Abstract
Multiple sclerosis (MS) is a demyelinating disease characterized by an unpredictable clinical course with intermittent relapses that lead over time to significant neurological disability. Clinical and radiological variables are limited in the ability to predict disease course. Peripheral blood genome scale analyses were used to characterize MS patients with different disease types, but not for prediction of outcome. Using complementary-DNA microarrays we studied peripheral-blood gene expression patterns in 53 relapsing–remitting MS patients. Patients were classified into good, intermediate and poor clinical outcome established after 2-year follow-up. A training set of 26 samples was used to identify clinical outcome differentiating gene-expression signature. Supervised learning and feature selection algorithms were applied to identify a predictive signature that was validated in an independent group of 27 patients. Key genes within the predictive signature were confirmed by quantitative reverse transcription–polymerase chain reaction in an additional 10 patients. The analysis identified 431 differentiating genes between patients with good and poor clinical outcome (change in neurological disability by the expanded disability status scale was −0·33 ± 0·24 and 1·6 ± 0·35, P = 0·0002, total number of relapses were 0 and 1·80 ± 0·35, P = 0·00009, respectively). An optimal set of 29 genes was depicted as a clinical outcome predictive gene expression signature and classified appropriately 88·9% of patients. This predictive signature was enriched by genes related biologically to zinc-ion binding and cytokine activity regulation pathways involved in inflammation and apoptosis. Our findings provide a basis for monitoring patients by prediction of disease outcome and can be incorporated into clinical decision-making in relapsing–remitting MS.
Keywords: gene expression, multiple sclerosis, pathways, prediction, regulation
Introduction
Multiple sclerosis (MS) is a central nervous system disease affecting young adults in which 85% of patients experience a relapsing–remitting (RR) clinical course [1]. Clinical outcome differs between patients, as the rate of disease progression and frequency of relapses vary along the disease course [2]. It has been suggested that age of disease onset below 35 years, rapid development and regression of initial symptoms, a single symptom at onset and visual loss as the initial symptom indicate a good prognosis. Brain magnetic resonance imaging (MRI) parameters have also been implicated as important in the evaluation of MS course by measuring disease load over time [3]. Brain atrophy has been reported to account for more variance than lesion burden in predicting cognitive impairment [4]. However, all these clinical and radiological variables are limited in their ability to predict disease outcome, especially during the early stages of the disease. Gene microarray technology, that analyses simultaneously the expression of thousands of genes [5], can be used as a comprehensive analysis method to correlate gene expression patterns with numerous clinical parameters related to patients' outcome. Attempts to correlate MS gene expression with disease activity disclosed that activity correlated with the frequency of CX3CR1-positive natural killer (NK) cells [6], and that MS expression profiling identified responder and non-responder phenotypes to interferon (IFN)-β treatment [7].
We have demonstrated previously that gene expression signature of peripheral blood mononuclear cells (PBMC) significantly differentiates RRMS patients from healthy subjects. Having also demonstrated that different gene signature characterizes MS disease stage (relapse versus remission) [8], in the current study we sought to evaluate whether gene expression profiling can differentiate RRMS patients according to their clinical course − either favourable or poor. Our idea was to use an informative subset of original training samples. This subset consists of only good-outcome RRMS patients who did not deteriorate neurologically within a 2-year period, and patients with poor outcome who increased their disability and demonstrated clinical disease activity within the same follow-up period. These extreme training samples yielded a clear platform from which to identify genes whose expression is related to clinical outcome. The discriminating genes were then integrated by a support vector machine (SVM) to build a prediction model, by which each validation sample was assigned a good or poor risk score for MS progression. We found that RRMS patients in high- and low-risk groups are clearly distinguishable. Our results indicate that gene expression profiles combined with carefully chosen learning algorithms can predict patient outcome and may be incorporated in individualized, tailored management of RRMS.
Methods
Patients
Sixty-three patients with definite RRMS (45 females, 18 males), aged 38·2 ± 3·9 years, disease duration 9·2 ± 2·8 years, annual relapse rate 1·1 ± 0·5 and neurological disability evaluated by the Expanded Disability Status Scale (EDSS) [9], 1·9 ± 0·6, were included in the study; 26 patients participated in the differentiating clinical outcome analysis, 27 patients in the validation process of prediction and 10 patients in the functional biological validation.
