Abstract
Host genetic factors may contribute to susceptibility to and outcome in infectious diseases. Recently polymorphisms in PARK2/PACRG, a gene cluster linked to ubiquitination and proteasome-mediated protein degradation, were found to be associated with manifest infection by M. leprae. Here, we address whether these polymorphisms are associated with susceptibility to infection with Salmonella typhi and S. paratyphi A, intracellular pathogens that upon infection of humans share with mycobacteria aspects of the hosts’ immune response. The polymorphisms of PARK_e01(−697), PARK2_e01(−2599), rs1333955 and rs1040079 were analysed by polymerase chain reaction and restriction fragment length polymorphism in a case-control study of typhoid and paratyphoid fever patients in an endemic area in Jakarta, Indonesia. For this study, samples were obtained from patients with blood culture-confirmed typhoid fever (n = 90), paratyphoid fever (n = 26) and fever controls (n = 337) in a passive, community-based surveillance and compared to those of randomly selected community controls (n = 322) from the same city area. The PARK2_e01(−2599) allele T was significantly associated with typhoid and paratyphoid fever (OR: 1·51, 95%CI: 1·02–2·23) but the other polymorphisms, PARK2_e01(−697), rs1333955 and rs1040079, were not associated. Although within the PARK2/PACRG gene cluster the PARK2_e01(−2599) allele T was most strongly associated with leprosy (OR∼ 3–5), the association with typhoid is much less strong. Our findings suggest that this polymorphism in PARK2/PACRG plays a small but significant role in susceptibility to the intracellular pathogens S. typhi and S. paratyphi.
Keywords: PARK2, PACRG, S. typhi, S. paratyphi, typhoid fever, gene polymorphism
Introduction
Typhoid fever constitutes a serious public health problem in the world, especially in the developing countries, claiming over 200 000 lives in 2000 [1]. Typhoid fever is a systemic infection caused by Salmonella enterica serotype typhi (S. typhi). Paratyphoid fever, caused by Salmonella paratyphi A, B or C, has a disease presentation highly similar to that of typhoid fever, but, at least in Jakarta, seems to follow a distinct route of transmission: whereas typhoid fever is spread predominantly within the household, paratyphoid fever is mainly transmitted outside the patient’s home [2]. The identification of such risk factors and the most relevant route of transmission of the disease are essential for the development of control strategies and the allocation of public health resources.
Several case-control studies have documented risk factors for typhoid fever at the community level, such as inadequate hygiene, lack of microbiologically safe drinking water or the consumption of street food [2]. Within this environmental context, an individual’s genetic makeup may predispose subjects to acquisition of typhoid fever or development of severe disease [3]. For instance, an association between the single nucleotide polymorphism (SNP) TNFA−308 and typhoid fever has been reported in Vietnam. Together with HLA-DRB1*0301/6/8 and HLA-DQB1*0201–3, the TNFA−308*A allele was thought to be associated with susceptibility to typhoid fever [4,5].
In families with members displaying increased susceptibility to infection with the intracellular pathogens salmonella and mycobacteria, defects in IL-12/IFNγ type-1 cytokine mediated activation of macrophages have been found [6], suggesting that there is considerable overlap in immune responses against these unrelated intracellular bacterial pathogens. Recently, variants in the shared PARK2 and PACRG regulatory region have been found to act as common risk factor for manifest infection by M. leprae: a strong association (i.e. OR of 3–5) was demonstrated between the PARK2_e01(−2599) polymorphism and leprosy [7]. Mutations in the PARK2 gene encoding Parkin, have been identified as the cause of autosomal recessive juvenile Parkinsonism [8]. Parkin is a E3 ubiquitin ligase that is required for polyubiquitination of proteins before degradation by the proteasome [9]. Parkin Co-Regulated Gene (PACRG) is a reverse strand gene located upstream of the PARK2 gene. The gene product, termed Glup, forms a large molecular chaperone complex containing heat shock proteins (Hsp) and chaperonin components. Glup binds Parkin via Hsp70, and this multicomponent aggregate may deal with bacterial proteins by breaking them down or turning them into harmless molecules [10]. PARK2 and PACRG share a common promoter regulating their expression [11]. Of interest here, variants in this shared regulatory region have been found to act as a common risk factor for the acquisition of leprosy and there was some evidence that the T allele of this polymorphism acts in a dominant fashion [7].
