Abstract
Periodontal disease is an inflammatory disorder characterized by the involvement of chemokines that are important for the recruitment of leucocytes. Several cytokines are involved in regulating levels of chemokines in periodontal disease. CXCL16 is a chemokine related to the migration of T helper 1 (Th1) cells and natural killer (NK) cells. In this study, we examined its expression in periodontal tissues. Moreover, we investigated the effects of cytokines on the production of CXCL16 by human gingival fibroblast (HGF). Reverse transcription–polymerase chain reaction (RT–PCR) analysis and immunohistochemistry revealed that CXCL16 and its receptor, CXCR6, were expressed at the mRNA and protein levels in diseased tissues. Proinflammatory cytokines [interleukin (IL)-1β, tumour necrosis factor (TNF)-α and interferon (IFN)-γ] increased the mRNA expression and release of CXCL16 in a dose-dependent manner. Moreover, treatment of HGFs with IFN-γ in combination with IL-1β had a synergistic effect on the production of CXCL16. On the other hand, IL-4 and IL-13 inhibited the IL-1β-induced CXCL16 production by HGFs. Inhibitors of A disintegrin and metalloprotease (ADAM)10 and ADAM17, a recently identified protease of CXCL16, reduced the amount of CXCL16 released from HGFs. These results suggest that the CXCL16 produced by HGFs may be involved in the migration of leucocytes into inflamed tissues, and provide evidence that CXCL16 production is controlled by cytokines in periodontal disease.
Keywords: CXCL16, CXCR6, human gingival fibroblasts, periodontal disease
Introduction
Periodontal disease is characterized by chronic inflammation associated with the presence of Gram-negative bacteria in the oral cavity [1, 2], resulting in the destruction of soft tissue and resorption of periodontal bone. The host-immune response to these bacteria has been suggested to be associated with pathological changes or disease progression [3, 4]. Diseased periodontal tissue shows an accumulation of T cells, B cells, macrophages and dendritic cells [5, 6]. T cells are reportedly involved in the pathogenesis of periodontal disease [7]. It has been reported that both T helper 1 (Th1) cells and Th2 cells infiltrate the inflamed in tissues, producing Th1 [interferon (IFN)-γ, tumour necrosis factor (TNF)-α] and [interleukin (IL)-4 and IL-13] cytokines [7]. However, it is unclear which cell types are most related to the pathogenesis of periodontal disease. Seymour et al. reported that Th1 cells are associated with stable areas in periodontal tissues, and with Th2 cells, with regions where the disease is progressing [8]. On the other hand, Taubman et al. reported that Th1 cells are involved in periodontal destruction, and Th2 cells abrogate periodontal disease symptoms [9]. Recently, Kawai et al. reported that Th1 cells were more effective at inducing bone resorption than Th2 cells in experimental periodontal disease [10]. This report suggests that Th1 cells are involved in the destruction of bone in diseased periodontal tissue. The migration of T cells into inflamed tissue is thought to contribute to the interaction between chemokines and chemokine receptors, and the roles of chemokines in periodontal disease have been analysed [7, 11–14].
Chemokines induce leucocytes to migrate and are generally classified as either type C, CC, CXC or CX3C, based on conserved cysteine motifs [15]. The majority of chemokines are secreted as small, soluble molecules. Two exceptions, fractalkine and CXCL16, are expressed on the cell surface as membrane-bound molecules [16–19]. Membrane-bound CXCL16 induces firm binding of cells that express CXCR6 [20], a unique receptor for CXCL16. The membrane-bound form of CXCL16 is cleaved by A disintegrin and metalloprotease (ADAM)10 [21, 22] and ADAM17 [TNF-α convertase (TACE)] [23], and the cleaved soluble form of CXCL16 induces migration of activated T cells [18]. It has been reported that CXCL16 is expressed by dendritic cells, macrophages, B cells, T cells, smooth muscle cells and endothelial cells [18, 19, 24–26]. CXCR6 is expressed on Th1 cells, natural killer (NK) cells, NK T cells and B cells [27–30]. CXCL16 is considered to play a pathogenic role in atherogenesis [31, 32], the development of inflammatory cardiac vascular disease [33], experimental autoimmune encephalomyelitis [34] and rheumatoid arthritis [35].
The aim of this study is to examine the expression of CXCL16 and CXCR6 in diseased periodontal tissues and to investigate whether human gingival fibroblasts (HGFs) produce CXCL16 when stimulated with proinflammatory cytokines, Th1 cytokines and Th2 cytokines.
Materials and methods
Gingival tissue biopsy and cell culture
Tissue samples were obtained at surgery from the inflamed gingiva of patients diagnosed with chronic periodontitis or from the gingivae of clinically healthy subjects. All gingival biopsy sites in the chronic periodontitis group exhibited radiographic evidence of bone destruction, as well as having clinical probing depths greater than 4 mm, with sulcular bleeding on probing, otherwise the patients were systemically healthy. We collected samples after basic periodontal therapy such as scaling. Samples of gingival tissues were obtained from 17 chronic periodontitis patients (four males and 13 females, average age: 61·0 ± 9·8, average probing depth: 6·33 ± 2·06, average attachment loss: 7·02 ± 2·26) and five healthy control subjects (five females, average age: 31·2 ± 9·8, average probing depth: 2·4 ± 0·54, average attachment loss: 2·7 ± 0·57). We used two clinically healthy gingival samples and eight chronic periodontitis samples for reverse transcription–polymerase chain reaction (RT–PCR), and two clinically healthy gingival samples and nine chronic periodontitis samples for immunohistochemical staining. We used HGF isolated from three clinically healthy gingivae during routine distal wedge surgical procedure. Gingival specimens were cut into small pieces and transferred to culture dishes. HGFs that grew from the gingivae were cultured on uncoated 100 mm2 plastic dishes in Dulbecco's modified Eagle medium (DMEM; Sigma, St Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) and antibiotics (penicillin G; 100 units/ml, streptomycin; 100 mg/ml) at 37°C humidified air with 5% C02. Confluent cells were transferred and cultured for use in the present study. After three to four subcultures by trypsinization, cultures contained homogeneous, slim, spindle-shaped cells growing in characteristic swirls. The cells were used for experiments after five passages. Informed consent was obtained from all subjects participating in this study. The study was performed with the approval and compliance of the Tokushima University Ethical Committee.
