Abstract
Regulatory T cells (Treg) are involved in the maintenance of peripheral tolerance by suppression of autoreactive lymphocytes that have avoided thymic depletion. The defective function of Treg cells has recently attracted attention in autoimmune diseases such as type 1 diabetes (T1D), rheumatoid arthritis and multiple sclerosis. Susceptibility to these diseases is associated with specific human leucocyte antigen (HLA) class II and cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) gene polymorphisms. This study aimed to investigate the relationship between HLA class II and CTLA +49 A/G polymorphisms associated with susceptibility to T1D and the number and characteristics of Treg cells in children. Samples from 47 5-year-old children who participated in the All Babies in South-east Sweden (ABIS) follow-up study were grouped according to the presence of the T1D risk-associated HLA genotype (DQA1*0501–DQB1*0201, DQA1*0301–DQB1*0302) or neutral HLA genotypes. Lower percentages of CD4+ T cells (P = 0·03) and CD4+ CD25high cells (P = 0·06) expressing intracellular CTLA-4 were detected in samples from children with CTLA-4 +49GG compared to children with the +49AA genotype. Similarly, lower percentages of CD4+ (P = 0·002) and CD4+ CD25high (P = 0·002) cells expressing CTLA-4 were observed in children positive for HLA DQA1*0501–DQB1*0201 and DQA1*0301–DQB1*0302 (P = 0·04 for CD4+ and P = 0·02 for CD4+ CD25high) risk haplotypes when compared to children without these alleles. The percentage of CD25high cells among CD4+ cells was correlated inversely with CTLA-4 mRNA expression in PBMC (r = −0·56, P = 0·03). Decreased levels of CTLA-4 in CD4+ and CD4+ CD25high cells in individuals with CTLA-4 and HLA class II alleles associated with T1D may contribute to the initiation and/or progression of autoimmune response.
Keywords: CTLA-4, HLA, regulatory T cells
Introduction
Regulatory T cells developed in the thymus are essential for halting autoimmune disease development by peripheral regulation of auto-reactive cells [1]. These CD4+ regulatory cells (Treg) are characterized by the expression of CD25 [Interleukin (IL)-2-receptor α chain] and more specifically, CD25 at high intensity [2,3], and by high mRNA expression of the forkhead/winged helix transcription factor, FOXP3 [4,5]. Removal of the thymus and depletion of CD4+ CD25+ cells in animal models lead to uncontrolled development of autoimmunity, characterized by an infiltration of lymphocytes following the destruction of internal organs [2,6]. Human diseases such as type 1 diabetes (T1D) have also been linked to a malfunctioning CD4+ CD25+ cell population [7–9].
T cells and Treg cells up-regulate cytotoxic T lymphocyte-associated antigen 4 (CTLA-4, CD152) from endosomal compartments upon stimulation [10,11]. CTLA-4 activates an inhibitory immune signal upon binding with a higher affinity than CD28 to CD80/86 molecules and has been shown to be essential for avoiding fatal lymphoproliferative syndrome [12–14]. Several single nucleotide polymorphisms (SNP) in the CTLA-4 gene, one being +49 A/G, have been shown to be associated with autoimmune diseases, although the mechanism remains under debate [15–17].
By an unclear mechanism of cell–cell contact, the CD4+ CD25+ cell population influences other T cells to down-regulate T cell reactivity [18,19]. This suppressive mechanism is associated with the inhibition of the IL-2R-α-chain (CD25) induced by the combined activity of CTLA-4 and membrane-bound transforming growth factor beta (TGF-β1), which is associated with the so-called latency-associated peptide (LAP) [1,20,21]. Some studies suggest that soluble molecules such as cytokines would exert a regulatory effect [22,23]. Altered expression of regulatory T cell surface proteins could indicate a susceptibility to autoimmune diseases [14,21].
It has been suggested that the development of the CD4+ CD25+ cell population is dependent upon major histocompatibility complex (MHC) class II-positive thymic epithelium [24]. Certain human leucocyte antigen (HLA) DQ-molecules, e.g. encoded by DQA1*0501–DQB1*0201 and DQA1*0301–DQB1*0302 allele combinations, show a strong association with autoimmune diseases such as T1D [25,26]. These alleles can be found in common HLA class II haplotypes DR3–DQ2 (DQA1*0501–DQB1*0201) and DR4–DQ8 (DQA1*0301–DQB1*0302), which are present in 95% of patients with T1D [27]. The DQB1*0602 allele has a protective effect on T1D even though it is associated with multiple sclerosis [28]. The possible interactions of these autoimmune disease-associated HLA class II genotypes and Treg cells are not yet fully understood.
In the present study we investigated the relationship between HLA class II and CTLA-4 +49 A/G polymorphisms and the expression of CTLA-4 and LAP (TGF-β1) protein in regulatory T cells obtained from blood samples from healthy 5-year-old children.
Materials and methods
Study population
The ‘All Babies in South-east Sweden’ (ABIS) is a large population-based follow-up study initiated to study T1D and other disease risk factors [29]. The parents of all 21 700 children born between October 1997 and October 1999 in South-east Sweden were asked to participate and 17 055 (78·6%) were included. Blood and other biological samples were collected at birth and later at defined intervals. In our study 68 samples from 5-year-old children were collected randomly during the period of July 2003 to February 2004.
