Abstract
P2X7 is a channel receptor gated by adenosine triphosphate (ATP) that is involved in the killing of intracellular mycobacteria. To explore further the role of P2X7 in immunity against Mycobacterium tuberculosis, we studied its expression and function in 19 patients with pulmonary tuberculosis (TB) and 19 healthy contacts. Flow cytometry analysis showed a similar and variable expression of P2X7 in TB patients and healthy subjects. In contrast, P2X7 mARN levels were significantly higher in TB patients. When the function of the P2X7 receptor in peripheral blood mononuclear cells (PBMC) was assessed by the effect of exogenous ATP on apoptosis, the uptake of the fluorescent marker Lucifer yellow or extracellular signal regulated kinase (ERK) phosphorylation, no significant differences were detected in patients and controls. However, mRNA macroarray analysis showed that upon stimulation with ATP, the PBMC from TB patients showed a significant induction of a higher number of cytokine genes (27 of 96), and a lower number of apoptosis genes (20 of 96) compared to healthy controls (17 and 76 genes, respectively). These results suggest that although the PBMC from TB patients do not show apparent abnormalities in the expression of P2X7, and the intracellular signals generated through it, the pattern of gene expression induced by ATP in these cells is different from that found in healthy contacts. This phenomenon suggests a defective function of P2X7 in the immune cells from TB patients, a condition that may contribute to the inability of these patients to eliminate the mycobacteria.
Keywords: apoptosis, lymphocytes, MAP kinases, P2X7, tuberculosis
Introduction
Tuberculosis (TB) is an infectious disease produced by Mycobacterium tuberculosis that, due to increasing antibiotic resistance and co-infection with HIV, constitutes an increasing world health problem with a high impact on morbidity and mortality [1]. The role of host genetic factors and the immune response in the outcome of infection with M. tuberculosis is well known [2]. Macrophages are the principal host cells for the intracellular replication of M. tuberculosis and, at the same time, they act as antigen-presenting cells (APCs) and play an important role in the killing of mycobacteria. In addition, protective acquired immunity to M. tuberculosis relies on cell-mediated immune response, mediated mainly by CD4+ and CD8+ T cells with the T helper type 1 (Th1) cytokine profile [3–5]. Thus, acquired resistance against TB seems to require the generation of a T cell-mediated immune response, the activation of infected macrophages and the formation of granuloma that prevent the dissemination of the mycobacteria [6,7]. However, the cellular and molecular interactions between mycobacteria and host immune cells have not been elucidated fully.
Apoptosis has been observed in monocytes and macrophages when they are infected with M. tuberculosis, either in vivo or in vitro[8]. In addition, different experimental data indicate that the programmed cell death of these cells has a protective role in human tuberculosis [9,10]. In this regard, it has been described previously that the induction of apoptosis of macrophages infected with M. bovis by adenosine triphosphate (ATP) significantly reduces the viability of this mycobacteria [11]. Similar results have been reported in M. tuberculosis infection [10].
P2X7, a member of the purinergic family of receptors, is gated by ATP, and seems to have an important role in the immune system [12]. This receptor is a ligand-gated cation channel with two transmembrane domains and a trimeric structure in the plasma membrane [13]. Activation of P2X7 by ATP results in cell permeabilization by opening cation-specific channels and non-specific pores that allow the passage of low-molecular-mass (< 900 Da) solutes [14,15]. In addition, the activation of phospholipase D, and an increase in intracellular calcium concentration when the killing of virulent M. tuberculosis is induced through P2X7 in infected human macrophages, has been observed [16,17]. Other downstream events observed after P2X7 activation and required for induction of apoptosis include the phosphorylation of mitogen-activated protein kinase (MAPK)/extracellular signal regulated kinase (ERK) 1/2 [18] as well as the activation of Src-family tyrosine kinases and PI3-K [18,19]. Additional studies have suggested that P2X7 may have a key role in the inflammatory response, mediating the release of interleukin (IL)-1β and tumour necrosis factor (TNF)-α[20,21]. Although the physiological role of P2X7 receptor-dependent cell death in immune cells has not been elucidated fully, its possible involvement in the resistance against M. tuberculosis infection is of interest [9].
In this study, we have hypothesized that there is a defective expression and/or function of P2X7 in immune cells from TB patients. We thus explored, in freshly isolated peripheral blood mononuclear cells (PBMC) from patients with pulmonary TB and healthy contacts, the expression and function of this receptor. We found a very variable expression of P2X7 with high levels of P2X7 mRNA in TB patients. When cells were stimulated with ATP, similar levels of apoptosis, pore formation and ERK1/2 phosphorylation were found in cells of TB patients and control subjects. However, cells from TB patients showed a very different pattern of gene expression in response to ATP compared to healthy subjects. These data suggest an altered function of P2X7 in immune cells from TB patients.
Materials and methods
Patients and healthy contacts
Nineteen patients with pulmonary tuberculosis from the Hospital Central ‘Dr Ignacio Morones Prieto’, San Luis Potosí, México were studied. Thirteen patients were male and six female; mean age 44·9 ± 20·3 years (range: 18–76). Diagnosis was based on clinical findings, chest X-ray examination and positive culture for M. tuberculosis. Ten patients had not received anti-tuberculous drugs, five had received them for less than 2 weeks before the study, and two had been receiving polychemotherapy for 2 months. Patients with systemic diseases such as immunodeficiency, diabetes mellitus, HIV infection or cirrhosis were excluded. Nineteen age- and sex-matched healthy contacts were studied as controls. Written informed consent was obtained from all individuals. The bioethical committee of the Hospital Central ‘Dr Ignacio Morones Prieto’ approved the study.
Cells and flow cytometry analysis
PBMC were isolated by a Ficoll-Hypaque density gradient centrifugation (Sigma Chemicals, St Louis, MO, USA). Cells were washed three times with phosphate-buffered saline (PBS) pH 7·3 and suspended at 1 × 106/ml in RPMI-1640 (HyClone, Laboratories, Inc., Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS), 100 μg/ml streptomycin, 100 U/ml penicillin and 2 mM l-glutamine (Sigma). To assess the expression of P2X7, cells (1 × 106) were incubated with a rabbit anti-P2X7 polyclonal antibody (2·0 μg/ml) or a negative control (1·0 μg/ml) (Chemicon International, Temecula, CA, USA) for 20 min at 4°C, followed by incubation with a fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit IgG (Sigma). Cells were then immunostained for CD14, CD19 or CD3 using phycoerythrin (PE)-labelled monoclonal antibodies (mAb) (BD-Pharmigen, San Diego, CA, USA). Cells were analysed in a FACSCalibur flow cytometer using CellQuest software (Becton Dickinson, San Jose, CA, USA).