The clinical and demographic variables were similar between groups (Table 1). In the differentiating clinical outcome group, 13 patients were receiving immunomodulatory treatments for at least 3 months prior to the gene expression study, and 13 patients were naive to immunomodulatory treatment. In the validation group, 11 patients were receiving immunomodulatory treatments for at least 3 months prior to the gene expression study, and 16 patients were naive to immunomodulatory treatments. Within up to 1 month from withdrawal of blood, all patients were treated with IFN-β1a. None of the patients had ever received cytotoxic treatments and all were free of steroid treatment for at least 30 days before blood was withdrawn. All patients had peripheral blood counts within the normal range. The study was approved by the Sheba Medical Center Institutional Review Board, and all patients gave written informed consent for participation.
Table 1.
Demographic and clinical variables of the study relapsing–remitting multiple sclerosis (RRMS) population.
| Validation | |||
|---|---|---|---|
| Variable | Differentiating clinical outcome group n = 26 | Clinical outcome classifier group n = 27 | Biological function by qRT–PCR group n = 10 |
| Age (years) | 40·2 ± 5·8 | 40·5 ± 1·6 | 36·1 ± 2·1 |
| F (M) | 21 (5) | 16 (11) | 8 (2) |
| Disease duration (years) | 9·9 ± 4·2 | 10·3 ± 1·6 | 8·5 ± 1·4 |
| Relapse rate | 1·3 ± 0·7 | 0·9 ± 0·2 | 0·9 ± 0·2 |
| EDSS | 2·0 ± 1·0 | 2·5 ± 0·2 | 1·9 ± 0·3 |
| Treated | 13 | 11 | None |
EDSS: Expanded Disability Status Scale; qRT–PCR: quantitative reverse transcription–polymerase chain reaction.
Clinical follow-up
Patients were followed-up prospectively for a period of 2 years. Neurological examination was performed once every 3 months and at the time of a suspected relapse, and EDSS assessment was completed accordingly. Relapse was defined as the onset of new objective neurological symptoms/signs or worsening of existing neurological disability, not accompanied by metabolic changes, fever or other signs of infection, lasting for a period of at least 48 h accompanied by objective change of at least 0·5 points in the EDSS score. For EDSS evaluations we used only stable EDSS scores that were confirmed at 3-month follow-up examinations. Confirmed relapses and EDSS scores were recorded consecutively.
Definition of clinical outcome
Clinical outcome was defined according to neurological disability as the primary criterion and total number of relapses as the secondary criterion.
Good outcome
Good outcome comprised patients who had not deteriorated in their neurological disability and had not experienced any relapse during the 24 months of follow-up.
Poor outcome
Poor outcome comprised patients who deteriorated in their neurological disability (ΔEDSS increased by at least 0·5 points) within the 24 months of follow-up, either with or without relapses.
Intermediate outcome
Intermediate outcome comprised patients who did not deteriorate in their neurological disability yet experienced at least one relapse during the 24 months of follow-up.
RNA isolation and microarray expression profiling
PBMC were separated on Ficol-Hypaque gradient, total RNA was purified, labelled, hybridized to a Genechip array (U95Av2 and HU-133A) and scanned (Hewlett Packard, GeneArray-TM scanner G2500A) according to the manufacturer's protocol (Affymetrix Inc, Santa Clara, CA, USA), as described previously [8].
Clinical outcome differentiating genes analysis
RMAExpress software was used to analyse the scanned arrays [10]. In order to be consistent with the ontology and array type, all the transcripts in U95Av2 microarray were converted to the corresponding transcripts in HU-133A using NetAffex comparison table. Probe sets that did not have a signal present in at least 90% of the samples were filtered. Noise effect was reduced by fitting a multiple effect model for each gene modelling the log-ratio measurement as a sum of contributions for age, gender, batch, subject state (naive or treated) and time from last steroid treatment.
Statistical methods
Statistical analysis was performed using the ScoreGenes software tools (http://compbio.cs.huji.ac.il/scoregenes/). Data were analysed by t-test, the threshold number of misclassifications (TNoM) method and the Info-test score. Differentiating genes were defined as genes whose expression was significantly higher or lower, with P < 0·05 in all three statistical tests. Overabundance analysis was used to compare between the number of observed and expected genes that differentiated between the good and poor clinical outcome under the null hypothesis that the classification of the samples was random [11,12]. To verify further the accuracy of the classification we used the leave-one-out cross-validation (LOOCV) statistical method [13]. LOOCV simulates removal of a single sample for every trial and trains on the rest. The procedure is repeated until each sample is left out once and the number of correct and incorrect predictions is counted. The demographic variables are presented as mean ± standard deviation (s.d.). Student's t-test was used to compare the difference in clinical variables between groups, and P < 0·05 was considered statistically significant.