Isolation of peripheral blood mononuclear cells (PBMC)
Venous blood samples were collected in 9 ml Vacuette sodium–heparin tubes (Greiner Bio-One, Kremsmünster, Austria) and analysed within 24 h. PBMC were isolated using the Ficoll Paque density gradient technique (Pharmacia Biotech, Sollentuna, Sweden) and then washed three times in RPMI-1640 (Gibco, Auckland, New Zealand). The cells were washed in phosphate buffered saline (PBS) (Medicago AB, Uppsala, Sweden) containing 0·1% bovine serum albumin (BSA) (Sigma-Aldrich, St Louis, MO, USA) and stained with antibodies for flow cytometry analysis.
Staining of PBMC
The cells were diluted in PBS–BSA 0·1% and divided into 200(µl aliquots in 5 ml plastic tubes prior to extracellular staining. Anti-human LAP (TGF-β1) goat IgG (R&D Systems, Minneapolis, MN, USA) was added to the tubes, followed by incubation for 30 min at 4°C in darkness. After washing and resuspension in 200 µl in PBS–BSA 0·1%, the cells were stained with fluorescein isothiocyanate (FITC)-conjugated anti-goat donkey IgG (R&D Systems), anti-CD8 (BD Biosciences, San Jose, CA, USA), anti-CTLA-4 (Pharmingen, San Jose, CA, USA), phycoerythrin (PE)-conjugated anti-CD4 (BD Biosciences) and CTLA-4 (Pharmingen); peridinin–chlorophyll–protein (PerCP)-conjugated anti-CD3, -CD4 and -CD8 (BD Biosciences) and allophycocyanin (APC)-conjugated anti-CD25 (BD Biosciences). Cells were then incubated for another 30 min at 4°C in darkness. After incubation, cells that were stained exclusively for extracellular marker expression were washed, resuspended in 300 ml PBS–BSA 0·1% and left at 4°C overnight in darkness until flow cytometry analysis.
To perform intracellular staining, 125 µl of 4% paraformaldehyde (PFA) (Merck, Darmstadt, Germany) was added to each of the remaining tubes. After 10 min of incubation in darkness at room temperature (RT), the tubes were centrifuged for 5 min at 500 g (GS-6R, Beckman AB, Bromma, Sweden). The supernatants were removed and the cells were resuspended in 500 µl of 1× Perm2 permeabilizing buffer (BD Biosciences). Cells were then incubated for another 10 min in darkness at RT, before washing in PBS–BSA 0·1%. The supernatants were removed and the cells were resuspended in 200 µl PBS–BSA 0·1%. Anti-human LAP (TGF-β1) goat IgG (R&D Systems) was added to selected tubes, and the cells were incubated for another 30 min at 4°C in darkness. After incubation, the cells were washed with PBS–BSA 0·1% and thereafter resuspended in 200 µl PBS–BSA 0·1% and stained with FITC-conjugated anti-goat donkey IgG (R&D Systems) and PE-conjugated CTLA-4 (Pharmingen). After the final 30 min of incubation at 4°C, the cells were washed in PBS–BSA 0·1%, resuspended in 300 µl PBS–BSA 0·1% and then left at 4°C in darkness overnight until flow cytometry analysis.
Isotype controls were included to estimate the amount of non-specific binding. Thus, cells were stained under identical conditions with FITC, PE, PerCP and APC fluorochrome-conjugated anti-mouse IgG. Autofluorescence was controlled with unstained cells. Four tubes containing cells stained with single antibodies marked with the four different fluorochoromes were used for compensation, to adjust for spectrally adjacent dye pairs.
Flow cytometry
A Becton Dickinson FACSCalibur (Immunocytometry Systems, San Diego, CA, USA) was used for four-colour cytometry. Cut-off for positivity was defined using isotype control. Data obtained from the fluorescence activated cell sorter (FACS) were analysed with CellQuest (Becton Dickinson Immunocytometry Systems). The resulting output was expressed as percentages of cells expressing each marker.
The lymphocyte gate was defined according to forward scatter (FSC) and side scatter (SSC). CD25high cells were gated using the CD25 intensity above that seen in CD8+ cells as a cut-off [3] (Fig. 1a,b). Percentages of cells expressing intracellular CTLA-4 were then analysed in these gates (Fig. 1c,d).
Fig. 1.