Apoptosis assay
PBMC (1 × 106) were exposed to 3·0 mM ATP, 100 μM 3′-O-(4-benzoylbenzoyl)-ATP (BzATP) (Sigma) or medium alone at 37°C for 30 min [18,22]. Cells were then washed with cold PBS, and apoptotic cells were detected by terminal deoxynucleotidyl transferase biotin-dUTP nick end labelling (TUNEL) using the Apo-Direct kit (BD-Pharmigen) according to the manufacturer's instructions. Cells were analysed in a FACSCalibur flow cytometer (Becton Dickinson).
Assay of pore formation induced by ATP
PBMC were incubated with 3·0 mM ATP or 100 μM BzATP or medium alone in the presence of the fluorescent dyes Lucifer yellow (0·5 mg/ml, Molecular Probes, Eugene, OR, USA) or ethidium bromide (250 μM, Sigma) at 37°C for 10 min. Cells were then washed three times with cold PBS and analysed using LaboPhot-2 epifluorescent microscopy (Nikon INC, Melville, NY, USA) using a 63× oil immersion objective.
Western blot assays
PBMC were activated with 3·0 mM ATP or 200 ng/ml phorbol myristate acetate (PMA) for 15 and 3 min, respectively. In some experiments the PBMC were pretreated with the MEK1/2 inhibitor PD098059 as a negative control for ERK1/2 phosphorylation. Cells were then treated with 100 μl of lysis buffer [25 mM HEPES, pH = 7·4, 150 mM NaCL, 1·5 mM MgCl2, 0·2 mM ethylenediamine tetraacetic acid (EDTA), 0·5% Triton X-100, 0·5 mM dithiothreitol, 10 mM β-glycerophosphate, 1 mM Na3VO4, 5 mM NaF, 4 mM phenylmethylsulphonyl fluoride and 1 μg/ml of aprotinin and leupeptin]. After 30 min of incubation in an ice bath, samples were centrifuged at 12 000 at 4°C for 10 min and cell lysates were run in a 10% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions. Then, separated proteins were transferred to nitrocellulose membranes (Millipore Corporation, Bedford, MA, USA), which were blocked for 1 h with 5% non-fat dry milk in Tris-buffered saline (TBS-T) (20 mM Tris, pH 7·5, 150 mm NaCL, 0·05% Tween 20) and incubated with an anti-p-ERK1/2-specific mAb. Membranes were washed three times with TBS-T and incubated with a peroxidase-conjugated affinity-purified secondary antibody for 1 h. After extensive washing, membranes were developed by enhanced chemiluminescence (ECL, Amersham, Pharmacia Biotech, Piscataway, NJ, USA), using the Kodak ID Station 440CF system (Eastman Kodak Co., Rochester, NY, USA).
Detection of P2X7 mRNA
Total RNA was isolated from PBMC (5 × 106/ml) using TRIzol (Invitrogen Life Technology, Carlsbad, CA, USA). Reverse transcription to obtain cDNA was performed using 1–2 μg of total RNA isolated and 0·5 μg/μl oligo-dT12−18 primer (Invitrogen), which were heated at 70°C for 10 min. Then, 0·5 mM dNTPs, 10 mM dithiothreitol, 300 U/μl of Moloney murine leukaemia virus reverse transcriptase (M-MLV RT) and 40 U of RNasin (Promega, Madison, WI, USA) were added and the reaction was incubated at 37°C for 90 min. To amplify the COOH terminal fragment of P2X7 receptor, the following primers were used: sense 5′-TCTGCAAGATGTCAAGGGC-3′, and anti-sense: 5′-TCACTCTTCGGAAACTCTTTCC-3′, which gave rise to a 538 base pairs (bp) product. Polymerase chain reaction (PCR) amplification was performed by 35 cycles of denaturation at 94°C for 5 min, annealing at 52°C for 45 s, and extension at 72°C for 10 min. PCR products were separated in 1% agarose gel containing ethidium bromide and a 100-bp DNA molecular weight ladder (Promega, Madison, WI, USA). β-Actin was used as a positive control of amplification in the PCR reaction. The densitometry analysis was performed using a Kodak Digital Science 1D Image Analysis Software (Eastman Kodak Co.).
Analysis of gene expression
To assess the expression of genes induced by ATP in PBMC, 192 apoptosis, cytokine and cytokine receptor genes were analysed using cDNA macroarray kits (GEAarray; SuperArray Bioscience Co., Frederick, MD, USA). Briefly, PBMC were stimulated or not with 3 mM ATP for 30 min at 37°C, and total RNA was then extracted using the Trizol reagent (Invitrogen). Afterwards, RNA was subject to reverse transcription using the MMLV reverse transcriptase and a mixture of non-specific primers. The corresponding cDNA was subject to linear polymerase replication (LPR), with a cocktail of gene-specific primers, biotin-16-dUTP (Roche Diagnostics GmbH, Penzberg, Germany) and a DNA polymerase. Then, the labelled cDNA probe was denaturized by heating at 94°C for 2 min, and chilling in ice. Array membranes were prehybridized at 60°C for 2 h with heat-denatured salmon sperm DNA (Invitrogen) under continuous shaking. Then, the denatured cDNA probe was added to the membranes and hybridized overnight at 60°C. Afterwards, membranes were washed twice, blocked with the GEA blocking solution and incubated with alkaline phosphatase-conjugated streptavidin for 10 min. Finally, membranes were washed and developed by enhanced chemiluminescence using the Kodak ID Station 440CF system (Eastman Kodak Co., Rochester, NY, USA).
Results were expressed as the intensity of the hybridization signal referred to a gene of constitutive expression (β-actin), which was considered as 1·0. A significant induction of gene expression by exogenous ATP was defined arbitrarily as a 1·5-fold or higher enhancement in gene expression in cells exposed to ATP compared to untreated cells. Conversely, a significant inhibition in gene expression was defined when ATP-treated cells showed at least a 25% diminution in gene expression compared to untreated cells.
Statistical analysis
Results were expressed as the arithmetic mean ± standard error (s.e.). Statistical analysis was performed using the Sigma stat program (Jandel Scientific Software, Chicago, IL, USA). Parametric analysis was performed using Student's paired t-tests and one way-analysis of variance (anova) and the Mann–Whitney U-test and Kruskal–Wallis one-way analysis were used when data were not normally distributed. Values of P < 0·05 were considered significant.