Given the overlap in immune response to Salmonellae and mycobacteria, and its proven role in leprosy, we hypothesized that PARK2/PACRG polymorphisms may be associated with clinical typhoid and paratyphoid fever. In addition, in vitro studies suggested a possible role for PARK2/PACRG regulated genes in Salmonella pathogenesis, since they link this pathway to intracellular bacterial evasion mechanisms [12–14]. Parkin, the protein encoded by PARK2 has ubiquitin ligase (E3) activity [15]. The ubiquitin-proteasome pathway is important in protein processing and degradation, and contributes to quality control of proteins within cells and antigen processing for cross-presentation [16]. Invasion of host cells by Salmonellae requires the reversible activation of Cdc42 and Rac1 by bacterial encoded SopE and SptP, which must exert their function at different times during uptake. Although both proteins are delivered into the host cell cytoplasm at approximately equivalent amounts, SopE is rapidly degraded through a proteosome-mediated pathway, while SptP exhibits much slower degradation kinetics. Stabilization of SopE by proteasome inhibition prevents cellular recovery after bacterial infection and therefore continuation of a permissive environment for the bacteria to replicate or evade host defences [12,13]. This mechanism is important in Salmonella interaction with its host cells, and we hypothesized that modification of its activity might result in an association between PARK2/PACRG polymorphism and typhoid and paratyphoid fever.
Given the above hypothesis on the possible role of ubiquitination and degradation of bacterial proteins in the cellular pathogenesis of these diseases [12–14,16], we investigated the role of PARK2 and PACRG polymorphisms as host-dependent risk factors for acquisition of Salmonella typhi and S. paratyphi infection.
Materials and methods
Study design
From June 2001 to February 2003 patients with blood culture-confirmed Salmonella typhi (n = 69) or Salmonella paratyphi A (n = 24) were identified in a prospective, community-based, case-controlled passive surveillance study in the Jatinegara district of Jakarta, Indonesia [2]. In this surveillance study, we enrolled 1019 consecutive individuals living in the study area who presented with fever lasting ≥ 3 days to one of 24 healthcare facilities in the district. The study was designed to address both environmental [2,17,18] and genetic determinants of susceptibility to enteric fever and was aimed at investigating new associations as well as identifying specific candidate genes that might reveal meaningful immunological insights [19]. Full details of the enrolment of patients have been described elsewhere [2].
Blood cultures were collected into Bactec bottles (aerobic) containing antibiotic absorbing resins (Becton Dickinson, Sparks, MD, USA) that were provided to the centres by the study group free of charge. Every second consecutive fever patient with a negative blood culture, or having a pathogen other than Salmonella cultured, was selected as a fever control. Also, during the surveillance community controls were randomly selected within a random household in every third rukun tetangga (RT) from a total of 1140 RTs in Jatinegara, RT being the smallest administrative unit of 40–60 households in the area. When a community control reported fever in the 30 days preceding the interview or refused participation, the house on alternating sides of the initially selected household was approached. The selection of both groups of controls was nonmatched for age, sex or neighbourhood to limit selection-bias and prevent overmatching. Four controls from both groups for every case of blood culture-confirmed enteric fever were selected in order to increase the statistical power. Furthermore, between March and October 2003, 4 participating centres and the Medistra Hospital adjacent to the study area contributed another 23 cases of enteric fever (i.e. 21 typhoid fever and 2 paratyphoid fever cases).
All cases and fever controls were visited at home within one month after the febrile episode that led to the blood culture. Community controls were visited randomly throughout the study period. This study was approved by the Indonesian National Institute of Health Research and Development (Litbangkes) and provincial authorities. Written informed consent was obtained from all participants or their guardians.
Household visits and sample collection
Cases, fever controls and randomly selected community controls were interviewed by trained medical school graduates using a validated, standardized questionnaire as described previously [2,20]. Three ml of blood was collected using an EDTA-containing vacutainer system (Becton Dickinson). Blood samples were stored in a cool box until processing in the Biomedical Laboratory, Faculty of Medicine, Catholic University of Atma Jaya. The plasma was separated and erythrocytes were lysed using lysis buffer (155 mM NH4Cl, 10 mM KHCO3, 1 mM EDTA, pH = 7·4). White blood cells were washed twice with phosphate buffered saline (PBS) and stored in a freezer until transport to the Laboratory of Infectious Diseases, Leiden University Medical Centre. Genomic DNA was isolated from the cells essentially as described by Sambrook and Russell [21]. Before assay, DNA was diluted to a suitable concentration and stored in microplates at −20 °C.