RNA extraction and RT–PCR analysis
Total RNA was prepared from biopsied gingival tissue or HGFs using the Rneasy total RNA isolation Kit (Qiagen, Hilden, Germany). Single-stranded cDNA for a PCR template was synthesized from 48 ng of total RNA using a primer, oligo(dT)12−18 (Invitrogen, Carlsbad, CA, USA) and superscript III reverse transcriptase (Invitrogen) under the conditions indicated by the manufacturer. Specific primers were designed from the cDNA sequences for CXCL16, CXCR6 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Each cDNA was amplified by PCR using Hot star Taq DNA polymerase (Qiagen). The sequences of the primers were as follows: CXCL16-F (5′-CGTCACTGGAAGTTGTTATTGTGGT-3′), CXCL16-R (5′-TGGTAGGAAGTAAATGCTTCTGGTG-3′), CXCR6-F (5′-CTGGTGGTGTTTGTCTGTGG-3′), CXCR6-R (5′-GGCTGACAAAGGCATAGAGC-3′), GAPDH-F (5′-TGAAGGTCGGAGTCA ACGGATTTGGT-3′) and GAPDH-R (5′-CATGTGGGCCATGAGGTCCACCAC-3′). The conditions for PCR were 1× (95°C, 15 min), 35× (94°C, 1 min, 59×, 1 min, 72°C, 1 min) and 1× (72°C, 10 min). The products were analysed on a 1·5% agarose gel containing ethidium bromide. The expected sizes of the PCR products for CXCL16, CXCR6 and GAPDH were 259 base pairs (bp), 762 bp and 985 bp, respectively. Signal intensities were quantified by densitometry using National Institutes of Health image software to obtain the CXCR6 : GAPDH or CXCL16 : GAPDH ratios.
Immunohistochemistry
Gingival tissue samples were embedded immediately in the optimal cutting temperature (OCT) compound (Miles Laboratories Inc., Elkhart, IN, USA) and quenched and stored in liquid nitrogen. The specimens were cut into 6-µm sections using a cryostat (SFS, Bright Instrumental Company, Huntingdon, UK) and collected on poly-l-lysine-coated slides. CXCL16 and CXCR6 expression was analysed with specific antibodies; rabbit anti-human CXCL16 antibody (Peprotech, London, UK; 1 µg/ml) and mouse anti-human CXCR6 antibody (R&D Systems, Minneapolis, MN, USA; 5 µg/ml), respectively. An isotype-matched control antibody was used as a negative control. The sections were reacted with specific antibodies overnight at 4°C. After being washed with phosphate-buffered saline (PBS), the sections were incubated with biotinylated anti-mouse and rabbit immunoglobulins (Universal Ab; Dako, Kyoto, Japan) for 20 min at room temperature and washed with PBS to remove any unreacted antibodies. The sections were then treated with peroxidase-conjugated streptavidin (Dako) for 10 min, and washed and reacted with 3,3-diamino-benzidine tetrahydrochrolide (DAB; Dako) in the presence of 3% H2O2 to develop colour. The sections were counterstained with haematoxylin and mounted with glycerol.
CXCL16 release by HGFs
HGFs were stimulated with IL-1β (Peprotech, Rocky Hill, NJ, USA), TNF-α (Peprotech), IFN-γ (Peprotech), IL-4 (Peprotech) and IL-13 (Peprotech) for 24 h. Supernatants from the cells were collected and the concentration of CXCL16 was measured in triplicate by enzyme-linked immunosorbent assay (ELISA). A CXCL16 ELISA development kit (Peprotech) was used for the determination. The range of detection for the ELISA was 32–4000 pg/ml. All assays were performed according to the manufacturer's instructions and cytokine levels were determined using a standard curve prepared for each assay. In selected experiments, HGFs were cultured for 1 h in the presence or absence of SB203580 (0·2–20 µM; Santa Cruz Biotechnology, Santa Cruz, CA, USA), PD98059 (0·2–20 µM; Calbiochem, La Jolla, CA, USA), SP600125 (0·2–20 µM; Sigma), LY294002 (0·2–20 µM; Calbiochem), MG-132 (0·2–20 µM; Calbiochem), TAPI2 (0·5–50 µM; Calbiochem) and GM-6001 (0·5–50 µM; Calbiochem), prior to their incubation with the various stimulants.
Statistical analysis
Statistical significance was analysed with Student's t-test. P-values < 0·05 were considered significant. We used statview software to perform the statistical analysis.