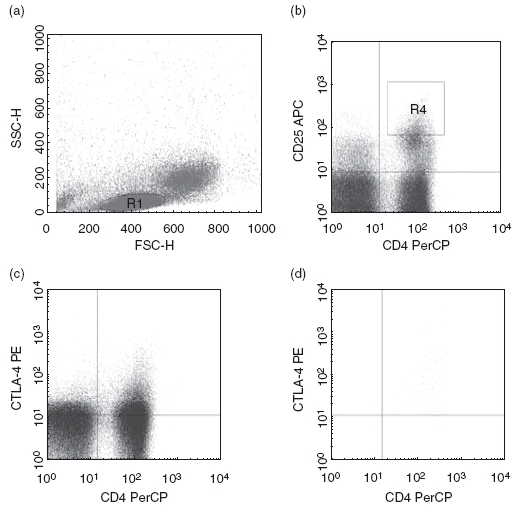
(a) Lymphocytes were gated based on forward scatter (FSC) and side scatter (SSC) (gate R1). (b) Regulatory CD4+CD25high+ T cell gate (gate R4) was defined as the CD25+ cells with intensity above that observed in CD8+ cells. (c) CD4+ cells co-expressing intracellular cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) in lymphocyte gate. (d) CD4+ cells co-expressing intracellular CTLA-4 in CD25high+ gate.
CTLA-4 +49 A/G polymorphism
All subjects were genotyped for the CTLA-4 +49 A/G polymorphism using an allele-specific hybridization assay with lanthanide-labelled probes (Perkin Elmer Analytical and Life Sciences, Turku, Finland), followed by time-resolved fluorometry [30].
HLA genotyping
HLA genotyping for DQB1 and DQA1 alleles was performed using an oligonucleotide hybridization assay, as described in detail elsewhere [26,31,32].
cDNA isolation
Total RNA was isolated from PBMC using the Sigma Genelute Mammalian Total RNA kit (Sigma Chemical Co., St Louis, MO, USA). Genomic DNA was eliminated using Roche DNase I (Roche Diagnostics, Mannheim, Germany). DNAse (2 U) was used for 200 ng of RNA and run in a PCR program for 30 min at 37°C and 5 min at 5°C. cDNA synthesis was carried out in the same tube using the TaqMan reverse transcription kit (Roche Diagnostics, Mannheim, Germany) and additional random hexamers (Applied Biosystems, CA, USA) in a total volume of 20 µl and in accordance with the manufacturer’s recommendations. PCR was then performed for 10 min at 25°C, 30 min at 48°C and 5 min at 95°C. cDNA samples were stored at −20°C until real-time PCR was performed.
Real-time PCR
6-Carboxy-fluorescein (FAM)-labelled TaqMan gene expression primers/probes were used to determine transcription levels of FOXP3, CTLA-4 and endogenous control (18 s) mRNA (all reagents from Applied Biosystems). Real-time PCR was carried out in a reaction volume of 25 µl, containing TaqMan predeveloped assay reagents (PDAR) Universal MasterMix, 1 × PDAR primers/probes for target genes and template cDNA. Ribosomal 18S was used as an endogenous control. The expression of each target was also measured from a home-made calibrator sample, which was prepared from unstimulated peripheral blood mononuclear cells of a healthy individual.
A volume of 1·8 µl of cDNA was used for each TaqMan triplicate measurement sample. ABI Prism 7700 was employed for sequence detection and programmed to carry out an initial step of 2 min at 50°C and 10 min at 95°C, followed by 50 thermal cycles of 15 s at 95°C and 1 min at 60°C.
A comparative threshold (CT) method was used to quantify the gene transcription in the samples. The CT of 18S was subtracted from target gene CT. This difference was determined as the ΔCT value. The ΔCT value was calculated as the difference of the ΔCT of the calibrator sample and the ΔCT of the unknown sample. The results were expressed as relative units based on calculation 2–ΔΔCT, which provided the relative amount of target normalized to the endogenous control (18S) and relative to the calibrator. The final relative amount of target mRNA was expressed as (2^ΔΔ) × 100.
Statistics
Because the percentage of cells expressing the cell markers was not normally distributed, unpaired analysis was performed with the Mann–Whitney U-test. Correlations were calculated using Spearman’s test. Data was analysed in spss for Windows (Chicago, IL, USA). A P-value equal to or less than 0·05 was considered to be statistically significant, and a P-value less than 0·1 was regarded as a tendency.
Ethics
The study was approved by the Ethics Committee for Human Research, Linköping University Hospital, Sweden as part of the ABIS-project.
Results
Relation of Treg cells to HLA and CTLA-4 polymorphisms
The results of HLA genotyping of 68 children were used to recognize the children with T1D-associated risk genotypes and children with the so-called neutral genotype (no risk allele associated with autoimmune diseases). Children with the HLA DQB*0602 allele were excluded due to the dual role of the allele in the risk of autoimmunity: HLA DQB*0602 is associated with the risk of multiple sclerosis but protects from T1D. The 47 children were divided into three groups: those positive for DR4–DQ8 (DQB1*0302) (n = 19, 40%), those positive for DR3–DQ2 (DQA1*05–DQB1*02) (n = 19, 40%), and neutral and those without either risk or DR2–DQ6 (DQB1*0602) haplotypes (n = 9, 19%).
Increased percentages of CD4+ cells expressing intracellular CTLA-4 were detected in samples from individuals carrying T1D neutral genotypes compared to DQA1*05–DQB1*02 (P = 0·002) and DQB1*0302-positive children (P = 0·04) (Fig. 2a).
Fig. 2.
