Abstract
The development of a statistical model based on simple immunological markers which could predict the response to tuberculosis treatment would facilitate clinical trials of new anti-tuberculosis drugs. We have examined the ability of immunological biomarkers, measured at diagnosis and after 4 weeks of treatment, to predict sputum smear status at week 8. Eighteen tuberculosis patients with positive Ziehl–Nielsen (ZN)-stained sputum smears 8 weeks after initiation of treatment (slow response) were matched for age, gender, sputum smear grade and extent of disease on chest radiograph to 18 patients with negative sputum smears at week 8 (fast response). In addition to total white blood cell (WBC) counts and absolute lymphocyte, monocyte and neutrophil numbers, concentrations of six serum markers were measured by enzyme-linked immunosorbent assay (ELISA) in all patients (soluble interleukin-2 receptor alpha (sIL-2Rα), granzyme B, soluble tumour necrosis factor alpha receptors 1 and 2 (sTNF-R1 and -2), nitrotyrosine and interferon-gamma (IFN-γ). At diagnosis, 4 biomarkers (sTNF-R1, total WBC, absolute monocyte and absolute neutrophil numbers) were significantly higher in slow response patients. At week 4, total WBC count and absolute monocyte and neutrophil numbers remained significantly higher in slow responders. Discriminant analysis of the diagnosis and week 4 data provided models for classification of slow response patients with 67% and 83% predictive accuracy. We suggest that treatment response phenotypes can be determined before the start of treatment. Reliable predictive models would allow targeted interventions for patients at risk for slow treatment response to standard tuberculosis therapy.
Keywords: discriminant analysis, predictive model, surrogate markers, tuberculosis
Introduction
The investigation of predictive biomarkers has become increasingly popular among tuberculosis (TB) researchers in recent years [1,2]. There have been reports of the ability of clinical, radiographic and microbiological data to predict the outcome of short-course treatment regimens with standard drugs [3]; however, there is currently no immunological surrogate marker which can predict response to TB therapy. Identifying immune markers measured soon after the initiation of therapy which accurately predetermine treatment outcome would benefit clinical trials of new anti-tubercular agents by determining proof of concept in phase II trials, thus expediting the progression to full phase III studies.
The International Union Against Tuberculosis and Lung Disease (IUATLD) recommends that sputum samples are taken from TB cases at the end of the 2-month intensive phase of treatment in order to monitor patient progress [4]. We have shown previously that 23% of TB patients in the greater Cape Town area remain smear positive at this time-point [5]. The primary aim of this study was to determine whether immunological biomarkers measured in serum at diagnosis and after 4 weeks of treatment could predict the smear status of TB patients after 2 months of standard intensive phase therapy. A wide variety of cell-mediated immunological factors have been suggested to play a role in the immune response to TB [6]. We selected interferon (IFN)-γ, soluble interleukin-2 receptor (sIL-2R)α, soluble tumour necrosis factor alpha receptors 1 and 2 (sTNF-R1, sTNF-R2), granzyme B and nitrotyrosine as markers for measurement in the serum of 18 TB patients with positive Ziehl–Nielsen (ZN)-stained sputum smears 8 weeks after initiation of treatment (slow responders) and 18 matched patients with negative sputum smears at the same time-point. In addition to the above serum markers, we had data available on immune cells through routine full blood count (FBC) and differential white cell count analysis. This included total white blood cell (WBC) counts and absolute numbers and percentages of lymphocytes, monocytes and neutrophils.