Predictive genes analysis and validation
To depict the predictive genes from the differentiating clinical outcome signature, the SVM in combination with Forward feature selection algorithm were applied (http://ro.utia.cz/fs/fs_algorithms.html) [14–16]. SVM generates a classifier based on a known labelled training set (19 of 26 RRMS patients with good or poor clinical outcome from the differentiating clinical outcome group). Then, the classification power of the generated classifier is evaluated by applying it to an independent test set (nine of 27 RRMS patients from the validation group). The feature selection algorithm finds a subset of predictive genes that enables the generated classifier to achieve the highest classification rate [14]. To validate the power of the predictive genes, the classifier was applied to an additional independent set (18 of 27 RRMS patients from the validation group). Additionally, to confirm independently the obtained expression profiling data, we performed quantitative reverse transcription–polymerase chain reaction (qRT–PCR) on PBMC samples from 10 MS patients for three key genes of the predictive signature.
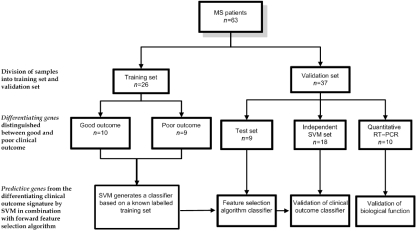
The study design is depicted in Fig. 1.
Fig. 1.
Flowchart of the study design. Overview of the strategy used for the identification and validation of predictive clinical outcome gene-expression signature in relapsing–remitting multiple sclerosis.
Biological functional analysis
Functional annotation of the clinical outcome differentiating and predictive gene signatures was performed using functional classification tools (FCT; David Bioinformatics Resources: http://david.abcc.ncifcrf.gov/home.jsp). Gene enrichment was defined as a group of genes associated highly with a specific biological function and measured statistically by one-tailed Fisher's exact probability value using the David system. Biological regulatory pathway reconstruction for the predictive gene signature was performed using: (1) PathwayArchitect software (http://www.stratagene.com) based on published data in the literature, and (2) Genomica software (http://genomica.weizmann.ac.il), based on Bayesian network methods taken from the field of machine learning, and was applied to our results of the differentiating gene microarray expression signature [17]. This evaluation was aimed to identify potentially target genes that share a common regulatory mechanism.
Results
Clinical classification of study patients
Patients were classified into three groups based on their clinical disease outcome: (1) patients with good outcome (n = 9, mean age 39·3 ± 3·3 years, disease duration 10·7 ± 3·4 years), (2) patients with intermediate outcome (n = 7, mean age 35·8 ± 5·4 years, disease duration 8·6 ± 4·7 years) and (3) patients with poor outcome (n = 10, mean age 46·3 ± 4·2 years, disease duration 10·3 ± 0·9). As the aim of our study was to evaluate the differences within the extremes, the analysis was performed between the good and the poor clinical outcome groups. The comparison demonstrated significant differences between patients with good and poor clinical outcomes. Change in neurological disability assessed by the EDSS was −0·33 ± 0·24 and 1·6 ± 0·35, P = 0·0002, and total number of relapses was 0 and 1·80 ± 0·35, P = 0·00009, respectively.
Differentiating clinical outcome gene expression signature
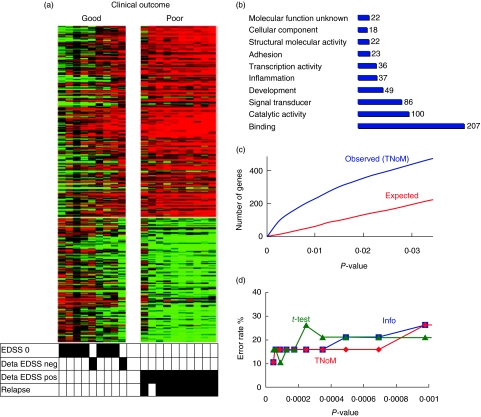
The distinctive clinical outcome gene expression pattern between patients with good and poor clinical outcomes included 431 differentiating genes which passed the three statistical tests, with P < 0·05 (Fig. 2a). Functional analysis disclosed genes associated with signal transduction, catalytic activity, adhesion and inflammation (Fig. 2b). Overabundance analysis of the observed compared with the expected number of genes that distinguished significantly between patients with good or poor clinical outcome was higher than expected (431 versus 200 genes at P = 0·03) (Fig. 2c). LOOCV resulted in a high classification rate of 90%, P < 0·0001 (Fig. 2d), suggesting that the differentiating genes signature is reliable and not related to spurious differences due to multiple testing.