Results
CXCL16 and CXCR6 expression in periodontal tissues
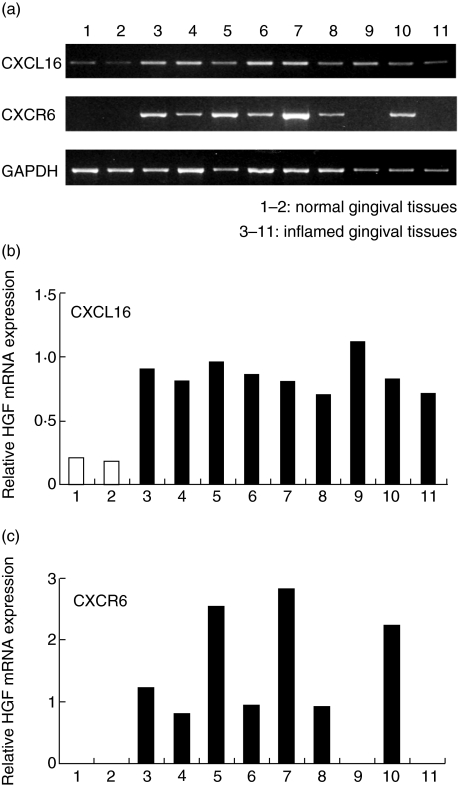
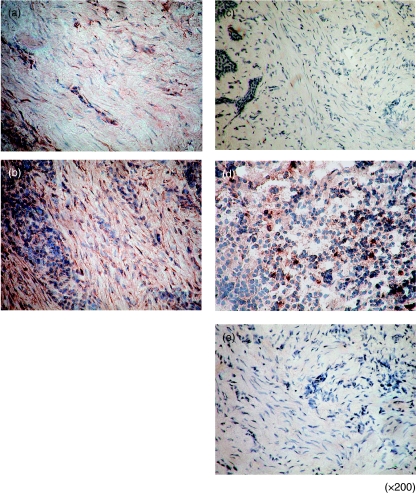
We first examined CXCL16 and CXCR6 mRNA expression in whole gingival tissue. CXCL16 mRNA was detected in all the samples used in this study. However, CXCL16 mRNA expression was weak in normal compared to diseased gingival tissues (Fig. 1a, b). CXCR6 mRNA was not detected in normal gingival tissues. We could detect CXCR6 mRNA in seven of nine samples from patients with periodontal disease (Fig. 1a,c). Next, we carried out immunohistochemical staining to investigate the expression of CXCL16 and CXCR6 in periodontal tissues (Fig. 2). Fibroblasts in diseased tissues strongly expressed CXCL16 (Fig. 2b) compared with those in normal periodontal tissues (Fig. 2a). We could see few CXCR6-positive cells in normal periodontal tissues (Fig. 2c). On the other hand, many CXCR6-positive cells were infiltrated in periodontally diseased tissues (Fig. 2d). CXCR6-positive cells were distributed mainly near the sulcular epithelium.
Fig. 1.
Reverse transcription–polymerase chain reaction (RT–PCR) analysis of CXCL16 and CXCR6 mRNA expression in diseased periodontal tissue. Total RNA was prepared from two clinically healthy gingival samples (pocket depth, 2 mm) and nine diseased gingival samples (pocket depth, 4–10 mm). The expression of CXCL16, CXCR6 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA in periodontal tissues was analysed by RT–PCR as described in the Methods. Bands were quantified further using National Institutes of Health image software (b,c). The results are expressed relative to GAPDH mRNA as an internal standard.
Fig. 2.
CXCL16 and CXCR6 immunostaining in periodontal tissues. Immunohistochemical analysis of normal gingival tissue (a,c) and diseased periodontal tissues (b,d). Immunohistochemical staining of human periodontal tissues with anti-CXCL16 antibody (a,b), anti-CXCR6 antibody (c,d) and control antibody (e). The original magnification for each photograph was ×200.
Cytokines differently modulated CXCL16 expression by HGFs
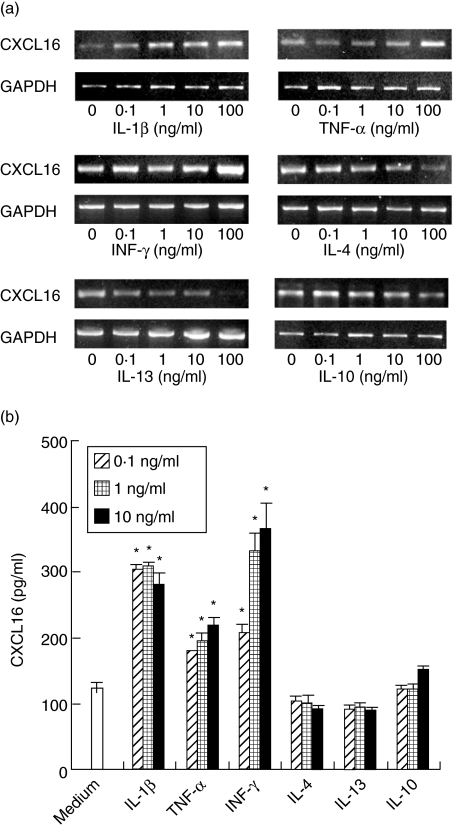
In order to examine the effects of cytokines on the expression of CXCL16, we used the RT–PCR and ELISA techniques. CXCL16 mRNA was expressed on unstimulated HGFs. Stimulation with IL-1β, TNF-α, IFN-γ up-regulated significantly the expression of CXCL16 mRNA (Fig. 3a). On the other hand, IL-4 and IL-13 decreased CXCL16 mRNA expression by HGFs in a concentration-dependent manner (Fig. 3a). IL-10 did not modulate CXCL16 mRNA expression by HGF. To investigate the release of CXCL16 by HGFs stimulated with cytokines, we used the ELISA. IL-1β, TNF-α, and IFN-γ treatment promoted the release of CXCL16 from HGFs. IL-1β and IFN-γ were the most potent inducers used in this study (Fig. 3b). IL-4 and IL-13 slightly inhibited the release of CXCL16 by HGFs. IL-10 did not modulate the release of CXCL16 from HGFs (Fig. 3b).
Fig. 3.
CXCL16 expression by human gingival fibroblasts (HGFs). (a) HGFs were treated with or without interleukin (IL)-1β (0·1–100 ng/ml), tumour necrosis factor (TNF)-α (0·1–100 ng/ml), interferon (IFN)-γ (0·1–100 ng/ml), IL-4 (0·1–100 ng/ml), IL-13 (0·1–100 ng/ml) and IL-10 (0·1–100 ng/ml) for 4 h. Total RNA was isolated and reverse transcription–polymerase chain reaction (RT–PCR) was carried out for CXCL16 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Similar results were obtained in three experiments. (b) Cells were stimulated with IL-1β (0·1–10 ng/ml), TNF-α (0·1–10 ng/ml), IFN-γ (0·1–10 ng/ml), IL-4 (0·1–10 ng/ml), IL-13 (0·1–10 ng/ml) and IL-10 (0·1–10 ng/ml) for 24 h. The expression levels of CXCL16 in the supernatants were measured with enzyme-linked immunosorbent assay. The results were calculated as the mean and standard deviation (s.d.) of one representative experiment performed in triplicate. Error bars show the s.d. of the values. *P < 0·01, significantly different from the medium.