IFN-γ has been measured extensively in studies of TB. The primary function of IFN-γ is macrophage activation [7]. A failure to generate IFN-γ in response to mycobacterial infection proves fatal in the mouse model [8], and IFN-γ receptor deficiency in humans results in increased susceptibility to mycobacterial infections [9]. The detection of sIL-2Rα indicates T cell activation and evidence suggests that its presence in serum may correlate with response to chemotherapy [10]. TNF-α has been implicated in both the protection and pathology of TB with evidence for a role in granuloma formation [11] and tissue damage [12]. Soluble TNF-RI (55 kDa) and sTNF-R2 (75 kDa) have been found in human body fluids [13,14] and are thought to regulate the bioavailability and limit the toxic effects of TNF-α in the body [15]. Increased levels of sTNF-R1 have been demonstrated previously in patients with active TB compared to uninfected subjects [16]. sTNF-R2 has been shown to serve as a marker of both innate and cell-mediated immunity in a mouse model of vesicular stomatitis virus [17]. Human CD8+ T cells have been shown to be cytolitic against Mycobacterium tuberculosis-infected target cells [18]. A primary cytolytic mechanism of lymphocytes involves the release of perforin [19] and granzyme [20] containing granules [21] upon contact with the target cell. Granzymes are proteases which enter target cells through perforin pores and augment cytolitic cell damage [20]. The production of inducible nitric oxide synthase (iNOS) is also a feature of TB [22,23]. Alveolar macrophages of TB patients, but not uninfected controls, have been shown to express iNOS [24] and IFN-γ-stimulated macrophages co-cultured with M. tuberculosis-primed lymphocytes exhibit bactericidal activity and nitric oxide synthase 2 (NOS2) expression [25]. Nitric oxide (NO) is produced readily by mouse macrophages infected with mycobacteria [26], whereas evidence for NO production by human macrophages has been more modest. One of the end results of NO production is the nitrogenization of proteins, therefore measurement of nitrotyrosine in serum serves as a marker of NO production [27]. We show that a combination of serum biomarkers and peripheral blood white cell counts holds promise to establish a predictive model of early treatment response as defined by sputum smear conversion at week 8 of treatment.
Methods
Participants
The subjects included were participants in a longitudinal study evaluating pulmonary TB cases during treatment [28,29]. All cases were new sputum smear positive (two sputum samples; ZN-stained) pulmonary TB cases, 18–65 years of age, HIV negative, not pregnant, had fully drug sensitive organisms, were adherent to treatment during the intensive phase (at least 32 of the prescribed 40 dosages) and had sputum results available after 8 weeks of treatment. Treatment was according to the directly observed therapy short-course (DOTS) strategy and the South African National Tuberculosis Program based on World Health Organization (WHO) guidelines. Smear positive cases received a fixed drug combination of isoniazid, rifampicin, ethambutol and pyrazinamide during the intensive phase (2 months), sputum samples were taken after 2 months of treatment and if negative the continuation phase of treatment was commenced (fixed drug combination of rifampicin and isoniazid for 4 months).
Chest X-rays and venous blood samples were taken at diagnosis and after 4 weeks of treatment. Blood for cell counts was collected in ethylenediamine tetraacetic acid (EDTA) and transported to the laboratory at room temperature. Blood for serum separation was collected without anti-coagulant and transported to the laboratory on ice, centrifuged at 4°C, aliquoted and stored at −80°C before testing. Chest radiographs were evaluated using a standardized method by a physician who had no prior knowledge of the patient's condition.
Patient characteristics
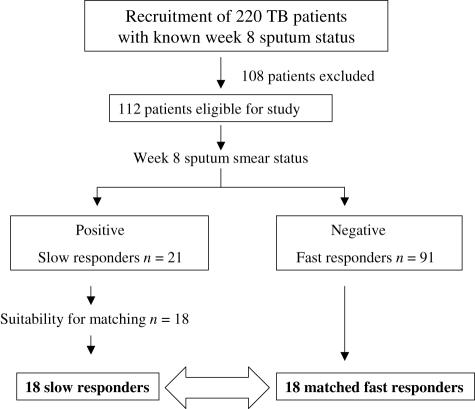
Of 220 patients enrolled, 112 met the inclusion criteria. Twenty-one had a positive sputum smear result at week 8 (‘slow responders’), of whom 18 were suitable for matching with ‘fast responders’ who had negative smears at week 8 (Fig. 1). Two sputum samples were collected at the 2-month time-point. Fourteen patients were female (seven in each group), 22 were male (11 in each group) and the mean age was 38 (range 21–54 years). Matching was performed on data collected at diagnosis and included age (within 5 years), gender and extent of disease on chest radiograph (total extent of alveolar disease involvement, lung areas containing consolidation and the amount of cavities present). Baseline sputum smear grades were regarded as being equal between the two groups. Clinical data of participants are shown in Table 1. Sixteen PPD skin test-positive people were also tested in this study, who acted as community controls. We chose skin-test positive controls as 76% of people in the study area have skin tests ≥ 10 mm [30].
Fig. 1.
Study profile. Of the 220 patients who were recruited, 112 met the inclusion criteria. Twenty-one had a positive sputum smear result at week 8 (‘slow responders’), of whom 18 were suitable for matching with ‘fast responders’ who had negative smears at week 8.
Table 1.