Fig. 2.
(a) Heatmap of clinical outcome differentiating genes. Heatmap of the 431 differentiating genes that distinguishes between patients with good and poor clinical outcome. Each row of the heatmap represents a gene and each column represents a patient sample. Genes with increased expression are shown in progressively brighter shades of red, and genes with decreased expression are shown in progressively darker shades of green. The bottom matrix shows corresponding clinical outcome attributes marked in black when positive. (b) Functional annotation histogram. Distribution of differentiating gene expression signature according to biologically relevant functional groups. (c) Overabundance analysis. Actual number of genes (blue line) is significantly more abundant than expected (red line) for threshold number of misclassifications (TNoM) statistical test. x-Axis denotes P-value; y-axis denotes number of genes. (d) Leave-one-out cross-validation (LOOCV) classification. Division of errors between patients with good and poor clinical outcome using TNoM, Info and t-test demonstrated high classification rate of 90% at P < 0·0001. x-Axis denotes P-value; y-axis denotes error rate in percentage.
Predictive clinical outcome gene expression signature
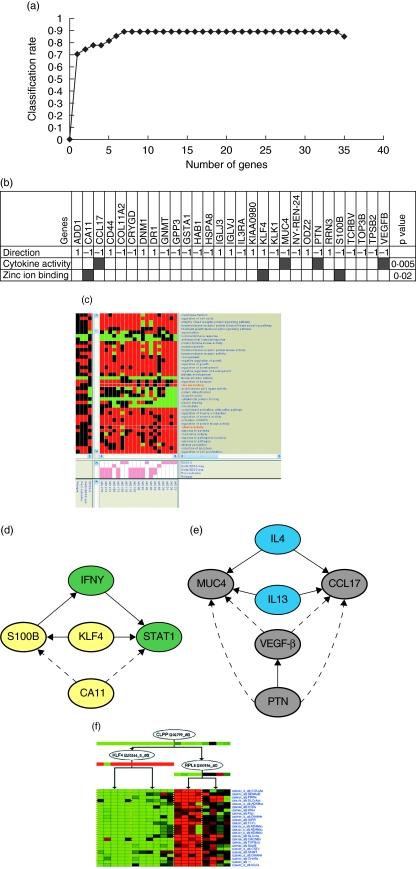
Application of the SVM on data from 19 of 26 patients with good (nine patients) or poor (10 patients) outcome as a training set, and nine of 27 additional patients from the validation group as test set, resulted in a high classification rate of 89%. This high classification was achieved by the Forward feature selection algorithm using 34 gene transcripts (29 genes), defined accordingly as predictive (Fig. 3a). Classification rate was 70·4% using only one gene (RRN3) and reached a rate of 85·2% using six genes (RRN3, KLF4, HAB1, TPSB2, IGLJ3, COL11A2). The addition of one or all of the remaining predictive genes resulted in maximal classification rate of 89·0%. This suggests that maximal predictive ability could be achieved using only seven genes. As there is no preference to any of the genes beyond the six predictive genes, we analysed the biological relevance of all 34 predictive genes.
Fig. 3.
(a) Predictive classification chart. The classification rate of 29 predictive genes is demonstrated. Highest classification rate is achieved using only seven genes, yet according to the feature selection algorithm, genes are added to the subset as long as the classification rate is not decreased. y-Axis denotes classification rate; x-axis denotes the number of genes. (b) Gene enrichment. Direction of an over-expressed (1) or down-expressed (− 1) gene is demonstrated in the enriched groups within the poor versus good outcome signature. (c) Heatmap of module analysis using differentiating clinical outcome genes. Enrichment of zinc-ion binding gene set for patients with relapses and cytokine activity gene set for patients with stable disease [no change in neurological disability, Expanded Disability Status Scale (EDSS) = 0] are demonstrated. The upper left graphical panel is a matrix of gene sets versus arrays, where a coloured entry indicates that the genes in the gene set had changed significantly in a co-ordinated fashion in the respective array (red, increased; green, decreased). The centre graphical panel shows individual clinical outcome attributes to which each array belongs. The bottom graphical panel demonstrates overall clinical outcome attributes in which gene sets were significantly enriched. (d) Reconstructed zinc-ion binding pathway. Pathway analysis performed using genes from the predictive signature (yellow circles) and genes brought into the pathway based on literature-known relationships according to PathwayArchitect software (green circles). Arrows indicate regulatory interactions confirmed by literature database, dashed arrows indicate suggested gene interactions. (e) Reconstructed cytokine activity pathway. Pathway analysis performed using genes from the predictive signature (grey circles) and genes brought into the pathway based on literature-known relationships according to PathwayArchitect software (blue circles). Arrows indicate regulatory interactions confirmed by literature database, dashed arrows indicate suggested gene interactions. (f) Gene expression regulatory network module. The single gene expression module from the gene expression regulatory network of 431 differentiating genes is demonstrated. Each node in the regulation tree represents a regulating gene. The expression of the regulating genes themselves is shown below their node. Cluster of gene expression profiles (rows represent genes, columns represent patients' arrays) arranged according to the regulation tree. Note that zinc-ion binding related genes KLF4 (regulating gene) and S100B (regulated gene) belong to the same regulatory module (black asterisks).
