interestingly, this is the region of the MHC that appears to be in closest contact to the TCR in the x-ray crystal structure (17). The TCR mutations from wild-type 2C to m6 are in the CDR3α, and we found in the 2C–H–2Ld–QL9 crystal structure that CDR3α primarily contacts the peptide, which we cannot see in NMR experiments because it is not labeled. Mutations in the CDR3α loop appear to have only a local structural effect, altering binding affinity but not the binding footprint. This observation suggests that the interaction between the variable CDR3α loop and the peptide does not act as a hinge upon which the whole TCR can rotate, but rather that the binding footprint is dictated by interaction between the other conserved CDR loops 1 and 2 and MHC residues, leaving the flexible CDR3α loop free to adapt to the peptide. The high-affinity mutant, m6, shows chemical shift changes that are relatively weak in 2C. Therefore, m6 is a good probe for the small chemical shifts given by 2C, amplifying the shifts observed in 2C and increasing our confidence in these measurements.
The complex experiments do not explain why peaks of the MHC residues 154–163 region are generally broad, giving poor signal in NMR experiments on the free pMHC. The NMR data suggest that the MHC structure in this region alternates between different conformations. In crystal structures, we only visualize the static conformations of the pMHC, which can be stabilized by crystal contacts. Therefore, the NMR experiments could provide insight into the conformational dynamics of the pMHC recognition surface that has so far not been appreciated.
Comparison of the Binding Orientation of 2C to H–2Kb and H–2Ld.
Through previously solved crystal structures, it is possible to verify the assignments and shift perturbations determined through NMR. First, we indicated the residues that change chemical shift upon complex formation on the known structure of the unliganded H–2Ld structure (Fig. 4 a and b). A clear footprint of 2C interaction is evident on the molecular surface (Fig. 4b). The NMR results were then compared with the crystal structure analyses that have been recently made between the complex structures of the 2C TCR with H–2Kb–dEV8 (16) or with H–2Ld–QL9 (17). Kb and Ld differ at 31 positions, and 13 of these residues are located in the α helices (5 in α1 and 8 in α2) (Fig. 3a) (21). Furthermore, peptides QL9 (QLSPFPFDL) and dEV8 (EQYKFYSV) are dramatically different, making these two complexes more distinct than any syngeneic/allogeneic TCR system that has been examined (22, 23). As the next step in this comparison, we highlighted the surface of the MHC that is within 6 Å of the 2C TCR in the 2C–H–2Kb–dEV8 structure to produce an approximation of an NMR shift map for this TCR–pMHC complex (Fig. 4c). This footprint was rotated more clockwise on its long axis (≈3 and 9 o'clock) from the NMR footprint we determined for 2C–H–2Ld–QL9, which is also diagonal but appears slightly more perpendicular (≈2 and 8 o'clock). In the case of the 2C–H–2Ld–QL9 interaction, residues identified by chemical-shift mapping as being part of the contact surface on H–2Ld–QL9 defines a footprint that accurately demarcates the binding footprint seen in the crystal structure of the 2.3-Å 2C–H–2Ld–QL9 complex (Fig. 4d). Our measurements suggest that the TCR is shifted more perpendicular to the peptide based on our shift-mapping results and mirrors the 44° crossing angle of the H–2Ld structure rather than the H–2Kb, which crosses the peptide with a 22° angle (16, 17).
Major differences between the 2C–H–2Kb–dEV8 and 2C–H–2Ld–QL9 footprints are evident by NMR mainly in helix α2 of the MHC (I142–E148). Here, chemical-shift changes upon complex formation are apparent in H–2Ld–QL9, but this region is not proximal to the TCR in the 2C–H–2Kb–dEV8 structure. Moreover, in the 2C–H–2Kb–dEV8 structure, the CDR1α loop of the TCR seems to be close to residues that do not shift upon binding in the Ld complex. The visual representation of these differences is striking if the expected contacts between 2C and H–2Kb–dEV8 (arbitrarily set at 6 Å) are compared with our chemical-shift mapping results (Fig. 4 c and d). By applying NMR analysis in the case of a known structure, we are able to confirm that two TCR–pMHC complexes can be distinguished when the crossing angle difference is >20°. At this point, we cannot rule out the possibility that NMR footprint mapping can identify even smaller differences in crossing angle. Because the basic footprint is maintained between the syngeneic (H–2Kb) and allogeneic MHC (H–2Ld), our results support the recent finding that allogeneic reactions are not the result of major shifts in peptide crossing angles and docking (17).