Matched pairs of slow and fast responders to intensive phase anti-tuberculosis therapy.
| Match | Group | Sex | Age | ZN | Consolidation (CXR) | Amount of cavities (CXR) | Extent of disease (CXR) |
|---|---|---|---|---|---|---|---|
| 1 | Slow | M | 24 | 1+† | LLL RUL | >4 | >RUL |
| 1 | Fast | M | 21 | 2++‡ | LING LUL | >4 | >RUL |
| 2 | Slow | M | 27 | 1+ | LLL LUL RLL RML RUL | >4 | >1 lung |
| 2 | Fast | M | 26 | 1+ | LLL LUL RLL RML RUL LING | >4 | >RUL |
| 3 | Slow | M | 50 | 3+++§ | LLL LUL RUL LING | >4 | >RUL |
| 3 | Fast | M | 54 | SCANTYII | LUL RUL LING | >4 | >RUL |
| 4 | Slow | M | 50 | 3+++ | RUL LING | 2 | < RUL |
| 4 | Fast | M | 48 | 1+ | RUL LUL | 2 | < RUL |
| 5 | Slow | M | 53 | 1+ | LUL LLL LING | >4 | >RUL |
| 5 | Fast | M | 49 | 1+ | LUL RLL RUL | >4 | >RUL |
| 6 | Slow | M | 29 | 2++ | LLL LUL RUL RLL RML | >4 | >1 lung |
| 6 | Fast | M | 26 | 2++ | LLL LUL RUL RLL | >4 | >RUL |
| 7 | Slow | F | 37 | 2++ | LLL LUL RLL RUL | 1 | >RUL |
| 7 | Fast | F | 36 | 2++ | RML RLL RUL | 1 | >RUL |
| 8 | Slow | F | 38 | 3+++ | LUL | >4 | < RUL |
| 8 | Fast | F | 41 | 1+ | RUL RLL | >4 | < RUL |
| 9 | Slow | F | 45 | 1+ | LUL RUL | 2 | < RUL |
| 9 | Fast | F | 42 | 1+ | LUL LING | 1 | < RUL |
| 10 | Slow | M | 28 | 3+++ | RUL | 2 | < RUL |
| 10 | Fast | M | 24 | 1+ | RML | 1 | < RUL |
| 11 | Slow | M | 40 | 1+ | LLL LING LUL RLL RML RUL | >4 | >1 lung |
| 11 | Fast | M | 45 | 3+++ | LING LUL RLL RML RUL | >4 | >1 lung |
| 12 | Slow | M | 41 | 1+ | RLL LUL RUL LING | >4 | >RUL |
| 12 | Fast | M | 45 | 1+ | LLL LUL RUL | >4 | >RUL |
| 13 | Slow | F | 42 | 2++ | LLL LUL RLL RML RUL LING | 1 | >RUL |
| 13 | Fast | F | 44 | 2++ | LLL LUL RLL RUL LING | 1 | >RUL |
| 14 | Slow | M | 38 | 1+ | LLL LUL RLL RUL | >4 | >1 lung |
| 14 | Fast | M | 39 | 3+++ | LLL LUL RLL RML RUL LING | >4 | >1 lung |
| 15 | Slow | M | 53 | 1+ | LLL LUL RLL RML RUL LING | 2 | >1 lung |
| 15 | Fast | M | 54 | 1+ | RML RUL | >4 | >RUL |
| 16 | Slow | M | 53 | 1+ | LLL LUL RLL RML RUL LING | 2 | >1 lung |
| 16 | Fast | M | 54 | 1+ | RML RUL | >4 | >RUL |
| 17 | Slow | F | 21 | 2++ | LUL RLL RUL LING | >4 | >RUL |
| 17 | Fast | F | 20 | 2++ | LLL LUL LING | >4 | >RUL |
| 18 | Slow | F | 32 | 1+ | LLL RLL RML RUL | 2 | >RUL |
| 18 | Fast | F | 29 | 1+ | LLL RUL | 2 | >RUL |
ZN, Ziehl–Nielsen; CXR, chest X-ray; LLL, left lower lobe; LING, lingua; LUL, left upper lobe; RLL, right lower lobe; RML, right middle lobe; RUL, right upper lobe; > greater then; < less than ZN-stained sputum result at diagnosis: II 1–9 acid fast bacilli per 100 oil immersion fields
10–99 acid fast bacilli per 100 oil immersion fields
1–10 acid fast bacilli per 1 oil immersion field
> 10 acid fast bacilli per one oil immersion field.
Written informed consent was obtained and the study was performed according to the Helsinki declaration of 1975, as revised in 1983 and International Conference on Harmonisation of Technical Requirements for Registration of Pharmaceuticals for Human Use (ICH) guidelines. Permission for the study was granted by the local health committees representing the people in the study communities, the Ethics Committee, Faculty of Health Sciences, Stellenbosch University and the Head of Health, City of Cape Town.