Independent validation of the predictive clinical outcome gene expression signature
Applying the resulting SVM-generated classifier, based on the 34 predictive genes to an additional data set of 18 of 27 patients from the validation group, maintained the high classification rate of 88·9%, P < 0·00001.
qRT–PCR performed in PBMC samples from 10 MS patients for three key genes of the predictive signature (S100B, KLF4 and RRN3) showed a perfect correlation of the expression levels of the candidate genes analysed with the mean expression levels obtained from the microarray experiments. S100B was lower by 1·47, P = 0·003; KLF4 was higher by 1·87, P = 0·044; and RRN3 was higher by 2·29, P = 0·003, in MS patients with poor outcome.
Biological regulation of the predictive clinical outcome gene expression signature
Functional annotation of the 34 predictive genes demonstrated that this group of genes was enriched significantly by zinc-ion binding protein genes (S100B, KLF4, CAII) and by genes with cytokine activity (CCL17, MUC4, PTN VEGFB), P = 0·02 and P = 0·005, respectively (Fig. 3b). The Genomica software confirmed enrichment by the zinc-ion binding gene family and by cytokine activity genes using all the 431 differentiating gene expression signature data (Fig. 3c). Using these enriched gene-families, regulatory pathways were reconstructed (Fig. 3d,e). These pathways suggest that apoptosis regulation through zinc-ion binding and cytokine activity is responsible for Th1/Th2 shift and may play a role in the clinical outcome of RRMS.
Genomica reconstruction of regulatory gene expression networks based on all 431 differentiating genes resulted in a regulation pathway in which the predictive zinc-ion binding gene KLF4, in association with CLPP and RRLP, mediate downstream genes including S100B (Fig. 3f). Other interesting functional groups in the 29 predictive genes include adhesion and cell migration such as CD44 and COL11A2, and T cell receptor genes such as TCRVB; all play an important role in MS pathogenesis.
Discussion
Prognostic modelling of patients with RRMS is a challenge in view of the unpredictable course of the disease. To the best of our knowledge, our study is the first that correlates gene expression profiles segregating RRMS patients into classes associated with clinical outcome. As MS has a winding course and the rate of disease progression differs between patients [18–20], it is of great importance to look for surrogate markers that will enable future envisaging of the disease process in an individual patient. Prediction of outcome in MS was reported to relate to different clinical variables such as age at disease onset, gender and the type of neurological symptomatology presented at onset. The major clinical determinants of more severe disease are male sex, relatively older age at onset, motor or cerebellar symptoms at onset and high annual relapse rate [21–23]. However, the ability of these variables to predict the clinical course is imperfect. This uncertainty in forecasting disease outcome means that some MS patients who need aggressive treatment do not receive it, while others are treated unnecessarily and as a result are exposed to the risk of side effects without a sound rationale [24]. In the current study we used a comprehensive approach to gain detailed understanding of the evolution of MS by analysing disease process-relevant gene signatures. Our rationale was to look for differentiating gene transcripts that are related to and predict clinical outcome and not − although it might be − are the causes for the change in clinical outcome. It is evident that the patients included in the study had a priori different clinical outcomes, but the ability to take a snapshot in time and identify a specific gene signature that characterizes good or poor clinical outcome is the major significance of our findings. Classifying patients into homogeneous groups based on the progression of neurological disability and number of relapses, and then sorting between different disease outcome groups according to gene expression anchors, enabled us to expand the knowledge of the disease phenotypes. The clinical outcome expression signature includes genes involved in proliferation, stimulation of T cell and T cell receptors, inflammation and adhesion. The reliability