IFN-γ, IL-4 and IL-13 differently modulated CXCL16 release enhanced by IL-1β
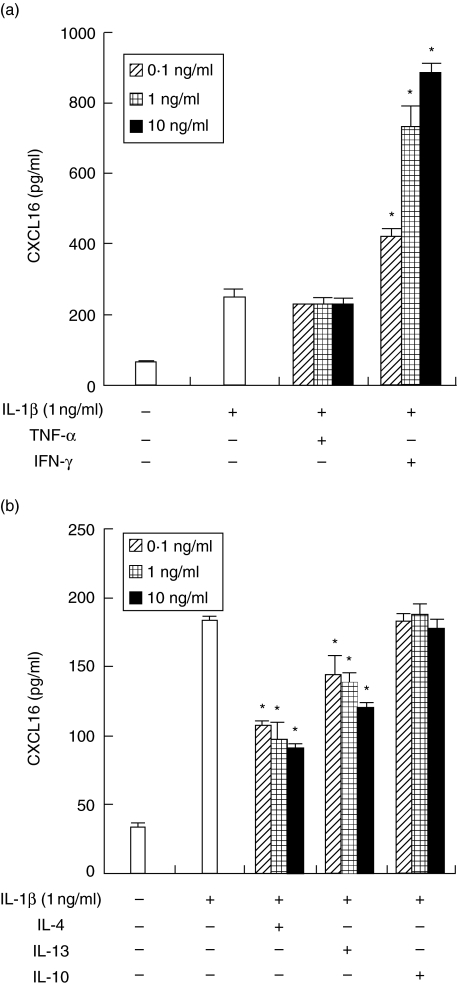
Next, we examined the effects of Th1 and Th2 cytokines on the release of CXCL16 induced by IL-1β because both Th1 and Th2 cells exist in diseased periodontal tissues. IFN-γ strongly enhanced the release of CXCL16 by HGFs. On the other hand, IL-4 and IL-13 decreased the IL-1β-induced release of CXCL16 by HGFs in a concentration-dependent manner. TNF-α and IL-10 did not affect the release induced by IL-1β (Fig. 4).
Fig. 4.
Interferon (IFN)-γ, interleukin (IL)-4 and IL-13 modulate the IL-1β-induced the release of CXCL16 by human gingival fibroblasts (HGFs). (a) HGFs were treated with IL-1β (1 ng/ml) with or without tumour necrosis factor (TNF)-α/IFN-γ (0·1, 1, 10 ng/ml), and the supernatants were collected after 24 h. The expression levels of CXCL16 in the supernatants were measured by enzyme-linked immunosorbent assay (ELISA). (b) HGFs were treated with IL-1β (1 ng/ml) with or without IL-4/IL-10/IL-13 (0·1, 1, 10 ng/ml), and the supernatants were collected after 24 h. The expression levels of CXCL16 in the supernatants were measured by ELISA. Data are representative of HGFs from three different donors. The results were calculated as the mean and standard deviation (s.d.) of one representative experiment performed in triplicate. Error bars show the s.d. of the values. *P < 0·01 significantly different from the IL-1β or IFN-γ-stimulated HGFs.
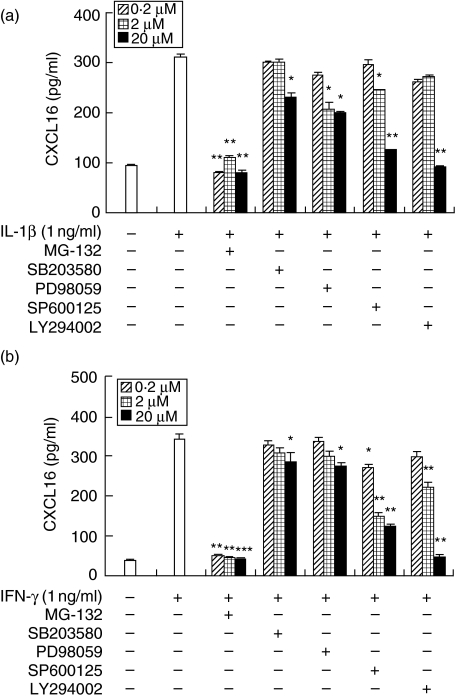
Involvement of p38 mitogen-activated protein kinase (MAPK), extracellular-regulated kinase (ERK), C-Jun amino terminal kinase (JNK), PI3K and nuclear factor (NF)-κB in the production of CXCL16 in HGFs stimulated with IL-1β and IFN-γ
To determine whether the activation of p38 MAPK, ERK, JNK, PI3K and NF-κB is required for the production of CXCL16 in response to IL-1β and IFN-γ, the effects of several inhibitors on the production of CXCL16 by HGFs were examined (Fig. 5). At a concentration of 20 µM, MG-132, a cell-permeable peptide–aldehyde protease inhibitor that blocks the activation of NF-κB by acting on the proteasome, completely prevented the production of CXCL16 in response to IL-1β and IFN-γ. SB203580, a selective inhibitor of p38 MAPK, binds with high affinity to p38 MAPK near the ATP-binding site, thus rendering p38 MAPK inactive. PD98059, a specific inhibitor that binds inactive forms of MEK and prevents their activation and phosphorylation resulting in inhibition of ERK and SP600125, a selective JNK inhibitor, partially inhibited CXCL16 production by HGF stimulated with IL-1β and IFN-γ. High concentration of LY294002, a selective PI3K inhibitor, almost completely inhibited CXCL16 release by HGF.
Fig. 5.