Single nucleotide polymorphism (SNP) analysis
SNP analysis was performed using polymerase chain reaction (PCR) amplification followed by restriction enzyme digestion. PCRs were performed using 100 ng of genomic DNA, 200 µM of each dNTP, 10 pmol of each primer, 50 mM KCl, 10 mM Tris-HCl (pH 9·0), 1% Triton X-100, 1·5 mM MgCl2, 0·5 U of Taq DNA polymerase (Promega, Madison, WI, USA) in a total volume of 25 µl. PCR cycling conditions were as follows: 94 °C for 5 min once, 30 cycles of 94 °C for 45 s, annealing temperature for 45 s, 72 °C for 45 s and 72 °C for 7 min once. Primers used for PCR are based on [9] with major modifications for PARK2_e01 (−697) and rs1040079.
Digestion reactions using Bsu36 I, HPyCH4 IV, Bsl I (New England Biolabs, Ipswich, MA, USA) and Bfm I (MBI Fermentas, Vilnius, Lithuania) were performed according to the manufacturers protocols.
We chose to study four SNPs within the shared 5′ regulatory region of PARK2 and PACRG, PARK2_e01(−2599), PARK2_e01(−697), rs1333955 and rs1040079 that were independently associated with an increased risk of leprosy [7].
The sequence of the primers, annealing temperatures and restriction enzymes used, the length of the products and the type of alleles are given in Table 1.
Table 1.
Tools to investigate PARK2/PACRG polymorphisms: primers, annealing temperature, and product length of SNPs.
| SNPs | Direction | Primer sequence* | Annealing temperature | Restriction enzyme | Product length | Allele |
|---|---|---|---|---|---|---|
| PARK2_e01(−697) | Forward | ACAGCCGCTCCCGGTGCAC | 62 °C | Bsu36 I | Uncut: 292 bp | Allele C |
| Reverse | ATGGGCAGAGTACATCACTTG | Cut: 139 and 153 bp | Allele T | |||
| PARK2_e01(−2599) | Forward | TTTAGCAGTATAGACTTCTCAGC | 60 °C | HpyCH4 IV | Uncut: 102 bp | Allele T |
| Reverse | GAGCATGAGGTTGCAATTAAGA | Cut: 45 and 57 bp | Allele C | |||
| rs1333955 | Forward | TTGGATTTTCAGGATTTTATAGC | 58 °C | Bsl I | Uncut: 153 bp | Allele T |
| Reverse | CTGGCCAGCCAGGTTTCTG | Cut: 66 and 87 bp | Allele C | |||
| rs1040079 | Forward | CCATGAGTATAGGAGGAACTGT | 52 °C 5× | Bfm I | Uncut: 103 bp | Allele G |
| Reverse | GGACTAAAGGGCATGGTGAG | 62 °C 25× | Cut: 23 and 80 bp | Allele A |
Primer sequences are based on [9] with major modifications for PARK2–01(−697) and rs1040079.
Statistical methods
Data from the questionnaires and polymorphisms were entered twice using EpiInfo 6·04b software (US Centers for Disease Control and Prevention, Atlanta, GA, USA), validated and imported into SPSS version 11·5 (SPSS Inc, Chicago, IL, USA) for statistical analysis. The Hardy–Weinberg equilibrium of each SNP was checked in the total population and in each group of respondents. For the comparisons of the proportion, either the Pearson’s χ2 test or Fisher’s exact test was used.
Results
Study population
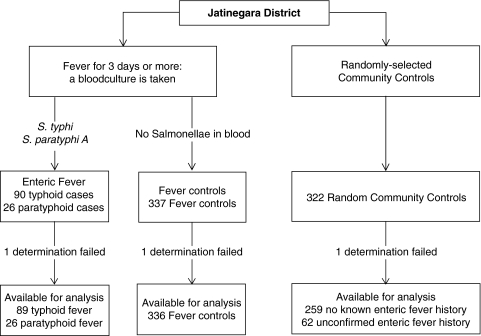
Of 1019 consecutive individuals living in the study area who presented with fever lasting ≥ 3 days, 116 individuals were enrolled with enteric fever and 337 as fever controls. In addition, 322 randomly selected community controls were included as detailed previously [2] and in Fig. 1. Of the 116 cases of enteric fever, 90 were caused by Salmonella typhi and 26 by Salmonella paratyphi A. Sixty-two community controls reported a possible history of enteric fever; in none of them had this past diagnosis been confirmed by (blood) culture.