of the differentiating 431 clinical outcome gene-expression pattern was validated further by the low error estimates using LOOCV as well as by the overabundance analysis, showing a significant difference between the expected and observed data. These results provided further evidence that the identified genes were indeed representative of true biological processes that lead to different clinical phenomena. We assumed that not all the genes that differentiated between good and poor clinical outcome were effective predictive genes. Accordingly, we applied the SVM method combined with the Forward feature selection algorithm to evaluate outcome predictive genes and validated further the predictive pattern in an additional group of RRMS patients. Classification rate was 70·4% using only one gene (RRN3), and reached a rate of 85·2% using six genes (RRN3, KLF4, HAB1, TPSB2, IGLJ3, COL11A2). In larger groups, the applicable methods would use much smaller groups of genes (six or 34). The clinical outcome predictive gene expression signature was enriched by zinc-ion binding and cytokine activity pathways. Activation of mononuclear cells in MS involves proinflammatory cytokines such as IFN-γ and tumour necrosis factor (TNF)-α that promote disease activity. Conversely, anti-inflammatory cytokines such as TGF-β, interleukin (IL)-4 and IL-10 decrease proinflammatory activation. The molecular transcripts we identified regulate the balance of these opposing effectors and are thus associated with clinical outcome prediction. The zinc-ion binding genes in the predictive signature include KLF4, known to be activated by (signal transucers and activators of transcription (STAT1), which is activated by S100B [25], and increases IFN-γ expression [26], a well-known proinflammatory cytokine involved in MS disease activity.
KLF4 is markedly induced in response to IFN-γ, lipolpolysaccharide (LPS) or TNF-α. Over-expression of KLF4 is associated with macrophage activation marker inducible nitric-oxide synthase and with TGF-β1 inhibition. KLF4 interacts with the NF-kB family member p65 (RelA), and has an important role as a regulator of key signalling pathways that control macrophage activation [27].
The S100B gene protein is known to be involved in intracellular and extracellular regulatory events within the central nervous system. S100B was found to be elevated in acute brain lesions of RRMS patients [28], and its plasma levels were reported elevated in RRMS patients responding to IFN-β treatment [29]. This is in accordance with our findings, demonstrating decreased S100B expression in patients with poor outcome. Additionally, a novel association of the zinc-ion binding gene CA11 was identified in the network reconstruction.
The balance between T helper 1 (Th1) and Th2 immune responses plays an important role in the pathogenesis of MS. In addition to the recognition of encephalitogenic epitopes, the ability to produce Th1 cytokines is an important functional requirement by which myelin-reactive T cells mediate the disease, while Th2 cells secreting IL-10 suppress the ongoing inflammation. The cytokine activity-enriched gene family identified in the prediction signature included CCL17, MUC4, PTN and VEGFB. CCL17 displays chemotactic activity for Th2 lymphocytes [30], and its activity is well known to be enhanced by the Th2-related cytokines IL-4 and IL-13, leading to inhibition of inflammation. MUC4 expression is dependent upon IL-4 and IL-13 levels [31–33]. PTN is involved in regulation of cell-mediated immunity [34], and negatively regulates VEGF activity [35]. These findings demonstrate that the poor clinical outcome predictive signature in RRMS is affected mainly by decreased Th2 cytokine activity and aberrant regulation of inflammation.
In conclusion, the predictive outcome gene expression signature is sensitive to RRMS evolution and as such provides a new perspective on disease progression. Moreover, our findings suggest that the co-stimulatory regulatory pathways of zinc-ion binding and cytokine activity-related genes within the predictive signature may serve as new targets for therapeutic interventions. Finally, the predictive signature may enable planning of tailored therapeutic strategies, and allow delineation of patients at high risk who may benefit from early therapy.