Effects of mitogen-activated protein kinase (MAPK) inhibitors, a PI3K inhibitor and a nuclear factor (NF)-κB inhibitor on the interleukin (IL)-1β or interferon (IFN)-γ-stimulated CXCL16 expression by human gingival fibroblasts (HGFs). Cells were preincubated with SB203580 (0·2, 2, 20 µM), PD98059 (0·2, 2, 20 µM), SP600125 (0·2, 2, 20 µM), LY294002 (0·2, 2, 20 µM) or MG-132 (0·2, 20, 20 µM) for 1 h and then incubated with IL-1β (1 ng/ml) (a) or IFN-γ (1 ng/ml) (b). After 24 h incubation, supernatants were collected and CXCL16 expression was measured by enzyme-linked immunosorbent assay. Data are representative of HGFs from three different donors. The results were calculated as the mean and standard deviation (s.d.) of one representative experiment performed in triplicate. Error bars show the s.d. of the values. *P < 0·05, **P < 0·01 significantly different from the IL-1β or IFN-γ-stimulated HGFs without inhibitors.
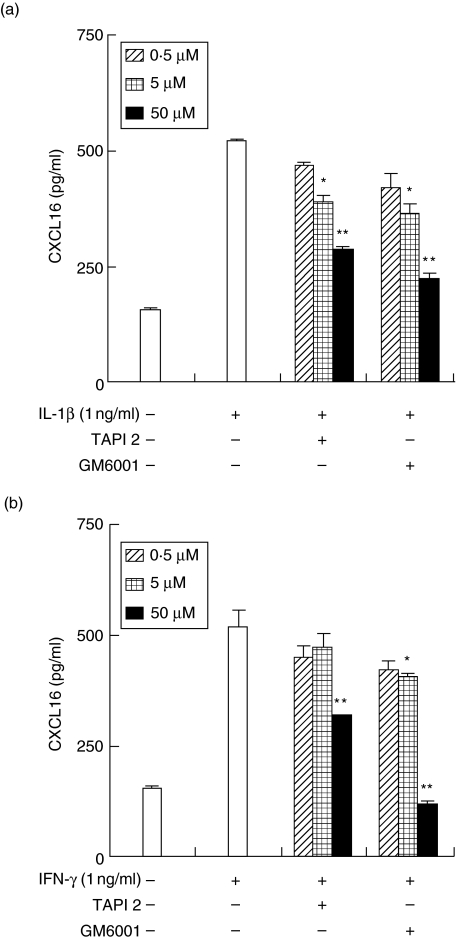
Effects of metalloproteinase inhibitors on CXCL16 release from HGFs
Next, we examined the role of ADAM10 and ADAM17 in the shedding of CXCL16 from HGFs using metalloproteinase inhibitors. The RT–PCR analysis revealed that HGFs expressed both ADAM10 and ADAM17 mRNA (data not shown). Therefore, we used GM6001 which inhibits the activity of metalloproteinases including ADAM10 and ADAM17, and TAPI2 which inhibits mainly only ADAM17. CXCL16 release from HGFs induced by IL-1β and IFN-γ was inhibited by GM6001 and TAPI2 in a dose-dependent manner (Fig. 6). GM6001 inhibited the release of CXCL16 more than did TAPI2.
Fig. 6.
Effects of metalloproteinase inhibitors on the release of CXCL16 from human gingival fibroblasts (HGFs). Cells were preincubated with GM6001 (0·5, 5, 50 µM) or TAPI2 (0·5, 5, 50 µM) for 1 h and then incubated with interleukin (IL)-1β (1 ng/ml) (a) or interferon (IFN)-γ (1 ng/ml) (b). After 24 h incubation, supernatants were collected and the CXCL16 expression was measured by enzyme-linked immunosorbent assay. Data are representative of HGFs from three different donors. The results were calculated as the mean and standard deviation (s.d.) of one representative experiment performed in triplicate. Error bars show the s.d. of the values. *P < 0·05, **P < 0·01 significantly different from the IL-1β or IFN-γ-stimulated HGF without inhibitors.
Discussion
Local inflammatory immune reactions to periodontal pathogens seem to be crucial protecting the host from infections, but may lead to pathological changes in host tissues [3, 4]. Selective migration of different cell types to gingival tissues is apparently related to the immunopathogenesis of periodontal disease [5]. We focused on CXCL16 among chemokines, as it has been reported to be involved in pathological damage in some diseases [31–35], and it is unknown whether or not CXCL16 is expressed in periodontal tissues.
CXCL16 is involved in the accumulation of Th1 cells [29]. Th1 cells are involved in bone resorption in periodontal disease [10]. Moreover, we found here that IFN-γ enhanced the production of CXCL16, and IL-4 and IL-13 inhibited CXCL16 release by HGFs stimulated with IL-1β. These results suggest that CXCL16 may control the migration of Th1 cells into diseased periodontal tissues. Furthermore, IFN-γ produced by Th1 cells induced the release of CXCL16 by HGFs, and increasingly shifting the Th1/Th2 balance in the Th1 direction. On the other hand, IL-4 and IL-13 produced by Th2 cells inhibit the expression of CXCL16 by HGFs in periodontal tissues, and the migration of Th1 cells might decrease. These results might explain why the production of CXCL16 is important for control of the Th1/Th2 balance in diseased periodontal tissues.
NK T cells infiltrate diseased periodontal tissue and are suggested to play an important role in the destruction of inflamed tissue [36]. It has been reported that NK T cells express CXCR6 [29]. Thus, the interaction of CXCR6 and CXCL16 may control the infiltration by NK T cells of diseased periodontal tissue. We found that CXCL16 is expressed by HGF in periodontally diseased tissue. These results suggest that the CXCL16 produced by HGFs in diseased periodontal tissue controls the migration of several types of inflammatory cells including Th1 cells and NK T cells.