Full blood count analysis
Venous blood (obtained in EDTA tubes) was analysed for automated routine FBC using a Bayer Advia 120 haematology analyser. Absolute cell counts were expressed as 109 cells per litre and to obtain a percentage value the following calculation was used: absolute cell count/total white blood cell count × 100%.
The FBC analysis included information on red blood cells, platelets, total and differential WBC which included neutrophils, eosinophils, basophils, monocytes and lymphocytes. We analysed all data from the WBC populations.
Enzyme-linked immunosorbent assays (ELISAs)
The average value of duplicate wells of serum samples was used in statistical analyses. Measurement of IFN-γ, sIL-2Rα and sTNF-R1 was carried out using commercially available paired antibodies (PharMingen, San Diego, California, USA and R&D systems, Abingdon, UK) according to standard methods [31]. Recombinant cytokines were used for the standard curves which ranged from 2000 pg/ml to 31 pg/ml. The enzyme and substrate used for the colorimetric reaction were streptavidin–horseradish–peroxidase conjugate and 3′,3′,5′5-tetramethylbenzidine (TMB) (Sigma Aldrich, Johannesburg, South Africa). Plates were read at 450 nm wavelength; the cubic curve fit and extrapolation options were used.
Nitrotyrosine, sTNF-R2 and granzyme B were measured in the sera using commercially available kits (Hycult Biotechnology, Uder, the Netherlands and CLB, Red Cross Blood Transfusion Centre, Amsterdam, the Netherlands). The standard curves ranged from 1500 nM to 2·1 nM (nitrotyrosine), 1000 pg/ml to 15·6 pg/ml (sTNF-R2) and 960 units/ml to 1·3 units/ml (granzyme B).
The serum was diluted 1:5 for the IL-2Rα, sTNF-R1 and granzyme B ELISAs. Nitrotyrosine and sTNFR2 were tested in serum diluted 1:10 according to the manufacturer's recommendations. The serum was undiluted for IFN-γ. Despite 1:5 and 1:10 dilution of the serum, the majority of patients had concentrations of sTNF-R1 and sTNF-R2 above the upper detection limit of the ELISA at both time-points, thus statistical calculations for these biomarkers were based upon extrapolated data. For all serum markers, any values that fell below the lowest point of detection of the assay were changed to zero. Levels of IFN-γ and nitrotyrosine were below the lowest limit of detection for the ELISA assay in all controls and in all subjects of both groups of TB patients at diagnosis and week 4 (results not shown).
Statistical analysis
Statistical analysis was carried out using statistica versions 6 and 7 (StatSoft, Inc. 2003, http://www.statsoft.com). The Mann–Whitney U-test was used for comparison between two treatment response groups at a single time-point, the Kruskal–Wallis anova test for comparisons between more than two groups and the Wilcoxon matched pairs test for data at different time-points [32]. A final value of P < 0·05 was considered significant.
The ability of the biomarkers to predict sputum smear status at week 8 was evaluated by fitting forward stepwise and best subsets discriminant analysis [33] models to the diagnosis and week 4 data sets, followed by training-test set and leave-one-out cross-validation, respectively. A restriction of a maximum of five variables was placed on the combinations for best subsets analysis. Due to the small sample size, results were verified by comprehensive bootstrap (resampling) analysis [34]. Five hundred random samples (with replacement) were drawn from the data, thus giving 500 pseudo-samples that are similar to but not the same as the original sample. A best subsets discriminant analysis was performed on each of the bootstrap samples. Persistently informative variables are characterized by a high number of inclusions in the 500 best subsets models. In order to compare the results of the discriminant analyses with a completely different method, the classification technique support vector machines (SVM) [33] was also applied to the data. A best subsets method was employed to determine optimal sets of predictor variables, and three kernel functions (linear, polynomial and radial) were used in the SVM analysis. The SVM results were generated using the r statistical programming language.