References
- 1.Confavreux C, Vukusic S. Natural history of multiple sclerosis: implications for counselling and therapy. Curr Opin Neurol. 2002;15:257–66. doi: 10.1097/00019052-200206000-00006. [DOI] [PubMed] [Google Scholar]
- 2.Trojano M, Paolicelli D, Bellacosa A, Cataldo S. The transition from relapsing–remitting MS to irreversible disability: clinical evaluation. Neurol Sci. 2003;24(Suppl. 5):S268–70. doi: 10.1007/s10072-003-0171-6. [DOI] [PubMed] [Google Scholar]
- 3.Simon JH. Contrast-enhanced MR imaging in the evaluation of treatment response and prediction of outcome in multiple sclerosis. J Magn Reson Imaging. 1997;7:29–37. doi: 10.1002/jmri.1880070106. [DOI] [PubMed] [Google Scholar]
- 4.Benedict RH, Weinstock-Guttman B, Fishman I, Sharma J, Tjoa CW, Bakshi R. Prediction of neuropsychological impairment in multiple sclerosis: comparison of conventional magnetic resonance imaging measures of atrophy and lesion burden. Arch Neurol. 2004;61:226–30. doi: 10.1001/archneur.61.2.226. [DOI] [PubMed] [Google Scholar]
- 5.Mantripragada KK, Buckley PG, de Stahl TD, Dumanski JP. Genomic microarrays in the spotlight. Trends Genet. 2004;20:87–94. doi: 10.1016/j.tig.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Infante-Duarte C, Weber A, Kratzschmar J, et al. Frequency of blood CX3CR1-positive natural killer cells correlates with disease activity in multiple sclerosis patients. FASEB J. 2005;19:1902–4. doi: 10.1096/fj.05-3832fje. [DOI] [PubMed] [Google Scholar]
- 7.Sturzebecher S, Wandinger KP, Rosenwald A, et al. Expression profiling identifies responder and non-responder phenotypes to interferon-beta in multiple sclerosis. Brain. 2003;126:1419–29. doi: 10.1093/brain/awg147. [DOI] [PubMed] [Google Scholar]
- 8.Achiron A, Gurevich M, Friedman N, Kaminski N, Mandel M. Blood transcriptional signatures of multiple sclerosis: unique gene expression of disease activity. Ann Neurol. 2004;55:410–17. doi: 10.1002/ana.20008. [DOI] [PubMed] [Google Scholar]
- 9.Kurtzke JF. Rating neurologic impairment in multiple sclerosis: an expanded disability status scale (EDSS) Neurology. 1983;33:1444–52. doi: 10.1212/wnl.33.11.1444. [DOI] [PubMed] [Google Scholar]
- 10.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA. 2001;98:31–6. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaminski N, Friedman N. Practical approaches to analyzing results of microarray experiments. Am J Respir Cell Mol Biol. 2002;27:125–32. doi: 10.1165/ajrcmb.27.2.f247. [DOI] [PubMed] [Google Scholar]
- 13.Ben-Dor A, Bruhn L, Friedman N, Nachman I, Schummer M, Yakhini Z. Tissue classification with gene expression profiles. J Comput Biol. 2000;7:559–83. doi: 10.1089/106652700750050943. [DOI] [PubMed] [Google Scholar]
- 14.Furey TS, Cristianini N, Duffy N, Bednarski DW, Schummer M, Haussler D. Support vector machine classification and validation of cancer tissue samples using microarray expression data. Bioinformatics. 2000;16:906–14. doi: 10.1093/bioinformatics/16.10.906. [DOI] [PubMed] [Google Scholar]
- 15.Statnikov A, Aliferis CF, Tsamardinos I, Hardin D, Levy S. A comprehensive evaluation of multicategory classification methods for microarray gene expression cancer diagnosis. Bioinformatics. 2005;21:631–43. doi: 10.1093/bioinformatics/bti033. [DOI] [PubMed] [Google Scholar]
- 16.Aha DW, Bankert RL. A comparative evaluation of sequential feature selection algorithms. In: Fisher D, Lenx JH, editors. Proceedings of the 5th International Workshop on Artificial Intelligence and Statistics. New York: Springer-Verlag; 1995. pp. 1–7. [Google Scholar]
- 17.Segal E, Shapira M, Regev A, et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat Genet. 2003;34:166–76. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 18.Goodkin DE, Hertsgaard D, Rudick RA. Exacerbation rates and adherence to disease type in a prospectively followed-up population with multiple sclerosis. Implications for clinical trials. Arch Neurol. 1989;46:1107–12. doi: 10.1001/archneur.1989.00520460093019. [DOI] [PubMed] [Google Scholar]