It has been reported that each cell type expresses different chemokine receptors. For example, Th1 cells express CCR5, CXCR3, CX3CR1 and CXCR6 [37], and NK cells express CCR1, CCR4, CCR7, CXCR3, CXCR4, CX3CR1 and CXCR6 [38]. Several chemokines are expressed in periodontal diseased tissues; for example, monocyte chemoattractant protein (MCP)-1 (CCR1 ligand), macrophage inflammatory protein (MIP)-1α (CCR1 and CCR5 ligand), regulated upon activation normal T cell expressed and secreted (RANTES) (CCR1 and CCR5 ligand), CCL20 (CCR6 ligand), interferon-inducible protein-10 (IP-10) (CXCR3 ligand), CXCL12 (CXCR4) and fractalkine (CX3CR1 ligand) [11–13, 39–41]. These previous reports and our results suggest that more than one chemokine controls one type of cell, so the chemokine system involved in periodontal disease is complex. Further investigations are necessary for clarifying the chemokine system in periodontal tissue.
We demonstrated that a JNK inhibitor and a PI3K inhibitor inhibited mainly CXCL16 production. Chandrasekar et al. reported that IL-18 induced the expression of CXCL16 by rat aortic smooth muscle cells (ASMC) via PI3K and JNK signalling [42]. Our results are consistent with their report. Moreover, we demonstrated here that a p38 MAPK inhibitor and an ERK inhibitor partially inhibit CXCL16 expression by HGFs. They also reported that IL-18 induces CXCL16 expression by ASMC to a limited extent via p38 MAPK and ERK. Our results in relation to signalling are very similar to those in Chandrasekar's article [42].
We found that the shedding of CXCL16 by HGFs involves a mechanism similar to that described for CX3CL1 which is cleaved by the activity of the two disintegrin-like metalloproteinases ADAM10 and ADAM17 [43, 44]. Both these protease mRNAs are expressed in HGFs (data not shown). ADAM10 and ADAM17 are involved in the shedding of TNF-α, intracellular adhesion molecule (ICAM)-1, receptor activator of nuclear factor-ligand ligand (RANKL), and so on [45–47]. This means that the expression of ADAM10 and ADAM17 by HGFs is related to an exacerbation of periodontal disease to shed molecules involved in inflammatory reactions. Further investigation of ADAM10 and ADAM17 related to periodontal disease will be necessary.
In conclusion, CXCL16 is expressed by HGFs in diseased periodontal tissues. The expression is controlled by various cytokines. The CXCL16 produced by HGFs may play a role in the attraction of T cells to diseased tissue and the exacerbation of periodontal disease. However, further study is needed to clarify the role of CXCL16 in the development of periodontal disease.
Acknowledgments
This study was supported by a grant-in-aid from the Ministry of Education, Science and Culture of Japan.
References
- 1.Dzink JL, Tanner AC, Haffajee AD, Socransky SS. Gram negative species associated with active destructive periodontal lesions. J Clin Periodontol. 1985;12:648–59. doi: 10.1111/j.1600-051x.1985.tb00936.x. [DOI] [PubMed] [Google Scholar]
- 2.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 3.Kjeldsen M, Holmstrup P, Bendtzen K. Marginal periodontitis and cytokines: a review of the literature. J Periodontol. 1993;64:1013–22. doi: 10.1902/jop.1993.64.11.1013. [DOI] [PubMed] [Google Scholar]
- 4.Wilson M, Reddi K, Henderson B. Cytokine-inducing components of periodontopathogenic bacteria. J Periodont Res. 1996;31:393–407. doi: 10.1111/j.1600-0765.1996.tb00508.x. [DOI] [PubMed] [Google Scholar]
- 5.Taubman MA, Valverde P, Han X, Kawai T. Immune response: the key to bone resorption in periodontal disease. J Periodontol. 2005;76:2033–41. doi: 10.1902/jop.2005.76.11-S.2033. [DOI] [PubMed] [Google Scholar]
- 6.Jotwani R, Cutler CW. Multiple dendritic cell (DC) subpopulations in human gingiva and association of mature DCs with CD4+ T-cells in situ. J Dent Res. 2003;82:736–41. doi: 10.1177/154405910308200915. [DOI] [PubMed] [Google Scholar]
- 7.Taubman MA, Kawai T. Involvement of T-lymphocytes in periodontal disease and in direct and indirect induction of bone resorption. Crit Rev Oral Biol Med. 2001;12:125–35. doi: 10.1177/10454411010120020301. [DOI] [PubMed] [Google Scholar]
- 8.Seymour GJ, Gemmell E, Reinhardt RA, Eastcott J, Taubman MA. Immunopathogenesis of chronic inflammatory periodontal diseases: cellular and molecular mechanisms. J Periodont Res. 1993;28:478–86. doi: 10.1111/j.1600-0765.1993.tb02108.x. [DOI] [PubMed] [Google Scholar]
- 9.Taubman MA, Haffajee AD, Socransky SS, Smith DJ, Ebersole JL. Longitudinal monitoring of human antibody in subjects with destructive periodontal diseases. J Periodont Res. 1992;27:511–21. doi: 10.1111/j.1600-0765.1992.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 10.Kawai T, Eisen-Lev R, Seki M, Eastcott J, Wilson ME, Taubman MA. Requirement of B7 costimulation for Th1-mediated inflammatory bone resorption in experimental periodontal disease. J Immunol. 2000;164:2102–9. doi: 10.4049/jimmunol.164.4.2102. [DOI] [PubMed] [Google Scholar]