Fig. 1.
Flow chart detailing the inclusion of typhoid and paratyphoid fever patients, and fever controls and the randomly selected community controls.
Demographic background of cases, fever controls and randomly selected community controls have been described elsewhere [2]. In short, the gender distribution of the typhoid and paratyphoid cases was about even (56 female from 116 cases); in the community controls this increased to 177 female from 322 cases. The median age of cases was 20 years (Interquartile range (IQR): 12–26·5) which was similar to that of fever controls, whereas both were significantly lower than that of the random community controls, i.e. 32 years (IQR: 18–49). Of note, the median age of the community controls that did not have a history of typhoid or paratyphoid fever was 32·5 years (IQR: 18–50), which was identical to that in community controls with a self-reported history of enteric fever. The age of typhoid fever cases did not differ significantly from that of paratyphoid fever cases. The population of Jatinegara is a mixture of, mainly, Indonesians from different islands of the archipelago and in individual cases it is not possible to designate a subject to one group or to exclude admixture with certainty. However, based on the sublocation in the area and the subjects’ names the ethnic makeup of the three study populations did not differ; no stratification with respect to possible admixture was made. In the nonenteric fever control group, patients could be infected with various bacterial and viral pathogens, each having distinct disease mechanisms. Therefore, the underlying genetic susceptibilities for this group could also be diverse. Although the inclusion of a fever control group would not provide a consistent reference group, we decided to present the findings in this group to further illustrate allelic frequencies in the population.
In 1 (1%) of 90 typhoid fever cases, 1 (0·3%) of 337 fever controls and 1 (0·3%) of 322 community controls, we could not determine the SNP alleles due to technical difficulties.
Hardy–Weinberg equilibrium calculation
The genotypes of PARK2_e01(−697), PARK2_e01(−2599), rs1333955 and rs1040079 were found to be in Hardy–Weinberg equilibrium in all cases, fever controls, community controls and in the total group of respondents (P > 0·7 in every group and in total for every SNP).
Genotyping PARK2/PACRG SNPs
The genotypic frequencies in cases, fever controls and randomly selected community controls are given in Table 2. In cases, fever controls and community controls alike, TT was found to be the most common genotype for PARK2_e01(−697) and PARK2_e01(−2599) (62% and 57%, respectively). CC was the most common genotype for rs1333955 (50%), whereas AA was the most common genotype for rs1040079 (63%). The most common allele for PARK2_e01(−697) and PARK2_e01(−2599) is the T-allele (proportion of 79% and 75%, respectively). For rs1333955, C is the most common allele with a proportion of 70%. A is the most common allele for rs1040079 with a proportion of 79%.
Table 2.
Genotypic frequencies in typhoid and paratyphoid cases, fever controls and randomly selected community controls.
| Typhoid cases (n = 89) | Paratyphoid cases (n = 26) | Community controls (n = 321) | Fever controls (n = 336) | |||||
|---|---|---|---|---|---|---|---|---|
| Locus/genotype | n | % | n | % | n | % | n | % |
| PARK2_e01(−697) | ||||||||
| CC | 6 | 7 | 2 | 8 | 14 | 5 | 16 | 5 |
| TC | 29 | 33 | 10 | 38 | 110 | 34 | 104 | 31 |
| TT | 54 | 60 | 14 | 54 | 197 | 61 | 216 | 64 |
| PARK2_e01(−2599) | ||||||||
| CC | 3 | 3 | 1 | 4 | 24 | 7 | 20 | 6 |
| TC | 29 | 33 | 7 | 27 | 121 | 38 | 124 | 37 |
| TT | 57 | 64 | 18 | 69 | 176 | 55 | 192 | 57 |
| rs1333955 | ||||||||
| CC | 49 | 55 | 16 | 61 | 154 | 48 | 167 | 50 |
| TC | 34 | 38 | 9 | 35 | 139 | 43 | 137 | 41 |
| TT | 6 | 7 | 1 | 4 | 28 | 9 | 32 | 9 |
| rs1040079 | ||||||||
| AA | 58 | 65 | 15 | 57 | 203 | 63 | 207 | 62 |
| AG | 27 | 30 | 9 | 35 | 106 | 33 | 115 | 34 |
| GG | 4 | 5 | 2 | 8 | 12 | 4 | 14 | 4 |
In a previous study demonstrating a strong association between the PARK2_e01(−2599) polymorphism and leprosy, there was some evidence that the T allele of this polymorphism acts in a dominant fashion. Because only 3 typhoid fever cases and 1 paratyphoid fever case were CC homozygotes, the present study has limited power to test such a hypothesis. Futhermore, the T allele appears to have a higher frequency in the two control populations in this study (75%) than in the Vietnamese and Brazilian population (67 and 61%, respectively) [9], possibly reflecting differences in ethnic background.