Results
Expression of P2X7 in PBMC from TB patients
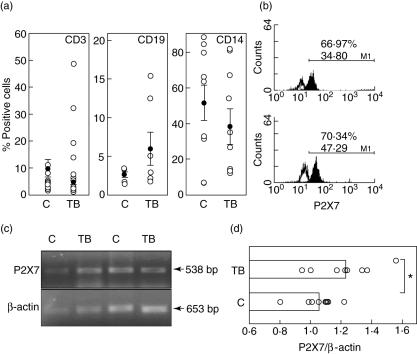
The surface expression of P2X7 by PBMC was measured by flow cytometry. We observed very variable levels of expression of this receptor in the three PBMC subsets studied (Fig. 1a,b). This variability was not associated significantly with anti-tuberculous therapy or the age of individuals studied (data not shown). Monocytes showed the highest and most variable expression of P2X7, which was three- to fourfold higher than those values found in B lymphocytes, both in controls and TB patients. Although levels of P2X7+ cells tended to be lower in monocytes from TB patients, no significant differences were found when compared with healthy contacts. Similar levels of expression of this receptor were detected in T and B lymphocytes from patients and controls. However, reverse transcription–polymerase chain reaction (RT–PCR) analysis indicated a significantly higher expression of P2X7 mRNA in TB patients than in controls (P < 0·05, Fig. 1c,d).
Fig. 1.
Expression of P2X7 in peripheral blood mononuclear cells from tuberculosis patients (TB) and healthy contacts (C). (a) Cells were immunostained for P2X7 and then for CD3, CD19 or CD14 and analysed by flow cytometry, as stated in Materials and methods. The arithmetic mean and s.d. of the percentage of positive cells are indicated. (b) Representative flow cytometry histograms of P2X7 expression in monocytes from a control (upper panel) and a TB patient (lower panel). Upper numbers correspond to the percentage of positive cells, and lower numbers to the mean fluorescence intensity. (c) Total mRNA isolated from peripheral blood mononuclear cells (PBMC) was reverse-transcribed to cDNA, and amplified by polymerase chain reaction using primers specific for P2X7 and β-actin. (d) Levels of P2X7 mRNA in PBMC from nine tuberculosis patients and nine healthy contacts. Data correspond to the arithmetic mean and s.e. of the densitometry values referred to β-actin signal (1·0). *P < 0·05.
P2X7 functional responses in PBMC from TB patients
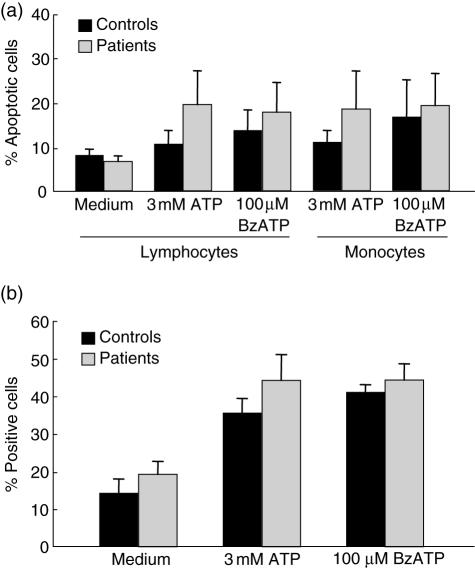
The function of P2X7 in PBMC was first assessed by analysing the effect of ATP on both the induction apoptosis (TUNEL and flow cytometry analysis) and the uptake of the low molecular weight fluorescent dye marker Lucifer yellow (fluorescence microscopy). As expected, P2X7-mediated apoptosis was observed in lymphocytes and monocytes from TB patients and controls when cells were exposed to ATP or BzATP for 30 min (Fig. 2a). Although cells from TB patients tended to show higher levels of apoptotic cells than healthy contacts, no significant differences were detected in all conditions tested. Similarly, when cells from TB patients or controls were incubated with ATP, a significant increase in cells stained with Lucifer yellow was detected (P < 0·02 in any case, Fig. 2b). However, no significant differences were detected when TB patients and healthy contacts were compared. Similar results were obtained when these assays were performed with ethidium bromide (data not shown).
Fig. 2.
Effect of adenosine triphosphate (ATP) on apoptosis and pore formation in cells from tuberculosis (TB) patients and healthy contacts. (a) Peripheral blood mononuclear cells were stimulated for 30 min with 3·0 mM ATP or 100 μM 3′-O-(4-benzoylbenzoyl)-ATP (BzATP). Then, apoptotic cells were detected by terminal deoxynucleotidyl transferase biotin-dUTP nick end labelling (TUNEL) and flow cytometry analysis. (b) Cells were stimulated or not for 10 min with 3·0 mM ATP or 100 μM BzATP in the presence of 0·5 mg/ml Lucifer yellow. The percentage of fluorescent cells was then determined by counting at least 200 cells in an epifluorescence microscopy. Data correspond to the arithmetic mean ± s.e.
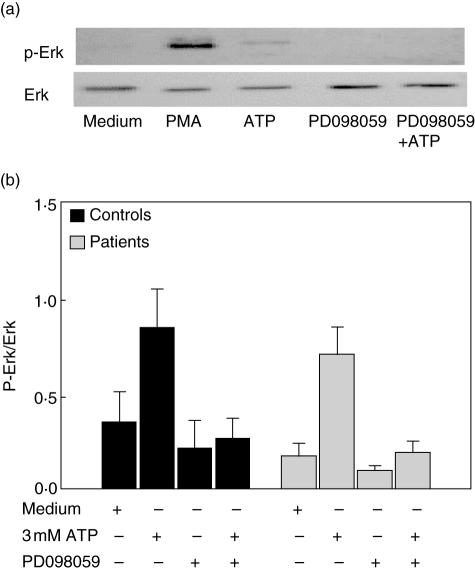
To explore further the functional status of P2X7 in TB patients, we then assessed the effect of ATP on the activation of ERK1/2 by their PBMC. As shown in Fig. 3, ATP induced a significant increase in the phosphorylation of ERK1/2 in cells from TB patients and healthy contacts. However, as in the case of other functional assays, no significant differences in ERK1/2 activation in response to ATP by cells from TB patients and healthy contacts were observed.
Fig. 3.
Induction of extracellular signal regulated kinase (ERK1/2) phophorylation by adenosine triphosphate (ATP) in cells from tuberculosis (TB) patients and healthy contacts. Peripheral blood mononuclear cells were stimulated or not with 3·0 mM ATP for 15 min, and then the phosphorylation of ERK1/2 was analysed by Western blot, as stated in Materials and methods. Data from a representative TB patient are shown in (a), and the ratio of the densitometry values of phosphorylated/total protein are shown in (b). The MAP kinase (MEK1/2) inhibitor, PD098059, was employed as a negative control for ERK1/2 phosphorylation.