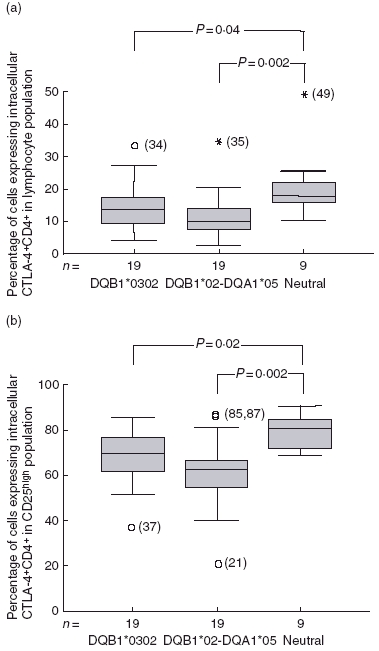
(a) Percentages of CD4+ lymphocytes co-expressing intracellular cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) were decreased in DQB1*0302 (P = 0.04) and DQA1*05–DQB1*02 (P = 0.002) positive individuals, compared to the neutral genotype group. Plots represent the percentages of CD4+ lymphocytes co-expressing intracellular CTLA-4 where samples are grouped as DQB1*0302 (n = 4), DQA1*05–DQB1*02 (n = 19) or neutral (n = 19) carrying neither risk nor DQB1*0602 genotypes. Percentages are illustrated in centile box plot (10th, 25th, 50th, 75th and 90th centiles) and outliers (1.5–3 box lengths from the box) shown as circles and extremes (> 3 box lengths from the box) as stars. (b) Percentages of CD4+CD25high cells co-expressing intracellular CTLA-4 were decreased in DQB1*0302 (P = 0.022) and DQB1*02-DQA1*05 (P = 0.002) genotype individuals compared to neutral genotypes.
Similar patterns were found in the CD4+ CD25high regulatory T cell population. Thus, higher percentages of intracellular CTLA-4 were detected in CD4+ CD25high cells from children with the neutral genotype compared to children with DQA1*05–DQB1*02 (P = 0·002) or DQB1*0302 (P = 0·02) haplotypes (Fig. 2b).
The 47 children included in our study were also grouped, based on CTLA-4 +49 A/G genotype distribution, into three groups: AA (n = 15. 32%), AG (n = 16, 34%) and GG (n = 16, 34%). Samples from 16 individuals were selected for mRNA analysis according to availability of the samples: three (19%) AA, 10 (63%) AG and three (19%) GG.
Samples from individuals with the CTLA-4 +49GG genotype had significantly lower percentages of intracellular CTLA-4 positive CD4+ cells (P = 0·03) (Fig. 3a) and tended to have lower percentages of intracellular CTLA-4 positive CD25high cells compared to AA-genotype individuals (P = 0·06) (Fig. 3b).
Fig. 3.
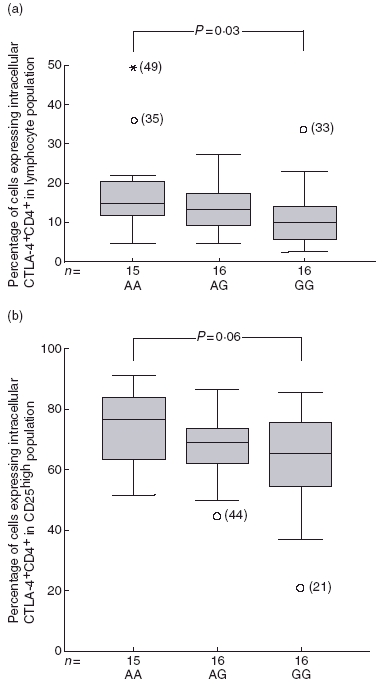
(a) Lower percentages of CD4+ lymphocytes co-expressing intracellular CTLA-4 was observed in the CTLA-4 +49GG genotype group compared to +49AA (P = 0.03). Percentages are illustrated in centile box plots (10th, 25th, 50th, 75th and 90th centiles and outliers). (b) Percentages of CD4+CD25high cells co-expressing intracellular CTLA-4 tended to be lower in +49GG genotype compared to AA genotype individuals (P = 0.06).
We found very low levels of extracellular LAP in resting T cells and did not consider these results reliable (data not shown).
Real-time PCR
A number of 47 samples were analysed for CTLA-4 and FOXP3 mRNA expression. The percentages of CD25high cells within the CD4+ population were correlated negatively with CTLA-4 mRNA expression (r = −0·56, P = 0·03) (Fig. 4). In contrast, the fraction of CD4+ cells expressing CD25high did not correlate with FOXP3 mRNA expression.
Fig. 4.
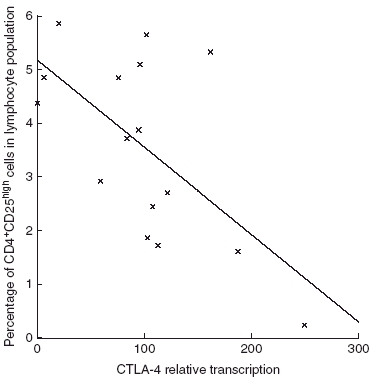
The percentages of CD4+CD25high cells of CD4+ lymphocytes correlated negatively with cytotoxic T lymphocyte-associated antigen 4 mRNA expression (r = -0.56, P = 0.03).