- 11.Hosokawa Y, Nakanishi T, Yamaguchi D, et al. Macrophage inflammatory protein 3alpha-CC chemokine receptor 6 interactions play an important role in CD4+ T-cell accumulation in periodontal diseased tissue. Clin Exp Immunol. 2002;128:548–54. doi: 10.1046/j.1365-2249.2002.01865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hosokawa Y, Nakanishi T, Yamaguchi D, Nakae H, Matsuo T. Expression of fractalkine (CX3CL1) and its receptor, CX3CR1, in periodontal diseased tissue. Clin Exp Immunol. 2005;139:506–12. doi: 10.1111/j.1365-2249.2005.02675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosokawa Y, Hosokawa I, Ozaki K, et al. CXCL12 and CXCR4 expression by human gingival fibroblasts in periodontal disease. Clin Exp Immunol. 2005;141:467–74. doi: 10.1111/j.1365-2249.2005.02852.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosokawa Y, Hosokawa I, Ozaki K, Nakae H, Matsuo T. Increase of CCL20 expression by human gingival fibroblasts upon stimulation with cytokines and bacterial endotoxin. Clin Exp Immunol. 2005;142:285–91. doi: 10.1111/j.1365-2249.2005.02912.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshie O, Imai T, Nomiyama H. Chemokines in immunity. Adv Immunol. 2001;78:57–110. doi: 10.1016/s0065-2776(01)78002-9. [DOI] [PubMed] [Google Scholar]
- 16.Bazan JF, Bacon KB, Hardiman G, et al. A new class of membrane-bound chemokine with a CX3C motif. Nature. 1997;385:640–4. doi: 10.1038/385640a0. [DOI] [PubMed] [Google Scholar]
- 17.Pan Y, Lloyd C, Zhou H, et al. Neurotactin, a membrane-anchored chemokine upregulated in brain inflammation. Nature. 1997;387:611–7. doi: 10.1038/42491. [DOI] [PubMed] [Google Scholar]
- 18.Matloubian M, David A, Engel S, Ryan JE, Cyster JG. A transmembrane CXC chemokine is a ligand for HIV-coreceptor Bonzo. Nat Immunol. 2000;1:298–304. doi: 10.1038/79738. [DOI] [PubMed] [Google Scholar]
- 19.Wilbanks A, Zondlo SC, Murphy K, et al. Expression cloning of the STRL33/BONZO/TYMSTRligand reveals elements of CC, CXC, and CX3C chemokines. J Immunol. 2001;166:5145–54. doi: 10.4049/jimmunol.166.8.5145. [DOI] [PubMed] [Google Scholar]
- 20.Shimaoka T, Nakayama T, Fukumoto N, et al. Cell surface-anchored SR-PSOX/CXC chemokine ligand 16 mediates firm adhesion of CXC chemokine receptor 6-expressing cells. J Leukoc Biol. 2004;75:267–74. doi: 10.1189/jlb.1003465. [DOI] [PubMed] [Google Scholar]
- 21.Gough PJ, Garton KJ, Wille PT, Rychlewski M, Dempsey PJ, Raines EW. A disintegrin and metalloproteinase 10-mediated cleavage and shedding regulates the cell surface expression of CXC chemokine ligand 16. J Immunol. 2004;172:3678–85. doi: 10.4049/jimmunol.172.6.3678. [DOI] [PubMed] [Google Scholar]
- 22.Abel S, Hundhausen C, Mentlein R, et al. The transmembrane CXC-chemokine ligand 16 is induced by IFN-gamma and TNF-alpha and shed by the activity of the disintegrin-like metalloproteinase ADAM10. J Immunol. 2004;172:6362–72. doi: 10.4049/jimmunol.172.10.6362. [DOI] [PubMed] [Google Scholar]
- 23.Ludwig A, Schulte A, Schnack C, et al. Enhanced expression and shedding of the transmembrane chemokine CXCL16 by reactive astrocytes and glioma cells. J Neurochem. 2005;93:1293–303. doi: 10.1111/j.1471-4159.2005.03123.x. [DOI] [PubMed] [Google Scholar]
- 24.Shimaoka T, Kume N, Minami M, et al. Molecular cloning of a novel scavenger receptor for oxidized low density lipoprotein, SR-PSOX, on macrophages. J Biol Chem. 2000;275:40663–6. doi: 10.1074/jbc.C000761200. [DOI] [PubMed] [Google Scholar]
- 25.Shashkin P, Simpson D, Mishin V, Chesnutt B, Ley K. Expression of CXCL16 in human T cells. Arterioscler Thromb Vasc Biol. 2003;23:148–9. doi: 10.1161/01.atv.0000043906.61088.4b. [DOI] [PubMed] [Google Scholar]
- 26.Hofnagel O, Luechtenborg B, Plenz G, Robenek H. Expression of the novel scavenger receptor SR-PSOX in cultured aortic smooth muscle cells and umbilical endothelial cells. Arterioscler Thromb Vasc Biol. 2002;22:710–1. doi: 10.1161/01.atv.0000012402.85056.45. [DOI] [PubMed] [Google Scholar]
- 27.Kim CH, Kunkel EJ, Boisvert J, et al. Bonzo/CXCR6 expression defines type 1-polarized T-cell subsets with extralymphoid tissue homing potential. J Clin Invest. 2001;107:595–601. doi: 10.1172/JCI11902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Motsinger A, Haas DW, Stanic AK, Van Kaer L, Joyce S, Unutmaz D. CD1d-restricted human natural killer T cells are highly susceptible to human immunodeficiency virus 1 infection. J Exp Med. 2002;195:869–79. doi: 10.1084/jem.20011712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas SY, Hou R, Boyson JE, et al. CD1d-restricted NKT cells express a chemokine receptor profile indicative of Th1-type inflammatory homing cells. J Immunol. 2003;171:2571–80. doi: 10.4049/jimmunol.171.5.2571. [DOI] [PubMed] [Google Scholar]
- 30.Sharron M, Pohlmann S, Price K, et al. Expression and coreceptor activity of STRL33/Bonzo on primary peripheral blood lymphocytes. Blood. 2000;96:41–9. [PubMed] [Google Scholar]