Effect of ATP on gene expression by PBMC from TB patients
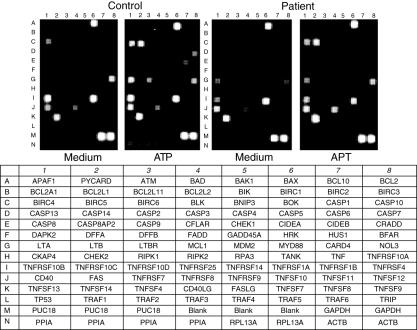
We then analysed the induction of gene expression by ATP in PBMC from TB patients. In contrast with the findings on the apparent similar expression and function of P2X7 in cells from TB patients and healthy contacts, we found a very different pattern of gene expression in response to ATP in these two groups when cells were incubated with ATP for 30 min or, in some experiments, for 15 min (Fig. 4, and data not shown). Regarding the 96 apoptosis genes analysed, we found that 20 of them were induced (1·5-fold or higher increase compared to non-stimulated cells) in PBMC from TB patients in response to ATP. These genes included those for TNF-α, CD30 and tumour necrosis factor-related apoptosis-inducing ligand treatment (TRAIL) receptor as well as several of the Bcl-2 family members (bak, bcl-2, blk), and some of the caspase and IAP families (Table 1). In contrast, ATP up-regulated 76 genes in PBMC from healthy controls, including those for TNF receptor and different TNF ligand family members (Table 1). In addition, 23 genes were down-regulated by the addition of ATP in cells from TB patients, including those encoding for TNF receptor 1 (R1) and TNFR2 as well as those for DR5, different caspases (14, 2, 8), bax proteins and Fas-associated death domain (FADD) (Table 1). In contrast, in cells from healthy contacts only bax and survivin genes showed a diminution in their expression after incubation with ATP. Finally, 13 genes showed a discordant regulation in cells from TB patients and controls (diminution of expression in TB with enhancement in control cells or the other way around, Table 1).
Fig. 4.
Analysis of gene induction by adenosine triphosphate (ATP) in cells from tuberculosis (TB) patients and healthy individuals. Peripheral blood mononuclear cells were stimulated or not with ATP for 30 min, and then the expression of 192 apoptosis, cytokine and cytokine receptor genes was analysed using a cDNA array test, as described in Materials and methods. Data from a representative patient and a healthy control are shown. The identity of genes analysed is indicated in the lower panel.
Table 1.
Analysis of induction of expression of apoptosis genes by adenosine triphosphate (ATP) in peripheral blood mononuclear cells from patients with tuberculosis and healthy contacts.
| Control | Patients | Fold induction | ||||
|---|---|---|---|---|---|---|
| Gene name | Medium | ATP | Medium | ATP | Control | Patients |
| CARD family | ||||||
| bcl-10/HuE10 | 0·59 ± 0·31* | 1·87 ± 0·13 | 2·89 ± 0·99 | 0·52 ± 0·12 | 3·15** | 0·18 |
| Nod/CARD4 | 0·26 ± 0·6 | 0·63 ± 0·10 | 0·20 ± 0·01 | 0·32 ± 0·07 | 2·37 | 1·65 |
| TNF ligand family | ||||||
| LT-β | 0·70 ± 0·26 | 1·12 ± 0·68 | 0·60 ± 0·01 | 0·32 ± 0·02 | 1·59 | 0·54 |
| TNF-α | 0·12 ± 0·03 | 0·49 ± 0·25 | 0·21 ± 0·03 | 0·32 ± 0·06 | 3·95 | 1·52 |
| TNFSF4/OX40L | 0·63 ± 0·50 | 1·44 ± 0·93 | 0·88 ± 0·80 | 0·73 ± 0·17 | 2·27 | 0·83 |
| CD40L/CD154 | 0·27 ± 0·02 | 1·09 ± 0·74 | 10·81 ± 3·2 | 15·60 ± 4·2 | 4·05 | 0·52 |
| Fas ligand | 1·59 ± 0·67 | 3·32 ± 0·27 | 0·81 ± 0·01 | 1·02 ± 0·58 | 2·09 | 1·27 |
| CD27L (CD70) | 0·45 ± 0·12 | 1·26 ± 0·94 | 0·34 ± 0·02 | 0·42 ± 0·21 | 2·77 | 1·22 |
| CD30L | 0·27 ± 0·01 | 1·37 ± 0·09 | 0·30 ± 0·07 | 0·40 ± 0·16 | 5·04 | 1·33 |
| TNFSF9/4–1BB | 0·13 ± 0·09 | 0·59 ± 0·04 | 0·11 ± 0·05 | 0·33 ± 0·15 | 4·46 | 2·93 |
| TNFSF11(TRANCE) | 0·35 ± 0·07 | 1·04 ± 0·69 | 0·30 ± 0·02 | 0·43 ± 0·22 | 2·96 | 1·42 |
| TNFSF12/APO3L | 0·09 ± 0·02 | 0·62 ± 0·09 | 0·12 ± 0·09 | 0·27 ± 0·85 | 6·53 | 2·32 |
| TNFSF13 (April) | 0·74 ± 0·34 | 2·65 ± 0·04 | 0·87 ± 0·08 | 1·26 ± 0·58 | 3·59 | 1·44 |
| TNFSF10 | 0·65 ± 0·04 | 1·51 ± 0·46 | 0·60 ± 0·19 | 0·49 ± 0·05 | 2·32 | 0·83 |
| TNF receptor family | ||||||
| TNFR1 | 10·49 ± 2·02 | 10·22 ± 2·97 | 13·61 ± 4·10 | 6·00 ± 2·95 | 1·03 | 0·44 |
| Fas (Apo-1) (CD95) | 2·10 ± 0·38 | 0·87 ± 0·45 | 1·32 ± 0·24 | 1·33 ± 0·20 | 2·40 | 1·01 |
| CD27 | 1·03 ± 0·72 | 0·41 ± 0·45 | 0·70 ± 0·66 | 0·72 ± 0·57 | 2·53 | 1·02 |
| CD30 | 4·90 ± 0·99 | 2·41 ± 0·42 | 3·04 ± 1·85 | 4·74 ± 1·57 | 2·03 | 1·56 |
| TRAIL-R | 1·09 ± 0·65 | 0·31 ± 0·52 | 0·24 ± 0·12 | 0·53 ± 0·26 | 3·49 | 2·17 |