- 19.Kurtzke JF, Beebe GW, Nagler B, Kurland LT, Auth TL. Early prognostic features of the later course of the illness. J Chronic Dis. 1977;30:819–30. doi: 10.1016/0021-9681(77)90010-8. Studies on the natural history of multiple sclerosis − 8. [DOI] [PubMed] [Google Scholar]
- 20.Weinshenker BG. The natural history of multiple sclerosis: update 1998. Semin Neurol. 1998;18:301–7. doi: 10.1055/s-2008-1040881. [DOI] [PubMed] [Google Scholar]
- 21.Weinshenker BG, Rice GPA, Noseworthy JH, Carriere W, Baskerville J, Ebers GC. The natural history of multiple sclerosis: a geographically bases study: 3. Multivariate analysis of predictive factors and models of outcome. Brain. 1991;114:1045–56. doi: 10.1093/brain/114.2.1045. [DOI] [PubMed] [Google Scholar]
- 22.Runmarker B, Andersen O. Prognostic factors in a multiple sclerosis incidence cohort with 25 years of follow-up. Brain. 1993;116:117–34. doi: 10.1093/brain/116.1.117. [DOI] [PubMed] [Google Scholar]
- 23.Kantarci OH, Weinshenker BG. Prognostic factors in multiple sclerosis. In: Cook DS, editor. Handbook of multiple sclerosis. 3. New York: Marcel Dekker; 2001. pp. 449–63. [Google Scholar]
- 24.Tremlett H, Paty D, Devonshire V. Disability progression in multiple sclerosis is slower than previously reported. Neurology. 2006;66:172–7. doi: 10.1212/01.wnl.0000194259.90286.fe. [DOI] [PubMed] [Google Scholar]
- 25.Zhang W, Geiman DE, Shields JM, et al. The gut-enriched Kruppel-like factor (Kruppel-like factor 4) mediates the transactivating effect of p53 on the p21WAF1/Cip1 promoter. J Biol Chem. 2000;275:18391–8. doi: 10.1074/jbc.C000062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen ZY, Shie JL, Tseng CC. STAT1 is required for IFN-gamma-mediated gut-enriched Kruppel-like factor expression. Exp Cell Res. 2002;281:19–27. doi: 10.1006/excr.2002.5633. [DOI] [PubMed] [Google Scholar]
- 27.Feinberg MW, Cao Z, Wara AK, Lebedeva MA, Senbanerjee S, Jain MK. Kruppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J Biol Chem. 2005;280:38247–58. doi: 10.1074/jbc.M509378200. [DOI] [PubMed] [Google Scholar]
- 28.Petzold A, Eikelenboom MJ, Gveric D, et al. Markers for different glial cell responses in multiple sclerosis: clinical and pathological correlations. Brain. 2002;125:1462–73. doi: 10.1093/brain/awf165. [DOI] [PubMed] [Google Scholar]
- 29.Petzold A, Brassat D, Mas P, et al. Treatment response in relation to inflammatory and axonal surrogate marker in multiple sclerosis. Mult Scler. 2004;10:281–3. doi: 10.1191/1352458504ms1021sr. [DOI] [PubMed] [Google Scholar]
- 30.Faffe DS, Whitehead T, Moore PE, et al. IL-13 and IL-4 promote TARC release in human airway smooth muscle cells: role of IL-4 receptor genotype. Am J Physiol Lung Cell Mol Physiol. 2003;285:L907–14. doi: 10.1152/ajplung.00120.2003. [DOI] [PubMed] [Google Scholar]
- 31.Dabbagh K, Takeyama K, Lee HM, Ueki IF, Lausier JA, Nadel JA. IL-4 induces mucin gene expression and goblet cell metaplasia in vitro and in vivo. J Immunol. 1999;162:6233–7. [PubMed] [Google Scholar]
- 32.Zhu Z, Lee CG, Zheng T, et al. Airway inflammation and remodeling in asthma. Lessons from interleukin 11 and interleukin 13 transgenic mice. Am J Respir Crit Care Med. 2001;164:S67–70. doi: 10.1164/ajrccm.164.supplement_2.2106070. [DOI] [PubMed] [Google Scholar]
- 33.Soto P, Price-Schiavi SA, Carraway KL. SMAD2 and SMAD7 involvement in the post-translational regulation of Muc4 via the transforming growth factor-beta and interferon-gamma pathways in rat mammary epithelial cells. J Biol Chem. 2003;278:20338–44. doi: 10.1074/jbc.M301886200. [DOI] [PubMed] [Google Scholar]
- 34.Achour A, Laaroubi D, Caruelle D, Barritault D, Courty J. The angiogenic factor heparin affin regulatory peptide (HARP) induces proliferation of human peripheral blood mononuclear cells. Cell Mol Biol. 2001;47:OL73–7. [PubMed] [Google Scholar]
- 35.Heroult M, Bernard-Pierrot I, Delbe J, et al. Heparin affin regulatory peptide binds to vascular endothelial growth factor (VEGF) and inhibits VEGF-induced angiogenesis. Oncogene. 2004;23:1745–53. doi: 10.1038/sj.onc.1206879. [DOI] [PubMed] [Google Scholar]