PARK2/PACRG alleles and risk of developing enteric fever
When comparing the frequencies of these alleles amongst the case group and randomly selected community controls, we found that the frequency of allele T of PARK2_e01(−2599) was significantly higher in enteric fever cases (P= 0·03). This difference was also significant when we excluded the community controls with an (unconfirmed) history of enteric fever (P= 0·02). We did not observe a significant difference in frequency of allele T of PARK2_e01(−2599) when we compared fever controls to community controls nor upon comparison of enteric fever cases and fever controls. The allele distribution of the other polymorphisms studied, PARK2_e01(−697), rs1333955 and rs1040079 was not significantly different in individuals with enteric fever when compared to those with fever due to other causes or randomly selected community controls.
Odds ratios (OR) were calculated on comparison of the alleles of PARK2_e01(−697), PARK2_e01(−2599), rs1333955 and rs1040079 in typhoid fever and paratyphoid fever with randomly selected community controls. Allele T PARK2_e01(−2599) was significantly but weakly associated with enteric fever (OR: 1·51, 95%CI: 1·02–2·23). This association became somewhat stronger when we compared the enteric fever cases to community controls without a history of enteric fever (OR: 1·58, 95%CI: 1·06–2·36). For PARK2_e01(−697), rs1333955 and rs1040079 polymorphisms, we did not observe an association of a particular allele or genotype with susceptibility to or resistance against typhoid fever and paratyphoid fever grouped as enteric fever, or typhoid fever alone.
Discussion
The main finding of this study is that the common allele T of the PARK2_e01(−2599) polymorphism is significantly but weakly associated with typhoid and paratyphoid fever patients as compared to randomly selected community controls. The same polymorphism, i.e. PARK2_e01(−2599) within PARK2 and PACRG, was, of all the polymorphisms in this gene region, most strongly associated with clinical leprosy in a Vietnamese population as well as a Brazilian population [7]. Alleles within this gene region that were less strongly but significantly associated with leprosy, i.e. PARK2_e01(−697), rs1333955 and rs1040079, were not found to be associated with typhoid and paratyphoid fever. The findings of the present study therefore, support a role for the PARK2 and PACRG genes in susceptibility to S. typhi and S. paratyphi as they do, more strongly, in susceptibility to M. leprae, and they suggest that the implicated mechanism linked to ubiquitination and proteasome-mediated protein degradation could be a common pathway in the intracellular fate of these intracellular pathogens [6,22].
To study the association of PARK2/PACRG polymorphisms and susceptibility to typhoid fever and paratyphoid fever, we compared the prevalence of the polymorphisms in blood culture-confirmed cases of typhoid fever and paratyphoid fever to those of randomly selected community controls from the same study area. Provisions taken to minimize misclassification of cases and controls have been described in detail elsewhere [2]. To get a robust estimate of the prevalence of PARK2/PACRG polymorphisms in the population, we included 4 controls for every typhoid or paratyphoid fever patient. Furthermore, fever controls that, similarly to the cases, presented with ≥ 3 days of fever, but from whom blood cultures showed either no growth or growth of bacteria other than Salmonellae, were recruited during the whole study period. Although the fever controls probably suffered from a divergent spectrum of diseases other than enteric fever, and therefore do not constitute a consistent reference group as the random community controls do, we decided to include the findings in this group to further illustrate the allelic frequencies found in the Indonesian population. Nineteen percent of the randomly selected community controls reported a possible episode of typhoid fever in the past. Probably, this percentage is an overestimation of the real number of cases since most fever patients are empirically treated in outpatient clinics without confirmatory diagnosis, but importantly, the distribution of polymorphisms in the community control group was not significantly different when these community controls were left out of the analysis. The age of the typhoid cases and the randomly selected community controls did differ, as the incidence of typhoid is higher in the age group < 20 years. In this respect, however, the distribution of polymorphisms in the community control group did not differ for different age cohorts, e.g. those < or > 20 years old. Of note, local HIV prevalence is low (e.g. 9 HIV positives were detected among 572 TB patients in a parallel study in Jakarta) and is unlikely to be an important confounder.