Similar expressions of FOXP3 and CTLA-4 mRNA were detected among the different CTLA-4 +49 and HLA genotype groups in the PBMC population.
Discussion
We found a decreased percentage of CD4+ cells expressing intracellular CTLA-4 in samples from children with the CTLA-4 +49GG, the T1D risk-associated genotype. Similarly, lower percentages of CD4+ and CD4+ CD25high cells expressing CTLA-4 were detected in children carrying T1D risk-conferring HLA class II haplotypes DQA1*0501–DQB1*0201 or HLA DQB1*0302 without protective haplotypes. A transition from adenine (A) to guanine (G) in the +49 position relative to the starting codon of the CTLA-4 gene results in a translation change from Thr to Ala. A correlation between the +49GG genotype and autoimmune diseases such as T1D has been observed [33,34]. CTLA-4 plays a key role in the CD4+ CD25+ T cell-mediated control of autoreactive T cells [35,36]. CTLA-4 is co-expressed with CD28, CD25 and CD45RO receptors, and has been reported to be expressed at higher levels on CD4+ CD25+ cells than in the CD4+ CD25– counterparts [37,38]. It has been established that CTLA-4 mRNA is not expressed in resting T cells, but is induced upon activation [39]. When CTLA-4 is endocytosed after T cell activation, it will be degraded if the cell is not stimulated repeatedly [40].
The risk-associated CTLA-4 +49GG genotype is associated with reduced cell surface CTLA-4 up-regulation on activated T cells [41,42]. In our material, we detected only very low levels of surface-bound CTLA-4 on unstimulated CD4+ cells from healthy children, and the data were not used for further statistical analysis. Instead, we studied the level of CTLA-4 as intracellular protein. According to our findings, the GG-genotype results in decreased production of CTLA-4 protein. The risk of autoimmune diseases associated with GG is probably affected by low CTLA-4, as suggested by some studies [41,42]. Recently, Anjos et al. [16] reported that there is no association between mRNA transcripts and the CLTA-4 polymorphism. The authors conclude that the mechanisms leading to autoimmunity do not involve modulation of steady-state mRNA in any of the known CLTA-4 isoforms. However, the authors emphasize that the polymorphisms may affect CTLA-4 expression under a physiological activation state of T cells not operating in the in vitro activation they employed. Our findings of low intracellular CTLA-4 protein levels in CD4+ and CD4+ CD25high cells fitted with the observation of an altered intracellular storage pool of CTLA-4, which could be due to interference with the signal peptide of CTLA-4 leading to defects in the up-regulation of CTLA-4 on the cell surface [41].
Atabani and co-authors [17] reported recently that individuals with the CT60/AA genotype, which is associated with a decreased risk of autoimmune diseases, showed increased numbers of Treg expressed as percentage of CD4+ cells. In agreement with our results, no association was found when the +49 polymorphism was studied. Individuals with the CT60/AA genotype are homozygous for +49/AA, but the results are probably explained by the fact that individuals with the +49/AA genotype are either CT60/AA or CT60/AG genotypes. Atabani et al. purified the population of Treg and did not find associations of CTLA-4 levels, either mRNA or protein, with the CTLA-4 genotypes. We studied the expression of intracellular CTLA-4 in CD4+ and CD4+ CD25high cells by flow cytometry without the step of immunomagnetic purification of the Treg, which may affect the stage of activation of the cells and further the expression of CTLA-4. Thus, methodological differences may explain the differences in the findings with regard to intracellular CTLA-4 expression.
The percentages of CD4+ and CD4+ CD25high cells expressing intracellular CTLA-4 were higher in samples from individuals without HLA class II alleles associated with autoimmune disease than in children with HLA risk genotypes. This is, to our knowledge, the first report of association of HLA genotype and CTLA-4 expression. It has been reported that HLA risk alleles for T1D are associated with the level of cytokine response [43]. It is tempting to speculate that the HLA genotype also somehow affects CTLA-4 expression and thus autoimmune risk, although our study does not reveal any such mechanism. CTLA-4 is transcribed and synthesized upon T cell receptor (TCR)-MHC stimulation [40], and it is possible that this interaction might be influenced by MHC class II alleles. Furthermore, the functional development of Treg cells in the thymus is dependent on the MHC class II molecules expressed on thymic epithelium [24].
We observed a negative correlation between the percentages of CD4+ CD25high cells and lymphocyte CTLA-4 mRNA expression. As the expression of CTLA-4 was measured in a PBMC population, the signals were not derived solely from Treg cells. Treg cells express CTLA-4 protein constitutively [35]. CTLA-4 mRNA, on the other hand, is expressed only upon upon T cell activation [39]. It could be speculated that lower expression of CTLA-4 mRNA can be found when the Treg cell pool in the periphery is active, bringing about an increased number of resting T cells with down-regulated CTLA-4 mRNA expression.