- 31.Minami M, Kume N, Shimaoka T, et al. Expression of SR-PSOX, a novel cell-surface scavenger receptor for phosphatidylserine and oxidized LDL in human atherosclerotic lesions. Arterioscler Thromb Vasc Biol. 2001;21:1796–800. doi: 10.1161/hq1001.096652. [DOI] [PubMed] [Google Scholar]
- 32.Wuttge DM, Zhou X, Sheikine Y, et al. CXCL16/SR-PSOX is an interferon-gamma-regulated chemokine and scavenger receptor expressed in atherosclerotic lesions. Arterioscler Thromb Vasc Biol. 2004;24:750–5. doi: 10.1161/01.ATV.0000124102.11472.36. [DOI] [PubMed] [Google Scholar]
- 33.Yamauchi R, Tanaka M, Kume N, et al. Upregulation of SR-PSOX/CXCL16 and recruitment of CD8+ T cells in cardiac valves during inflammatory valvular heart disease. Arterioscler Thromb Vasc Biol. 2004;24:282–7. doi: 10.1161/01.ATV.0000114565.42679.c6. [DOI] [PubMed] [Google Scholar]
- 34.Fukumoto N, Shimaoka T, Fujimura H, et al. Critical roles of CXC chemokine ligand 16/scavenger receptor that binds phosphatidylserine and oxidized lipoprotein in the pathogenesis of both acute and adoptive transfer experimental autoimmune encephalomyelitis. J Immunol. 2004;173:1620–7. doi: 10.4049/jimmunol.173.3.1620. [DOI] [PubMed] [Google Scholar]
- 35.Nanki T, Shimaoka T, Hayashida K, Taniguchi K, Yonehara S, Miyasaka N. Pathogenic role of CXCL16-CXCR6 pathway in rheumatoid arthritis. Arthritis Rheum. 2005;52:3004–14. doi: 10.1002/art.21301. [DOI] [PubMed] [Google Scholar]
- 36.Amanuma R, Nakajima T, Yoshie H, Yamazaki K. Increased infiltration of CD1d and natural killer T cells in periodontal disease tissues. J Periodont Res. 2006;41:73–9. doi: 10.1111/j.1600-0765.2005.00837.x. [DOI] [PubMed] [Google Scholar]
- 37.Sallusto F, Mackay CR, Lanzavecchia A. The role of chemokine receptors in primary, effector, and memory immune responses. Annu Rev Immunol. 2000;18:593–620. doi: 10.1146/annurev.immunol.18.1.593. [DOI] [PubMed] [Google Scholar]
- 38.Inngjerdingen M, Damaj B, Maghazachi AA. Expression and regulation of chemokine receptors in human natural killer cells. Blood. 2001;97:367–75. doi: 10.1182/blood.v97.2.367. [DOI] [PubMed] [Google Scholar]
- 39.Hanazawa S, Kawata Y, Takeshita A, et al. Expression of monocyte chemoattractant protein 1 (MCP-1) in adult periodontal disease. increased monocyte chemotactic activity in crevicular fluids and induction of MCP-1 expression in gingival tissues. Infect Immun. 1993;61:5219–24. doi: 10.1128/iai.61.12.5219-5224.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gemmell E, Carter CL, Seymour GJ. Chemokines in human periodontal disease tissues. Clin Exp Immunol. 2001;125:134–41. doi: 10.1046/j.1365-2249.2001.01511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gamonal J, Acevedo A, Bascones A, Jorge O, Silva A. Levels of interleukin-1 beta-8, and -10 and RANTES in gingival crevicular fluid and cell populations in adult periodontitis patients and the effect of periodontal treatment. J Periodontol. 2000;71:1535–45. doi: 10.1902/jop.2000.71.10.1535. [DOI] [PubMed] [Google Scholar]
- 42.Chandrasekar B, Mummidi S, Valente AJ, et al. The pro-atherogenic cytokine interleukin-18 induces CXCL16 expression in rat aortic smooth muscle cells via MyD88, interleukin-1 receptor-associated kinase, tumor necrosis factor receptor-associated factor 6, c-Src, phosphatidylinositol 3-kinase, Akt, c-Jun N-terminal kinase, and activator protein-1 signaling. J Biol Chem. 2005;280:26263–77. doi: 10.1074/jbc.M502586200. [DOI] [PubMed] [Google Scholar]
- 43.Garton KJ, Gough PJ, Blobel CP, et al. Tumor necrosis factor-alpha-converting enzyme (ADAM17) mediates the cleavage and shedding of fractalkine (CX3CL1) J Biol Chem. 2001;276:37993–8001. doi: 10.1074/jbc.M106434200. [DOI] [PubMed] [Google Scholar]
- 44.Hundhausen C, Misztela D, Berkhout TA, et al. The disintegrin-like metalloproteinase ADAM10 is involved in constitutive cleavage of CX3CL1 (fractalkine) and regulates CX3CL1-mediated cell–cell adhesion. Blood. 2003;102:1186–95. doi: 10.1182/blood-2002-12-3775. [DOI] [PubMed] [Google Scholar]
- 45.Zheng Y, Saftig P, Hartmann D, Blobel C. Evaluation of the contribution of different ADAMs to tumor necrosis factor alpha (TNFalpha) shedding and of the function of the TNFalpha ectodomain in ensuring selective stimulated shedding by the TNFalpha convertase (TACE/ADAM17) J Biol Chem. 2004;279:42898–906. doi: 10.1074/jbc.M403193200. [DOI] [PubMed] [Google Scholar]
- 46.Tsakadze NL, Sithu SD, Sen U, English WR, Murphy G, D'Souza SE. Tumor necrosis factor-alpha-converting enzyme (TACE/ADAM-17) mediates the ectodomain cleavage of intercellular adhesion molecule-1 (ICAM-1) J Biol Chem. 2006;281:3157–64. doi: 10.1074/jbc.M510797200. [DOI] [PubMed] [Google Scholar]
- 47.Pan B, Farrugia AN, To LB, et al. The nitrogen-containing bisphosphonate, zoledronic acid, influences RANKL expression in human osteoblast-like cells by activating TNF-alpha converting enzyme (TACE) J Bone Miner Res. 2004;19:147–54. doi: 10.1359/jbmr.2004.19.1.147. [DOI] [PubMed] [Google Scholar]