Discussion
Structural biology has identified many of the important characteristics of the interaction of TCR with pMHC. Despite many studies, two fundamental questions remain unanswered: What are the structural rules for TCR recognition of antigenic peptides in the context of MHC molecules and how can a limited set of TCRs respond to a diverse group of foreign peptides? Because structural information on a large number of TCR–pMHC complexes may help answer these questions, a rapid method for detection and characterization of TCR–pMHC interaction is needed. Here we report the use of NMR chemical-shift mapping to define the binding footprint of the 2C TCR on QL9/Ld. The approach proved successful despite technical challenges for NMR spectroscopy including large molecular weight, low protein concentration, sample aggregation, and weak binding interaction. We believe that the methodology we present can be used to construct a database of TCR–pMHC complex footprints, furthering our understanding of the rules for receptor ligation.
The method described here satisfies several requirements to study a number of TCR–pMHC interactions. The first step in this process is assignment of the NMR backbone resonances for the free protein of interest. Although here we assigned the MHC, assignments could also be done for an scTCR, and shifts could be measured when MHC was added. Following assignment of one component, chemical-shift mapping of related complexes can be achieved with simple HSQC experiments lasting 15 min to a few hours, depending on sample concentration; the data can then be analyzed in a single day. All protein components used in this experiment express at a level of several milligrams per liter in E. coli and so an NMR sample of 0.2 mM can be reliably produced in a few days with a 3 L bacterial culture. Although isotopic 15N-labeling of the protein of interest is needed for NMR, the source of 15N for minimal media is inexpensive. Engineering the platform MHC (18) and assigning its NMR spectrum was the first necessary step and a major hurdle.
The technique can now be used to probe the binding of additional TCRs that are restricted by H–2Ld. By studying a panel of TCRs, it will be possible to test whether TCRs recognize peptide/H–2Ld complexes with grossly different footprints. Mutagenesis studies by Connolly and coworkers have suggested that a group of residues (G69, Q72, V76, Y155, and R157) are involved in recognition by a panel of different T cells that are restricted by H–2Ld (24). These residues are among those identified in the present study (Fig. 3a), supporting the notion that NMR can be used to define the complete footprint of diverse TCRs. Should such an approach prove successful, it could be expanded to other MHC molecules, thereby determining the structural solutions represented by many different TCR–pMHC complexes.
The role of protein intra and intermolecular dynamics in TCR–MHC recognition remains unknown. NMR probes molecular dynamics over a range of time scales from nanoseconds to seconds (25). The experimental system outlined here will allow NMR dynamics measurements on the MHC–TCR complexes. Such results cannot be assessed directly by crystallography, and conformational dynamics may be important in the mechanisms by which TCRs engage their ligands and transmit signals to the cell (26).
Materials and Methods
H–2Ld Protein Expression and Purification.
Ld was cloned into pET28a (Novagen, Madison, WI) and expressed in BL21 (DE3) Codon Plus E. coli (Stratagene, La Jolla, CA). Starter cultures grown in LB media containing 30 μg/ml kanamycin were diluted into labeled M9 minimal media and grown to an OD600 of 1.0. Cultures were then induced with 1 mM IPTG for 3.5 h. Minimal media was produced containing 15N alone or 15N and 13C as sole source of nitrogen and glucose, as required, and 50% D2O when sample deuteration was desired. Selectively labeled 15N tyrosine protein was prepared by adding 50 mg/liter of each unlabeled amino acid except tyrosine and 50 mg/liter 15N-labeled tyrosine, while removing any other source of nitrogen from the media. After induction, bacteria were passed through a French press and inclusion bodies were washed in 10 mM Tris/1 mM EDTA, pH 8.0 (TE) supplemented with 0.1% Triton-X until they formed a homogeneous pellet and washed twice again in TE alone. The yield of inclusion bodies was generally 150 mg per liter of bacterial culture. Washed inclusion bodies were solubilized in 8 M urea/50 mM Mes, pH 6.5 at 25°C overnight while shaking. For the refolding reaction, 8 mg of purified QL9 peptide was added to 400 ml of refolding buffer (100 mM Tris·HCl/400 mM l-arginine/0.5 mM PMSF/0.5 mM oxidized/5 mM reduced glutathione, pH 8.0). Over a 24-h period, 200 mg of urea-solubilized Ld–m31 inclusion bodies were added in three equal additions. The refold mixture was filtered; dialyzed against 20 mM Tris·HCl/150 mM NaCl, pH 8.0; concentrated to 500 μl in each of four Vivaspin 20 10 K cutoff (VivaScience, Stonehouse, U.K.); and subjected to size-exclusion chromatography (Superdex 75; Amersham Biosciences, Piscataway, NJ). The yield of purified, refolded protein from a 400-ml refold was 2–4 mg.