No association between FOXP3 mRNA expression and the percentages of CD4+ CD25high cells in the lymphocyte population could be found. This might seem surprising, as it is established that FOXP3 expression is a useful marker for regulatory T cell activity [4]. Our results could, however, be explained by the fact that the mRNA measurement was performed using the total lymphocyte population. Because the regulatory T cell population constitutes only 1–2% of the total lymphocyte population, variations in regulatory T cell mRNA levels are hard to detect in the peripheral blood mononuclear cell population. However, expression of FOXP3 mRNA might still correlate with the regulatory function of the cells, even though it did not reveal any correlation with the low amount of regulatory T cells in the lymphocyte population. We observed similar mRNA expression of FOXP3 and CTLA-4 in the different CTLA-4 and HLA genotype groups, the results being similar to previous studies [16,17,42].
It has been reported that TGF-β1 propagates a cell contact-dependent inhibitory signal similar to that of CTLA-4, which is also enhanced by CTLA-4 co-stimulation [21,44]. Surface-bound TGF-β is believed to be present in high amounts in its latent form bound to the LAP on Treg cells until activation, and thereafter mediate the regulatory action [21,45,46]. The very low expression of LAP–TGF-β1 in our samples could be due to the resting condition of the T cells.
We were not able to perform functional studies of Treg cells, e.g. inhibition of T cell proliferation or cytokine profiles of Treg cells, due to the limits of blood volume available from healthy children.
Recently, Lindley and associates reported that individuals with T1D (mean age 32·3 years) have significantly elevated percentages of CD4+ and Treg cells expressing intracellular CTLA-4 [47]. The +49 A/G polymorphism was also studied, but revealed no significant associations. Similar to our results, very low levels of surface CTLA-4 were observed and the authors also speculate that the Treg population constitutes a resting T cell population.
We conclude that intracellular expression of CTLA-4 is decreased in peripheral blood CD4+ and CD4+ CD25+ cell populations in individuals with CTLA-4 +49 and HLA class II polymorphism associated with autoimmune diseases. Low levels of CTLA-4 may explain, at least partly, the risk of autoimmune diseases mediated by these genetic factors.
Acknowledgments
This study was funded by the Swedish Research Council, the Swedish Child Diabetes Foundation (Barndiabetesfonden), Schelin’s Foundation, the Juvenile Diabetes Research Foundation and the Swedish Diabetes Association.
References
- 1.Annunziato F, Cosmi L, Liotta F, et al. Phenotype, localization, and mechanism of suppression of CD4(+) CD25(+) human thymocytes. J Exp Med. 2002;196:379–87. doi: 10.1084/jem.20020110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol. 1995;155:1151–64. [PubMed] [Google Scholar]
- 3.Baecher-Allan C, Brown JA, Freeman GJ, Hafler DA. CD4+CD25high regulatory cells in human peripheral blood. J Immunol. 2001;167:1245–53. doi: 10.4049/jimmunol.167.3.1245. [DOI] [PubMed] [Google Scholar]
- 4.Fontenot JD, Gavin MA, Rudensky AY. FOXP3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol. 2003;4:330–6. doi: 10.1038/ni904. [DOI] [PubMed] [Google Scholar]
- 5.Walker MR, Kasprowicz DJ, Gersuk VH, et al. Induction of FOXP3 and acquisition of T regulatory activity by stimulated human CD4+CD25– T cells. J Clin Invest. 2003;112:1437–43. doi: 10.1172/JCI19441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asano M, Toda M, Sakaguchi N, Sakaguchi S. Autoimmune disease as a consequence of developmental abnormality of a T cell subpopulation. J Exp Med. 1996;184:387–96. doi: 10.1084/jem.184.2.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roncarolo MG, Levings MK. The role of different subsets of T regulatory cells in controlling autoimmunity. Curr Opin Immunol. 2000;12:676–83. doi: 10.1016/s0952-7915(00)00162-x. [DOI] [PubMed] [Google Scholar]
- 8.Bach JF, Chatenoud L. Tolerance to islet autoantigens in type 1 diabetes. Annu Rev Immunol. 2001;19:131–61. doi: 10.1146/annurev.immunol.19.1.131. [DOI] [PubMed] [Google Scholar]
- 9.Kukreja A, Cost G, Marker J, et al. Multiple immuno-regulatory defects in type-1 diabetes. J Clin Invest. 2002;109:131–40. doi: 10.1172/JCI13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bluestone JA. Is CTLA-4 a master switch for peripheral T cell tolerance? J Immunol. 1997;158:1989–93. [PubMed] [Google Scholar]