We hypothesized that PARK2/PACRG polymorphisms might be associated with typhoid and paratyphoid fever, given the overlap in immune responses to salmonella and mycobacteria [6], the finding that PARK2/PACRG polymorphisms are strongly associated with clinical leprosy [7] and in vitro studies that suggest a possible role for PARK2/PACRG regulated genes in Salmonella pathogenesis and link this pathway to intracellular bacterial evasion mechanisms and antigen processing for cross-presentation [12–14,16]. The study was powered to discern a similar strong association as described for leprosy, i.e. an OR of 3–5. To have the necessary power to confirm that the T allele of the PARK2_e01 (−2599) polymorphism acts in a dominant fashion, a study with a much larger sample size would be required, because only three typhoid and one paratyphoid case were CC homozygotes. In addition, the T allele appears to have a higher frequency in the Indonesian population (i.e. about 75%) than that previously found in the Vietnamese and Brazilian populations (about 65%) [7]. Given the weak but significant association between a PARK2 polymorphism and typhoid and paratyphoid fever compared to randomly selected community controls, future studies of larger typhoid cohorts in different populations should elucidate to what extent these processes may play a role in the complex host defence mechanisms against Salmonella.
Acknowledgments
Financial support was provided by the Royal Netherlands Academy of Arts and Sciences (KNAW). We thank the physicians, nurses and technicians of the participating health centres in Jakarta for their cooperation: Mitra International Hospital and Microbiology Laboratory, Budhi Asih, St. Carolus, Medistra and Persahabatan Hospitals, all puskesmas’ in Jatinegara and local private practitioners. We thank Adriëtte W. de Visser who isolated DNA from the blood samples, and our research assistants in Jakarta: Billy Hunsinger, Ferry Kandaw, Rinny Listyani, Meily, Vea Noveria, Carmelita Ridwan, Min Ali Sugiharto, Lidwina Sutikno, Mariana Tasman and Lily Yaputra. We also thank Leo G. Visser, and Henry A.G.H. van Asten, for their contribution to the project.
References
- 1.Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. Bull World Health Org. 2004;82:346–53. [PMC free article] [PubMed] [Google Scholar]
- 2.Vollaard AM, Ali S, van Asten HA, et al. Risk factors for typhoid and paratyphoid fever in Jakarta, Indonesia. J Am Med Assoc. 2004;291:2607–15. doi: 10.1001/jama.291.21.2607. [DOI] [PubMed] [Google Scholar]
- 3.Parry CM, Hien TT, Dougan G, White NJ, Farrar JJ. Typhoid fever. N Engl J Med. 2002;347:1770–82. doi: 10.1056/NEJMra020201. [DOI] [PubMed] [Google Scholar]
- 4.Dunstan SJ, Stephens HA, Blackwell JM, et al. Genes of the class II and class III major histocompatibility complex are associated with typhoid fever in Vietnam. J Infect Dis. 2001;183:261–8. doi: 10.1086/317940. [DOI] [PubMed] [Google Scholar]
- 5.Dharmana E, Joosten I, Tijssen HJ, et al. HLA-DRB1*12 is associated with protection against complicated typhoid fever, independent of tumour necrosis factor alpha. Eur J Immunogenet. 2002;29:297–300. doi: 10.1046/j.1365-2370.2002.00318.x. [DOI] [PubMed] [Google Scholar]
- 6.Ottenhoff THM, Verreck FA, Lichtenauer-Kaligis EG, Hoeve MA, Sanal O, van Dissel JT. Genetics, cytokines and human infectious disease: lessons from weakly pathogenic mycobacteria and salmonellae. Nat Genet. 2002;32:97–105. doi: 10.1038/ng0902-97. [DOI] [PubMed] [Google Scholar]