| Trail receptor (DR5) | 10·88 ± 3·84 | 8·56 ± 2·51 | 10·36 ± 3·82 | 4·63 ± 1·63 | 1·27 | 0·45 |
| TRAIL-R3 (DcR1) | 1·53 ± 0·62 | 0·66 ± 0·32 | 0·66 ± 0·28 | 0·40 ± 0·13 | 2·32 | 0·61 |
| TRAIL-R4 (DcR2) | 0·70 ± 0·55 | 0·25 ± 0·08 | 0·44 ± 0·25 | 0·46 ± 0·16 | 2·74 | 1·04 |
| DR3 (Apo3) | 0·49 ± 0·67 | 0·26 ± 0·41 | 0·34 ± 0·03 | 0·49 ± 0·15 | 1·90 | 1·46 |
| TNFRSF14 | 0·92 ± 0·04 | 0·51 ± 0·18 | 0·44 ± 0·04 | 0·41 ± 0·12 | 1·81 | 0·91 |
| Bcl-2 family | ||||||
| Bak | 0·64 ± 0·23 | 1·11 ± 0·68 | 0·16 ± 0·07 | 0·26 ± 0·02 | 1·73 | 1·65 |
| Bax | 17·13 ± 3·84 | 10·23 ± 2·65 | 17·58 ± 2·96 | 10·99 ± 3·91 | 0·60 | 0·63 |
| bcl-10/HuE10 | 0·59 ± 0·31 | 1·87 ± 0·13 | 2·89 ± 1·01 | 0·52 ± 0·19 | 3·15 | 0·18 |
| bcl-2 | 0·27 ± 0·065 | 1·72 ± 0·14 | 0·12 ± 0·01 | 0·35 ± 0·09 | 6·35 | 3·04 |
| bfl-1 | 0·30 ± 0·06 | 0·87 ± 0·59 | 0·33 ± 0·18 | 0·37 ± 0·21 | 2·92 | 1·14 |
| bcl-x | 0·42 ± 0·11 | 0·85 ± 0·07 | 0·45 ± 0·62 | 0·48 ± 0·04 | 2·03 | 1·06 |
| biml | 0·85 ± 0·29 | 1·90 ± 0·63 | 0·53 ± 0·08 | 0·49 ± 0·22 | 2·24 | 0·94 |
| bcl-w | 0·48 ± 0·09 | 1·51 ± 0·85 | 1·19 ± 0·35 | 0·48 ± 0·08 | 3·11 | 0·40 |
| bik | 0·33 ± 0·13 | 1·18 ± 0·91 | 0·12 ± 0·01 | 0·12 ± 0·04 | 3·61 | 0·99 |
| blk | 0·32 ± 0·16 | 1·14 ± 0·90 | 0·14 ± 0·04 | 0·22 ± 0·10 | 3·53 | 1·62 |
| Nip3 | 0·33 ± 0·02 | 1·21 ± 0·62 | 0·08 ± 0·03 | 0·17 ± 0·04 | 3·69 | 2·11 |
| MCL-1 | 0·24 ± 0·07 | 0·62 ± 0·05 | 0·24 ± 0·02 | 0·33 ± 0·21 | 2·53 | 1·36 |
| Caspase family | ||||||
| Caspase-1 (ICE) | 0·87 ± 0·06 | 2·55 ± 0·46 | 0·30 ± 0·07 | 0·43 ± 0·19 | 2·94 | 1·42 |
| Caspase-10 (mch4) | 0·28 ± 0·09 | 1·54 ± 0·12 | 0·21 ± 0·05 | 0·32 ± 0·12 | 5·43 | 1·56 |
| Caspase 13 | 0·55 ± 0·10 | 1·26 ± 0·16 | 0·34 ± 0·02 | 0·26 ± 0·03 | 2·29 | 0·78 |
| Caspase 14 | 0·75 ± 0·14 | 0·59 ± 0·07 | 0·37 ± 0·06 | 0·21 ± 0·06 | 0·78 | 0·56 |
| Caspase-2 | 0·65 ± 0·04 | 1·01 ± 0·39 | 0·40 ± 0·14 | 0·24 ± 0·06 | 1·54 | 0·59 |
| Caspase-3 (cpp32) | 0·90 ± 0·06 | 2·81 ± 0·83 | 0·52 ± 0·26 | 0·67 ± 0·07 | 3·12 | 1·29 |
| Caspase 4(Ich-2) | 0·19 ± 0·01 | 1·31 ± 0·25 | 0·05 ± 0·01 | 0·17 ± 0·09 | 6·85 | 3·14 |
| Caspase-5 | 2·60 ± 0·98 | 2·18 ± 0·15 | 0·14 ± 0·02 | 0·29 ± 0·19 | 0·84 | 2·03 |
| Caspase-6 (mch2) | 0·29 ± 0·08 | 1·05 ± 0·18 | 0·06 ± 0·01 | 0·16 ± 0·04 | 3·57 | 2·57 |
| Caspase 9 (Mch6) | 0·32 ± 0·05 | 0·92 ± 0·07 | 0·27 ± 0·03 | 0·23 ± 0·08 | 2·90 | 0·85 |
| IAP family | ||||||
| NAIP/BIRC1 | 1·42 ± 0·07 | 3·08 ± 0·98 | 0·27 ± 0·02 | 0·47 ± 0·09 | 2·18 | 1·77 |
| IAP-2 | 0·26 ± 0·11 | 1·80 ± 0·13 | 0·12 ± 0·01 | 0·19 ± 0·02 | 6·96 | 1·59 |
| IAP-1 | 0·21 ± 0·10 | 1·80 ± 0·12 | 0·12 ± 0·03 | 0·27 ± 0·12 | 8·46 | 2·26 |
| survivin (API4) | 5·69 ± 2·50 | 2·02 ± 0·68 | 3·23 ± 1·20 | 6·28 ± 2·72 | 0·35 | 1·95 |
| Apollon/Bruce | 0·49 ± 0·06 | 0·96 ± 0·25 | 0·40 ± 0·04 | 0·32 ± 0·01 | 1·96 | 0·79 |
| TRAF family | ||||||
| TANK (I-TRAF) | 0·55 ± 0·22 | 1·33 ± 0·92 | 0·57 ± 0·09 | 0·49 ± 0·12 | 2·54 | 0·87 |
| TRAF1 | 1·14 ± 0·84 | 2·81 ± 0·74 | 2·43 ± 0·45 | 0·79 ± 0·15 | 2·45 | 0·32 |
| TRAF2 | 0·49 ± 0·26 | 1·47 ± 0·27 | 0·51 ± 0·06 | 0·42 ± 0·07 | 3·02 | 0·82 |
| TRAF-4 | 0·38 ± 0·03 | 1·40 ± 0·91 | 0·47 ± 0·03 | 0·39 ± 0·09 | 3·71 | 0·83 |
| TRAF5 | 0·04 ± 0·01 | 0·99 ± 0·04 | 0·35 ± 0·18 | 0·31 ± 0·15 | 8·17 | 0·88 |
| TRAF6 | 0·66 ± 0·14 | 2·48 ± 0·48 | 1·09 ± 0·11 | 1·02 ± 0·36 | 3·74 | 0·94 |
| Trip | 0·35 ± 0·13 | 1·31 ± 0·65 | 0·88 ± 0·05 | 0·87 ± 0·31 | 3·71 | 0·98 |
| Death domain family | ||||||
| FADD | 0·99 ± 0·46 | 0·13 ± 0·09 | 0·32 ± 0·17 | 0·18 ± 0·01 | 7·82 | 0·58 |
| MyD88 | 1·41 ± 0·13 | 0·58 ± 0·04 | 0·49 ± 0·03 | 0·36 ± 0·09 | 2·41 | 0·74 |
| CIDE domain family | ||||||
| CIDE-A | 0·33 ± 0·03 | 1·08 ± 0·42 | 0·20 ± 0·06 | 0·13 ± 0·07 | 3·31 | 0·66 |
| DFF40 (CAD) | 0·19 ± 0·01 | 0·82 ± 0·03 | 0·37 ± 0·04 | 0·26 ± 0·17 | 4·37 | 0·71 |
| p53 and ATM pathway | ||||||
| GADD45 | 0·22 ± 0·01 | 0·80 ± 0·06 | 0·33 ± 0·02 | 0·25 ± 0·08 | 3·54 | 0·75 |
| p63 | 0·45 ± 0·24 | 1·47 ± 0·10 | 0·48 ± 0·16 | 0·48 ± 0·17 | 3·2 | 1·00 |
| p53 | 0·11 ± 0·03 | 1·00 ± 0·07 | 0·37 ± 0·05 | 0·35 ± 0·06 | 9·09 | 0·95 |
| Hus1 | 0·31 ± 0·07 | 0·58 ± 0·35 | 0·31 ± 0·02 | 0·58 ± 0·03 | 1·87 | 1·87 |
Peripheral blood mononuclear cells from TB patients and healthy contacts were stimulated with ATP, and then gene expression was assessed with a cDNA macroarray kit, as indicated in Materials and methods.