- 11.Fallarino F, Grohmann U, Vacca C, Bianchi R, Fioretti MC, Puccetti P. CD40 ligand and CTLA-4 are reciprocally regulated in the Th1 cell proliferative response sustained by CD8(+) dendritic cells. J Immunol. 2002;169:1182–8. doi: 10.4049/jimmunol.169.3.1182. [DOI] [PubMed] [Google Scholar]
- 12.Lindsten T, Lee KP, Harris ES, et al. Characterization of CTLA-4 structure and expression on human T cells. J Immunol. 1993;151:3489. [PubMed] [Google Scholar]
- 13.Brunner MC, Chambers CA, Chan FK, Hanke J, Winoto A, Allison JP. CTLA-4-mediated inhibition of early events of T cell proliferation. J Immunol. 1999;162:5813–20. [PubMed] [Google Scholar]
- 14.Tivol EA, Boyd SD, McKeon S, et al. CTLA4Ig prevents lymphoproliferation and fatal multiorgan tissue destruction in CTLA-4-deficient mice. J Immunol. 1997;158:5091–4. [PubMed] [Google Scholar]
- 15.Ueda H, Howson JM, Esposito L, et al. Association of the T cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature. 2003;423:506–11. doi: 10.1038/nature01621. [DOI] [PubMed] [Google Scholar]
- 16.Anjos SM, Shao W, Marchand L, Polychronakos C. Allelic effects on gene regulation at the autoimmunity-predisposing CTLA4 locus: a re-evaluation of the 3′+6230G>A polymorphism. Genes Immun. 2005;6:305–11. doi: 10.1038/sj.gene.6364211. [DOI] [PubMed] [Google Scholar]
- 17.Atabani SF, Thio CL, Divanovic S, et al. Association of CTLA4 polymorphism with regulatory T cell frequency. Eur J Immunol. 2005;35:2157–62. doi: 10.1002/eji.200526168. [DOI] [PubMed] [Google Scholar]
- 18.Gribben JG, Freeman GJ, Boussiotis VA, et al. CTLA4 mediates antigen-specific apoptosis of human T cells. Proc Natl Acad Sci USA. 1995;92:811–15. doi: 10.1073/pnas.92.3.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheipers P, Reiser H. Fas-independent death of activated CD4(+) T lymphocytes induced by CTLA-4 crosslinking. Proc Natl Acad Sci USA. 1998;95:10083–8. doi: 10.1073/pnas.95.17.10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thornton AM, Shevach EM. CD4+CD25+ immunoregulatory T cells suppress polyclonal T cell activation in vitro by inhibiting interleukin 2 production. J Exp Med. 1998;188:287–96. doi: 10.1084/jem.188.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakamura K, Kitani A, Strober W. Cell contact-dependent immunosuppression by CD4(+)CD25(+) regulatory T cells is mediated by cell surface-bound transforming growth factor beta. J Exp Med. 2001;194:629–44. doi: 10.1084/jem.194.5.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piccirillo CA, Letterio JJ, Thornton AM, et al. CD4(+)CD25(+) regulatory T cells can mediate suppressor function in the absence of transforming growth factor beta1 production and responsiveness. J Exp Med. 2002;196:237–46. doi: 10.1084/jem.20020590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bach JF. Regulatory T cells under scrutiny. Nat Rev Immunol. 2003;3:189–98. doi: 10.1038/nri1026. [DOI] [PubMed] [Google Scholar]
- 24.Bensinger SJ, Bandeira A, Jordan MS, Caton AJ, Laufer TM. Major histocompatibility complex class II-positive cortical epithelium mediates the selection of CD4(+)25(+) immunoregulatory T cells. J Exp Med. 2001;194:427–38. doi: 10.1084/jem.194.4.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thorsby E, Ronningen KS. Particular HLA-DQ molecules play a dominant role in determining susceptibility or resistance to type 1 (insulin-dependent) diabetes mellitus. Diabetologia. 1993;36:371–7. doi: 10.1007/BF00402270. [Review]. [DOI] [PubMed] [Google Scholar]
- 26.Sjöroos M, Iitiä A, Ilonen J, Reijonen H, Lövgren T. Triple-label hybridization assay for type-1 diabetes-related HLA alleles. Biotechniques. 1995;18:870–7. [PubMed] [Google Scholar]
- 27.Atkinson MA, Maclaren NK. The pathogenesis of insulin-dependent diabetes mellitus. N Engl J Med. 1994;331:1428–36. doi: 10.1056/NEJM199411243312107. [Review]. [DOI] [PubMed] [Google Scholar]
- 28.Lobnig BM, Chantelau E, Vidgren G, Van Landeghem AA, Kinnunen L, Tuomilehto-Wolf E. HLA-patterns in patients with multiple sclerosis and type I diabetes mellitus: evidence for possible mutual exclusion of both diseases. Diabetes Metab. 2002;28:217–21. [PubMed] [Google Scholar]
- 29.Wahlberg J, Fredriksson J, Vaarala O, Ludvigsson J. Vaccinations may induce diabetes-related autoantibodies in one-year-old children. Ann NY Acad Sci. 2003;1005:404–8. doi: 10.1196/annals.1288.068. ABIS Study Group. [DOI] [PubMed] [Google Scholar]