2C TCR Expression and Purification.
Protein was expressed from pET-22b (Novagen) in BL21 (DE3) Codon Plus E. coli (Stratagene) and purified as described in ref. 27. Briefly, cultures were grown overnight at 30°C in LB media plus 200 μg/ml ampicillin. Cells were then transferred to fresh LB media plus ampicillin, grown 1 h at 30°C and induced with 1 mM IPTG. After 4 h, cells were harvested and subjected to an osmotic shock procedure. Osmotic shock supernatant was dialyzed in 10 mM Tris·HCl/200 mM NaCl, pH 8.0, and subjected to affinity chromatography with Ni-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen,Valencia, CA) and size-exclusion chromatography (Superdex 75, Amersham Biosciences, Piscataway, NJ).
NMR Experiments.
NMR spectra were collected at 25°C or 35°C on 600 and 800 MHz (Varian Inova, Palo Alto, CA) or 500 MHz (Bruker AMX, Rheinstetten, Germany) with cryoprobe, processed with NMRPipe (28), and analyzed with SPARKY (29). HNCA, HNCACB, CBCACONH, and HCCH-TOCSY experiments were recorded on a uniformly 13C, 15N platform MHC sample. 15N-HSQC-NOESY experiments were collected on a uniformly 15N-labeled, 50% nominally 2D-labeled sample. The NMR buffer was 10 mM NaPO4, 100 mM NaCl, 0.2% NaN3, 10% D2O, pH 7.0. Sample concentration was typically 0.3 mM for 3D experiments and 0.1 mM for HSQCs. pMHC and TCR samples were individually purified, mixed in equimolar amounts, and concentrated in a Vivaspin 500 10-kDa cutoff (VivaScience) to the desired concentration.
Supplementary Material
Acknowledgments
We thank Phil Holler and Susan Brophy (University of Illinois at Urbana–Champaign) for generation of the m6 TCR and the m31 Ld mutants, respectively; and Herman Eisen (Massachusetts Institute of Technology, Cambridge, MA) for his advice and previous studies on the 2C system. This work was supported by National Institutes of Health grants (to K.C.G., J.D.P., and D.M.K.). L.A.C. was supported by a National Science Foundation predoctoral fellowship, J.D.P. was supported by the David and Lucile Packard Foundation, and K.C.G. was supported by the W. M. Keck Foundation, Pew Scholars, and the Howard Hughes Medical Institute.
Abbreviations
- TCR
T cell receptor
- pMHC
peptide-MHC
- CDRs
complementarity-determining regions
- HSQC
heteronuclear sequential quantum correlation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0703702104/DC1.
References
- 1.Rudolph MG, Stanfield RL, Wilson IA. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 2.Davis MM, Bjorkman PJ. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 3.Ely LK, Kjer-Nielsen L, McCluskey J, Rossjohn J. IUBMB Life. 2005;57:575–582. doi: 10.1080/15216540500215556. [DOI] [PubMed] [Google Scholar]
- 4.Bankovich AJ, Garcia KC. Immunity. 2003;18:7–11. doi: 10.1016/s1074-7613(02)00517-4. [DOI] [PubMed] [Google Scholar]
- 5.Housset D, Malissen B. Trends Immunol. 2003;24:429–437. doi: 10.1016/s1471-4906(03)00180-7. [DOI] [PubMed] [Google Scholar]
- 6.Huseby E, Kappler J, Marrack P. Eur J Immunol. 2004;34:1243–1250. doi: 10.1002/eji.200425000. [DOI] [PubMed] [Google Scholar]
- 7.Sherman LA, Chattopadhyay S. Annu Rev Immunol. 1993;11:385–402. doi: 10.1146/annurev.iy.11.040193.002125. [DOI] [PubMed] [Google Scholar]