Arithmetic mean ± s.d. of the hybridization signal referred to β-actin, which was considered as 1·0.
Fold induction of each gene referred to non-stimulated cells. ATM: ataxia telangiectasia mutated; CARD: caspase recruitment domain; CIDE: cell death-inducing DFFA-like effector; FADD: Fas-associated death domain; IAP: inhibitor of apoptosis protein; TNF: tumour necrosis factor; TRAF: TNF receptor-associated factor; TRAIL: tumour necrosis factor-related apoptosis-inducing ligand treatment.
The induction of cytokine genes by ATP was also different in PBMC from TB patients and controls. In healthy contacts, we found a significant induction of 17 cytokine genes (of 96 analysed), including three of chemokine receptors (CCR2, CCR5 and CCR6), four of different cytokines [interleukin (IL)-10, -13, -18, -25], and three of cytokine receptors (IL-10Rα, IL-12Rβ, IL-2Rα). In contrast, 27 cytokine genes were up-regulated by ATP in cells from TB patients, including five of chemokine receptors (CCR1,CCR3, CCR6, CX3CR1, CXCR4), seven of different cytokines [IL-1α, IL-1β, IL-6, IL-9, IL-16, TNF-α, transforming growth factor (TGF)-β3] and 11 of cytokine receptors (IL-1R1, IL-2Rβ, IL-2Rγ, IL-6Rα, IL-9Rα, IL-9/p40, IL-12Rβ1, IL-13Rα1, IL-13Rα2, IL-18R1, gp130). In addition, 20 genes were down-regulated upon ATP stimulation of PBMC from TB patients, including those encoding for several chemokines/cytokines [eotaxin, stromal cell-derived factor (SDF)1, SDF2, TGF-β1, IL-12B, IL-13, IL-18] and for some chemokine receptors (CCR4, CCR7, CCR8). In control cells, ATP down-regulated a different set of 23 genes, including genes encoding for chemokine receptors (CCR8, CCR9). Accordingly, a discordant response to ATP by TB and control cells was observed for five cytokine/cytokine receptor genes (IL-13, IL-18, IL-2Rα, SDF-2 and TNFR2) of 96 tested.
Discussion
In this study, we have explored the expression and function of P2X7 in immune cells from TB patients. In agreement with previous work conducted with cells from normal individuals and patients with chronic lymphocytic leukaemia [23,24], we found a very variable expression of this receptor in both tuberculosis patients and healthy contacts. Because we have assessed the expression of P2X7 in only T and B cells and in monocytes, it is feasible that our results on the overall expression of this receptor in PBMC could be affected by other cell subsets, mainly natural killer (NK) and NKT lymphocytes [25,26]. Therefore, we believe that it would be of interest to assess the expression of P2X7 in these cells, both in healthy individuals and tuberculosis patients.
Although the factors that regulate the expression of P2X7 have not been elucidated fully, it has been reported that different cytokines, including IL-2, IL-6 and TNF-α, and proinflammatory stimuli (e.g. bacterial lipopolysaccharides) up-regulate its synthesis [27]. In this regard, it is very likely that the elevated levels of TNF-α seen in TB patients [28,29], or that some mycobacterial-derived molecules (e.g. lipoarabinomannans) may affect the expression of this receptor in these patients. However, it is worth mentioning that in this study we did not find any apparent correlation between the level of expression of P2X7 and the clinical or laboratory parameters of our TB patients (data not shown). Thus, elucidation of the factors that determine the variable levels of expression of P2X7 in both healthy and diseased individuals remains a relevant point to be studied. In this regard, it is of interest that the PBMC from our TB patients showed higher levels of P2X7 mRNA than healthy contacts. This finding suggests that although there are no apparent differences in P2X7 protein expression, the post-transcriptional regulation of this receptor is different in PBMC from TB patients and healthy individuals.
It has been reported that P2X7 is able to mediate the apoptosis of immune cells, including monocytes, macrophages and lymphocytes [22]. In addition, this receptor is also involved in macrophage fusion, lymphoid cell proliferation and release of proinflammatory cytokines [30–34]. Interestingly, it has been found that the programmed cell death of infected macrophages mediated by ATP through P2X7 induces the killing of mycobacteria [10]. Therefore, this phenomenon seems to have an important role as a mechanism of resistance against M. tuberculosis[9,17]. We did not detect apparent defects in the induction of apoptosis by ATP in PBMC from TB patients, suggesting a normal function of P2X7. This point is supported by similar levels of induction of pore formation by ATP in cells from TB patients and controls. The lack of apparent abnormalities in ERK1/2 activation induced by ATP in PBMC from TB patients further supports a normal function of the P2X7 receptor in individuals infected with M. tuberculosis. However, our results on the induction of gene expression by ATP are not in agreement with this conclusion.