- 7.Mira MT, Alcais A, Nguyen VT, et al. Susceptibility to leprosy is associated with PARK2 and PACRG. Nature. 2004;427:636–40. doi: 10.1038/nature02326. [DOI] [PubMed] [Google Scholar]
- 8.Kitada T, Asakawa S, Hattori N, et al. Mutations in the Parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–8. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 9.Shimura H, Hattori N, Kubo S, et al. Familial parkinson disease gene product, Parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25:302–5. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 10.Imai Y, Soda M, Murakami T, Shoji M, Abe K, Takahashi R. A Product of the human gene adjacent to Parkin is a component of Lewy bodies and suppresses pael receptor-induced cell death. J Biol Chem. 2003;278:1901–10. doi: 10.1074/jbc.M309655200. [DOI] [PubMed] [Google Scholar]
- 11.West AB, Lockhart PJ, O’Farell C, Farrer MJ. Identification of a novel gene linked to Parkin via a bi-directional promoter. J Mol Biol. 2003;326:11–9. doi: 10.1016/s0022-2836(02)01376-1. [DOI] [PubMed] [Google Scholar]
- 12.Kubori T, Galan JE. Temporal regulation of salmonella virulence effector function by proteasome-dependent protein degradation. Cell. 2003;115:333–42. doi: 10.1016/s0092-8674(03)00849-3. [DOI] [PubMed] [Google Scholar]
- 13.Fu Y, Galan JE. A salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature. 1999;401:293–7. doi: 10.1038/45829. [DOI] [PubMed] [Google Scholar]
- 14.Neish AS, Gewirtz AT, Zeng H, et al. Prokaryotic regulation of epithelial responses by inhibition of IkappaB-alpha ubiquitination. Science. 2000;289:1560–3. doi: 10.1126/science.289.5484.1560. [DOI] [PubMed] [Google Scholar]
- 15.Imai Y, Soda M, Takahashi R. Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J Biol Chem. 2000;275:35661–4. doi: 10.1074/jbc.C000447200. [DOI] [PubMed] [Google Scholar]
- 16.Houde M, Bertholet S, Gagnon E, et al. Phagosomes are competent organelles for antigen cross-presentation. Nature. 2003;425:402–6. doi: 10.1038/nature01912. [DOI] [PubMed] [Google Scholar]
- 17.Vollaard AM, Ali S, Widjaja S, Asten HA, Visser LG, Surjadi C, van Dissel JT. Identification of typhoid fever and paratyphoid fever cases at presentation in outpatient clinics in Jakarta, Indonesia. Trans R Soc Trop Med Hyg. 2005;99:440–50. doi: 10.1016/j.trstmh.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 18.Vollaard AM, Ali S, van Asten HA, Ismid IS, Widjaja S, Visser LG, Surjadi C, van Dissel JT. Risk factors for transmission of foodborne illness in restaurants and street vendors in Jakarta, Indonesia. Epidemiol Infect. 2004;132:863–72. doi: 10.1017/s0950268804002742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van de Vosse E, Ali S, de Visser AW, Surjadi C, Widjaja S, Vollaard AM, van Dissel JT. Susceptibility to typhoid fever is associated with a polymorphism in the cystic fibrosis transmembrane conductance regulator (CFTR) Hum Genetics. 2005;118:138–40. doi: 10.1007/s00439-005-0005-0. [DOI] [PubMed] [Google Scholar]
- 20.Gasem MH, Dolmans WM, Keuter MM, Djokomoeljanto RR. Poor food hygiene and housing as risk factors for typhoid fever in Semarang, Indonesia. Trop Med Int Health. 2001;6:484–90. doi: 10.1046/j.1365-3156.2001.00734.x. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Russell DW. Molecular Cloning: a Laboratory Manual. 3. Cold Spring Harbor: Cold Spring. Harbor Laboratory Press; 2001. [Google Scholar]
- 22.Sanal O, Turul T, De Boer T, et al. Presentation of interleukin-12/-23 receptor beta1 deficiency with various clinical symptoms of salmonella infections. J Clin Immunol. 2006;26:1–6. doi: 10.1007/s10875-006-7830-3. [DOI] [PubMed] [Google Scholar]