- 30.Haller K, Kisand K, Nemvalts V, Laine AP, Ilonen J, Uibo R. Type 1 diabetes is insulin-2221 MspI and CTLA-4 +49 A/G polymorphism dependent. Eur J Clin Invest. 2004;34:543–8. doi: 10.1111/j.1365-2362.2004.01385.x. [DOI] [PubMed] [Google Scholar]
- 31.Nejentsev S, Sjöroos M, Soukka T, et al. Population based genetic screening for type 1 diabetes risk in Finland: selective genotyping of the markers in the HLA-DQB1, -DQA and -DRB1 loci. Diabet Med. 1999;16:985–92. doi: 10.1046/j.1464-5491.1999.00186.x. [DOI] [PubMed] [Google Scholar]
- 32.Laaksonen M, Pastinen T, Sjoroos M, et al. HLA class II associated risk and protection against multiple sclerosis − a Finnish family study. J Neuroimmunol. 2002;122:140–5. doi: 10.1016/s0165-5728(01)00456-8. [DOI] [PubMed] [Google Scholar]
- 33.Donner H, Rau H, Walfish PG, et al. CTLA4 alanine-17 confers genetic susceptibility to Graves’ disease and to type 1 diabetes mellitus. J Clin Endocrinol Metab. 1997;82:143–6. doi: 10.1210/jcem.82.1.3699. [DOI] [PubMed] [Google Scholar]
- 34.Mochizuki M, Amemiya S, Kobayashi K, et al. Association of the CTLA-4 gene 49 A/G polymorphism with type 1 diabetes and autoimmune thyroid disease in Japanese children. Diabetes Care. 2003;26:843–7. doi: 10.2337/diacare.26.3.843. [DOI] [PubMed] [Google Scholar]
- 35.Takahashi T, Tagami T, Yamazaki S, et al. Immunologic self-tolerance maintained by CD25(+)CD4(+) regulatory T cells constitutively expressing cytotoxic T lymphocyte-associated antigen 4. J Exp Med. 2000;192:303–10. doi: 10.1084/jem.192.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Read S, Malmstrom V, Powrie F. Cytotoxic T lymphocyte-associated antigen 4 plays an essential role in the function of CD25(+)CD4(+) regulatory cells that control intestinal inflammation. J Exp Med. 2000;192:295–302. doi: 10.1084/jem.192.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Linsley PS, Greene JL, Tan P, et al. Coexpression and functional cooperation of CTLA-4 and CD28 on activated T lymphocytes. J Exp Med. 1992;176:1595–604. doi: 10.1084/jem.176.6.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levings MK, Sangregorio R, Roncarolo MG. Human CD25(+)CD4(+) T regulatory cells suppress naive and memory T cell proliferation and can be expanded in vitro without loss of function. J Exp Med. 2001;193:1295–302. doi: 10.1084/jem.193.11.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perkins D, Wang Z, Donovan C, et al. Regulation of CTLA-4 expression during T cell activation. J Immunol. 1996;156:4154–9. [PubMed] [Google Scholar]
- 40.Saito T, Yamasaki S. Negative feedback of T cell activation through inhibitory adapters and costimulatory receptors. Immunol Rev. 2003;192:143–60. doi: 10.1034/j.1600-065x.2003.00022.x. [DOI] [PubMed] [Google Scholar]
- 41.Mäurer M, Loserth S, Kolb-Maurer A, et al. A polymorphism in the human cytotoxic T-lymphocyte antigen 4 (CTLA4) gene (exon 1 +49) alters T cell activation. Immunogenetics. 2002;54:1–8. doi: 10.1007/s00251-002-0429-9. Epub 12 March 2002. [DOI] [PubMed] [Google Scholar]
- 42.Ligers A, Teleshova N, Masterman T, Huang WX, Hillert J. CTLA-4 gene expression is influenced by promoter and exon 1 polymorphisms. Genes Immun. 2001;2:145–52. doi: 10.1038/sj.gene.6363752. [DOI] [PubMed] [Google Scholar]
- 43.Petrovsky N, Harrison LC. HLA class II-associated polymorphism of interferon-gamma production. Implications for HLA-disease association. Hum Immunol. 1997;53:12–16. doi: 10.1016/S0198-8859(96)00271-6. [DOI] [PubMed] [Google Scholar]
- 44.Chen W, Wahl SM. TGF-beta: the missing link in CD4(+)CD25(+) regulatory T cell-mediated immunosuppression. Cytokine Growth Factor Rev. 2003;14:85–9. doi: 10.1016/s1359-6101(03)00003-0. [DOI] [PubMed] [Google Scholar]
- 45.Oida T, Zhang X, Goto M, et al. CD4+CD25– T cells that express latency-associated peptide on the surface suppress CD4+CD45RBhigh-induced colitis by a TGF-beta-dependent mechanism. J Immunol. 2003;170:2516–22. doi: 10.4049/jimmunol.170.5.2516. [DOI] [PubMed] [Google Scholar]
- 46.Oklu R, Hesketh R. The latent transforming growth factor beta binding protein (LTBP) family. Biochem J. 2000;352:601–10. [PMC free article] [PubMed] [Google Scholar]
- 47.Lindley S, Dayan CM, Bishop A, Roep O, Peakman M, Tree TI. Defective suppressor function in CD4(+)CD25(+) T cells from patients with type 1 diabetes. Diabetes. 2005;54:92–9. doi: 10.2337/diabetes.54.1.92. [DOI] [PubMed] [Google Scholar]