To explore further the functional status of P2X7 in cells from TB patients, we performed a cDNA array analysis of 192 apoptosis, cytokine and cytokine receptor genes upon stimulation of PBMC with ATP. Interestingly, we found a very different pattern of induction and repression of gene expression induced by ATP in PBMC from TB patients and healthy contacts. Although the ultimate cause of this phenomenon remains to be determined, it could be related to either genetic polymorphisms of the P2X7 gene, which affects the function of this receptor [35], or to the infectious process by itself [36]. In addition, it is feasible that our results of gene expression could be affected by the induction of apoptosis, which tended to be higher in patients compared to controls. In any case, we consider that it would be interesting to corroborate our data of macroarray analysis by RT–PCR or Western blot analysis, and to investigate further these findings to explore their physiological consequences. In addition, we think that our results further reveal the complexity of the pathogenesis of TB infection and the intricacy of the factors that seem to determine the susceptibility to this infectious disease. Furthermore, these results show our incomplete knowledge of the physiology of P2X7 in immune cells and its role in the pathogenesis of M. tuberculosis infection. However, it could be speculated that the different pattern of gene induction by ATP in PBMC from TB patients may play a role in the inability of the immune system of these individuals to eliminate the mycobacteria. In this regard, it is feasible that the up- and down-regulation of certain genes (e.g. TGF-β and IL-18, respectively) by ATP may alter the ability of these cells, mainly monocytes/macrophages, to act as both antigen-presenting cells, inducing an effective cell-mediated immune response, and as an important effector mechanism in killing the mycobacteria. However, it is also possible that P2X7 does not play a relevant role in the control of pulmonary TB in humans [37].
In summary, we have found that the PBMC from TB patients show an abnormal profile of gene expression in response to ATP, with no apparent defects in the expression of P2X7 or in different functions induced by ATP (apoptosis, pore formation, ERK1/2 phosphorylation). We believe that these results indicate that P2X7 has a particular functional behaviour in the PBMC from patients with pulmonary TB, a phenomenon that may contribute to the complex pathogenesis of this infectious disease.
Acknowledgments
This work was supported by the grants 36198-M (to D. P.-P.), and G35943-M (to R. G.-A.) from CONACYT, México. S. F.-M. and P. N.-M. were recipients of scholarships from CONACYT, México.
References
- 1.World Health Organization. Geneva: WHO.; Global tuberculosis programme. Tuberculosis factsheet. Available at: http://www.who.int/gtb/publication/factsheet/index.html. (Last accessed 2005) [Google Scholar]
- 2.Kato-Maeda M, Bifani PJ, Kreiswirth BN, Small PM. The nature and consequence of genetic variability within Mycobacterium tuberculosis. J Clin Invest. 2001;107:533–7. doi: 10.1172/JCI11426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chackerian AA, Perera TV, Behar SM. Gamma interferon-producing CD4+ T lymphocytes in the lung correlate with resistance to infection with Mycobacterium tuberculosis. Infect Immun. 2001;69:2666–74. doi: 10.1128/IAI.69.4.2666-2674.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serbina NV, Lazarevic V, Flynn JL. CD4(+) T cells are required for the development of cytotoxic CD8(+) T cells during Mycobacterium tuberculosis infection. J Immunol. 2001;167:6991–00. doi: 10.4049/jimmunol.167.12.6991. [DOI] [PubMed] [Google Scholar]
- 5.Zhang M, Lin Y, Lyer DV, Gong J, Abrams JS, Barnes PF. T-cell cytokine responses in human infection with Mycobacterium tuberculosis. Infect Immun. 1995;63:3231–4. doi: 10.1128/iai.63.8.3231-3234.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Algood HM, Lin PL, Flynn JL. Tumor necrosis factor and chemokine interactions in the formation and maintenance of granulomas in tuberculosis. Clin Infect Dis. 2005;41:S189–93. doi: 10.1086/429994. [DOI] [PubMed] [Google Scholar]
- 7.Lazarevic V, Nolt D, Flynn JL. Long-term control of Mycobacterium tuberculosis infection is mediated by dynamic immune responses. J Immunol. 2005;175:1107–17. doi: 10.4049/jimmunol.175.2.1107. [DOI] [PubMed] [Google Scholar]
- 8.Keane J, Balcewicz-Sablinska MK, Remold HG, et al. Infection by Mycobacterium tuberculosis promotes human alveolar macrophage apoptosis. Infect Immun. 1997;65:298–304. doi: 10.1128/iai.65.1.298-304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mancino G, Placido R, Di Virgilio F. P2X7 receptors and apoptosis in tuberculosis infection. J Biol Regul Homeost Agents. 2001;15:286–93. [PubMed] [Google Scholar]
- 10.Lammas DA, Stober C, Harvey CJ, Kendrick N, Panchalingam S, Kumararatne DS. ATP-induced killing of mycobacterium by human macrophages is mediated by purinergic P2Z (P2X7) receptor. J Immun. 1997;7:433–44. doi: 10.1016/s1074-7613(00)80364-7. [DOI] [PubMed] [Google Scholar]
- 11.Molloy A, Laochumroonvorapong P, Kaplan G. Apoptosis, but not necrosis, of infected monocytes is coupled with killing of intracellular bacillus Calmette–Guérin. J Exp Med. 1994;180:1499–509. doi: 10.1084/jem.180.4.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Di Virgilio F, Chiozzi P, Ferrari D, et al. Nucleotide receptors: an emerging family of regulatory molecules in blood cells. Blood. 2001;97:587–00. doi: 10.1182/blood.v97.3.587. [DOI] [PubMed] [Google Scholar]
- 13.North RA. Molecular physiology of P2X7 receptors. Physiol Rev. 2002;82:1013–67. doi: 10.1152/physrev.00015.2002. [DOI] [PubMed] [Google Scholar]
- 14.Lajdova I, Chorvat D, Jr, Spustova V, Chorvatova A. 4-Aminopyridine activates calcium influx through modulation of the pore-forming purinergic receptor in human peripheral blood mononuclear cells. Can J Physiol Pharmacol. 2004;82:50–6. doi: 10.1139/y03-128. [DOI] [PubMed] [Google Scholar]