- 8.Berman H, Henrick K, Nakamura H. Nat Struct Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 9.Gao G, Williams JG, Campbell SL. Methods Mol Biol. 2004;261:79–92. doi: 10.1385/1-59259-762-9:079. [DOI] [PubMed] [Google Scholar]
- 10.Hare BJ, Wyss DF, Osburne MS, Kern PS, Reinherz EL, Wagner G. Nat Struct Biol. 1999;6:574–581. doi: 10.1038/9359. [DOI] [PubMed] [Google Scholar]
- 11.Schmitt L, Boniface JJ, Davis MM, McConnell HM. J Mol Biol. 1999;286:207–218. doi: 10.1006/jmbi.1998.2463. [DOI] [PubMed] [Google Scholar]
- 12.Gardner KH, Kay LE. Annu Rev Biophys Biomol Struct. 1998;27:357–406. doi: 10.1146/annurev.biophys.27.1.357. [DOI] [PubMed] [Google Scholar]
- 13.Chen J, Eisen HN, Kranz DM. Microbes Infect. 2003;5:233–240. doi: 10.1016/s1286-4579(03)00016-9. [DOI] [PubMed] [Google Scholar]
- 14.Speir JA, Garcia KC, Brunmark A, Degano M, Peterson PA, Teyton L, Wilson IA. Immunity. 1998;8:553–562. doi: 10.1016/s1074-7613(00)80560-9. [DOI] [PubMed] [Google Scholar]
- 15.Garcia KC, Degano M, Stanfield RL, Brunmark A, Jackson MR, Peterson PA, Teyton L, Wilson IA. Science. 1996;274:209–219. [PubMed] [Google Scholar]
- 16.Garcia KC, Degano M, Pease LR, Huang M, Peterson PA, Teyton L, Wilson IA. Science. 1998;279:1166–1172. doi: 10.1126/science.279.5354.1166. [DOI] [PubMed] [Google Scholar]
- 17.Colf LA, Bankovich AJ, Hanick NA, Bowerman NA, Jones LL, Kranz DM, Garcia KC. Cell. 2007;129:135–146. doi: 10.1016/j.cell.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 18.Jones LL, Brophy SE, Bankovich AJ, Colf LA, Hanick NA, Garcia KC, Kranz DM. J Biol Chem. 2006;281:25734–25744. doi: 10.1074/jbc.M604343200. [DOI] [PubMed] [Google Scholar]
- 19.Hoo WF, Lacy MJ, Denzin LK, Voss EW, Jr, Hardman KD, Kranz DM. Proc Natl Acad Sci USA. 1992;89:4759–4763. doi: 10.1073/pnas.89.10.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holler PD, Holman PO, Shusta EV, O'Herrin S, Wittrup KD, Kranz DM. Proc Natl Acad Sci USA. 2000;97:5387–5392. doi: 10.1073/pnas.080078297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen T, Balendiran G, Solheim J, Ostrov D, Nathenson S. Immunol Today. 2000;21:83–88. doi: 10.1016/s0167-5699(98)01426-1. [DOI] [PubMed] [Google Scholar]
- 22.Luz JG, Huang M, Garcia KC, Rudolph MG, Apostolopoulos V, Teyton L, Wilson IA. J Exp Med. 2002;195:1175–1186. doi: 10.1084/jem.20011644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reiser JB, Darnault C, Guimezanes A, Gregoire C, Mosser T, Schmitt-Verhulst AM, Fontecilla-Camps JC, Malissen B, Housset D, Mazza G. Nat Immunol. 2000;1:291–297. doi: 10.1038/79728. [DOI] [PubMed] [Google Scholar]
- 24.Hornell TM, Solheim JC, Myers NB, Gillanders WE, Balendiran GK, Hansen TH, Connolly JM. J Immunol. 1999;163:3217–3225. [PubMed] [Google Scholar]
- 25.Wand AJ. Nat Struct Biol. 2001;8:926–931. doi: 10.1038/nsb1101-926. [DOI] [PubMed] [Google Scholar]
- 26.Krogsgaard M, Li QJ, Sumen C, Huppa JB, Huse M, Davis MM. Nature. 2005;434:238–243. doi: 10.1038/nature03391. [DOI] [PubMed] [Google Scholar]
- 27.Maynard J, Adams EJ, Krogsgaard M, Petersson K, Liu CW, Garcia KC. J Immunol Methods. 2005;306:51–67. doi: 10.1016/j.jim.2005.07.022. [DOI] [PubMed] [Google Scholar]
- 28.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 29.Goddard TD, Kneller DG. SPARKY 3. San Francisco, CA: University of California; 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.