- 15.Virginio C, MacKenzie A, North RA, Surprenant A. Kinetics of cell lysis, dye uptake and permeability changes in cells expressing the rat P2X7 receptor. J Physiol. 1999;519:335–46. doi: 10.1111/j.1469-7793.1999.0335m.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fairbairn IP, Stober CB, Kumararatne DS, Lammas DA. ATP-mediated killing of intracellular Mycobacterium by macrophages is a P2X7-dependent process inducing bacterial death by phagosome–lysosome fusion. J Immunol. 2001;167:3300–7. doi: 10.4049/jimmunol.167.6.3300. [DOI] [PubMed] [Google Scholar]
- 17.Kusner DJ, Barton JA. ATP stimulates human macrophages to kill intracellular virulent Mycobacterium tuberculosis via calcium-dependent phagosome–lysosome fusion. J Immunol. 2001;167:3308–15. doi: 10.4049/jimmunol.167.6.3308. [DOI] [PubMed] [Google Scholar]
- 18.Humphreys BD, Rice J, Kertesy SB, Dubyak GR. Stress-activated protein kinase/JNK activation and apoptotic induction by the macrophage P2X7 nucleotide receptor. J Biol Chem. 2000;275:26792–8. doi: 10.1074/jbc.M002770200. [DOI] [PubMed] [Google Scholar]
- 19.Auger R, Motta I, Benihoud K, Ojcius DM, Kanellopoulos JM. A role for mitogen-activated protein kinase (Erk1/2) activation and non-selective pore formation in P2X7 receptor-mediated thymocyte death. J Biol Chem. 2005;280:28142–51. doi: 10.1074/jbc.M501290200. [DOI] [PubMed] [Google Scholar]
- 20.Kahlenberg JM, Dubyak GR. Mechanisms of caspase-1 activation by P2X7 receptor-mediated K+ release. Am J Physiol Cell Physiol. 2004;286:C1100–8. doi: 10.1152/ajpcell.00494.2003. [DOI] [PubMed] [Google Scholar]
- 21.Labasi JM, Petrushova N, Donovan C, et al. Absence of the P2X7 receptor alters leukocyte function and attenuates an inflammatory response. J Immunol. 2002;168:6436–45. doi: 10.4049/jimmunol.168.12.6436. [DOI] [PubMed] [Google Scholar]
- 22.Saunders BM, Fernando SL, Sluyter R, Britton WJ, Wiley JS. A loss-of-function polymorphism in the human P2X7 receptor abolishes ATP-mediated killing of Mycobacterium. J Immunol. 2003;171:5442–6. doi: 10.4049/jimmunol.171.10.5442. [DOI] [PubMed] [Google Scholar]
- 23.Gu BJ, Zhang WY, Bendall LJ, et al. Expression of P2X7 purinoceptors on human lymphocytes and monocytes: evidence for nonfunctional P2X7 receptors. Am J Physiol Cell Physiol. 2000;279:C1189–97. doi: 10.1152/ajpcell.2000.279.4.C1189. [DOI] [PubMed] [Google Scholar]
- 24.Miller JS, Cervenka T, Lund J, Okazaki IJ, Moss J. Purine metabolites suppress proliferation of human NK cells through a lineage-specific purine receptor. J Immunol. 1999;162:7376–82. [PubMed] [Google Scholar]
- 25.Kawamura H, Aswad F, Minagawa M, Govindarajan S, Dennert G. P2X7 receptors regulate NKT cells in autoimmune hepatitis. J Immunol. 2006;176:2152–60. doi: 10.4049/jimmunol.176.4.2152. [DOI] [PubMed] [Google Scholar]
- 26.Wiley JS, Dao-Ung LP, Gu BJ, et al. A loss-of-function polymorphic mutation in the cytolytic P2X7 receptor gene and chronic lymphocytic leukemia: a molecular study. Lancet. 2002;359:1114–9. doi: 10.1016/S0140-6736(02)08156-4. [DOI] [PubMed] [Google Scholar]
- 27.Zhang XJ, Zheng GG, Ma XT, Lin YM, Song YH, Wu KF. Effects of various inducers on the expression of P2X7 receptor in human peripheral blood mononuclear cells. Sheng Li Xue Bao. 2005;57:193–8. [PubMed] [Google Scholar]
- 28.Fiorenza G, Rateni L, Farroni MA, Bogue C, Dlugovitzky DG. TNF-alpha, TGF-beta and NO relationship in sera from tuberculosis (TB) patients of different severity. Immunol Lett. 2005;98:45–8. doi: 10.1016/j.imlet.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 29.Olobo JO, Geletu M, Demissie A, et al. Circulating TNF-alpha, TGF-beta, and IL-10 in tuberculosis patients and healthy contacts. Scand J Immunol. 2001;53:85–91. doi: 10.1046/j.1365-3083.2001.00844.x. [DOI] [PubMed] [Google Scholar]
- 30.Adinolfi E, Callegari MG, Ferrari D, et al. Basal activation of the P2X7 ATP receptor elevates mitochondrial calcium and potential, increases cellular ATP levels, and promotes serum-independent growth. Mol Biol Cell. 2005;16:3260–72. doi: 10.1091/mbc.E04-11-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baricordi OR, Melchiorri L, Adinolfi E, et al. Increased proliferation rate of lymphoid cells transfected with the P2X(7) ATP receptor. J Biol Chem. 1999;274:33206–8. doi: 10.1074/jbc.274.47.33206. [DOI] [PubMed] [Google Scholar]
- 32.Chiozzi P, Sanz JM, Ferrari D, et al. Spontaneous cell fusion in macrophage cultures expressing high levels of the P2Z/P2X7 receptor. J Cell Biol. 1997;138:697–06. doi: 10.1083/jcb.138.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Virgilio F, Falzoni S, Chiozzi P, Sanz JM, Ferrari D, Buell GN. ATP receptors and giant cell formation. J Leukoc Biol. 1999;66:723–6. doi: 10.1002/jlb.66.5.723. [DOI] [PubMed] [Google Scholar]
- 34.Ke HZ, Qi H, Weidema AF, et al. Deletion of the P2X7 nucleotide receptor reveals its regulatory roles in bone formation and resorption. Mol Endocrinol. 2003;17:1356–67. doi: 10.1210/me.2003-0021. [DOI] [PubMed] [Google Scholar]
- 35.Shemon AN, Sluyter R, Fernando SL, et al. A Thr357 to Ser polymorphism in homozygous and compound heterozygous subjects causes absent or reduced P2X7 function and impairs ATP-induced mycobacterial killing by macrophages. J Biol Chem. 2006;281:2079–86. doi: 10.1074/jbc.M507816200. [DOI] [PubMed] [Google Scholar]
- 36.Blumenthal A, Lauber J, Hoffmann R, et al. Common and unique gene expression signatures of human macrophages in response to four strains of Mycobacterium avium that differ in their growth and persistence characteristics. Infect Immun. 2005;73:3330–41. doi: 10.1128/IAI.73.6.3330-3341.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Myers AJ, Eilertson B, Fulton SA, Flynn JL, Canaday DH. The purinergic P2X7 receptor is not required for control of pulmonary Mycobacterium tuberculosis infection. Infect Immun. 2005;73:3192–5. doi: 10.1128/IAI.73.5.3192-3195.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]