Results
Serum biomarkers
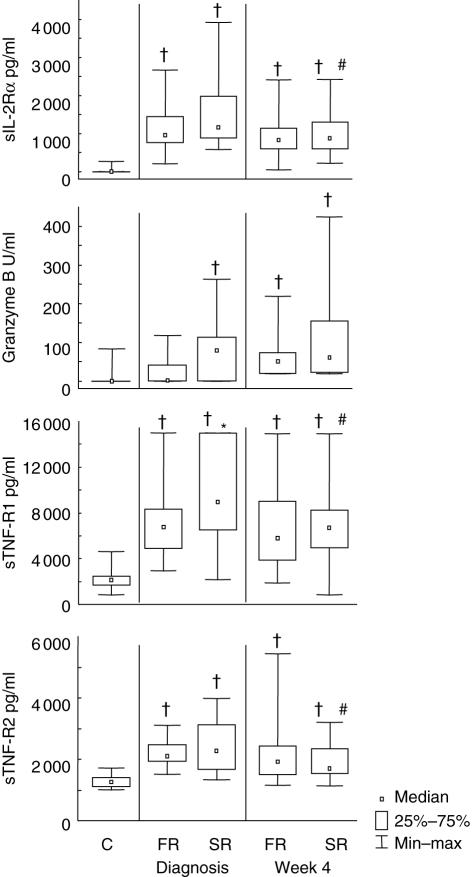
Concentrations of serum biomarkers were measured at diagnosis and after 4 weeks of treatment (Fig. 2). At diagnosis, sTNF-R1 was significantly higher in slow compared to fast responders (P < 0·01). After 4 weeks of treatment there were significant (P < 0·05) decreases in levels of sIL-2Rα, sTNF-R1 and sTNF-R2 in slow treatment responders. None of the biomarkers showed differences between the two response groups at week 4. Granzyme B was not detectable (< 1·3 units/ml) in nine fast responders and six slow responders at diagnosis, or five fast responders and four slow responders at week 4. Community controls had significantly lower levels of sIL-2Rα, sTNF-R1 and sTNF-R2 than both the responder groups at both time-points. Granzyme B levels were also significantly lower in controls than in slow responders at diagnosis and lower than both of the responder groups at week 4.
Fig. 2.
Serum biomarker concentrations in community controls and in tuberculosis (TB) patients with fast and slow responses to TB treatment. The median serum levels, 25th and 75th percentiles and minimum and maximum values of four biomarkers as determined by enzyme-linked immunosorbent assay (ELISA) at one time-point in controls and at diagnosis and after 4 weeks of TB treatment in fast and slow responders to treatment are shown. Soluble interleukin-2 receptor alpha (sIL-2Rα) (a), granzyme B (b), soluble tumour necrosis factor alpha receptor 1 (sTNF-R1) (c) and sTNF-R2 (d) are represented. C = community controls; FR = fast responder TB patients [negative Ziehl–Nielsen (ZN)-stained sputum smear after 8 weeks of therapy]; SR = slow responder TB patients. †Statistically significant differences between patient groups and controls; *statistically significant differences between FR and SR groups at a single time-point; #significant differences between diagnosis and the 4-week time-point within a treatment response group. The data for interferon (IFN)-γ and nitrotyrosine are not shown, as the mean values were all below the lower limit of detection for the assay at all the time-points.
Peripheral white blood cell counts
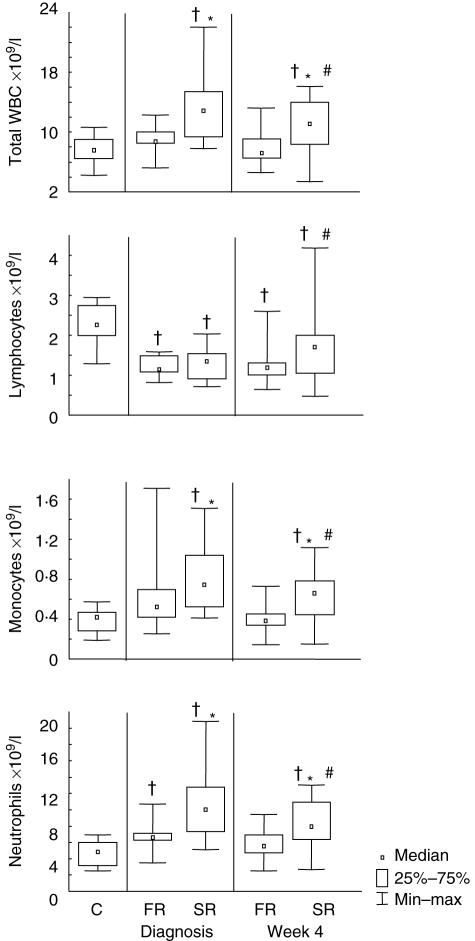
Peripheral white blood cell counts were carried out at diagnosis and 4 weeks after initiation of treatment (Fig. 3). One fast responder had no cell count data available at diagnosis. Total WBC counts, absolute monocyte counts and absolute neutrophil counts were significantly higher in slow responders than fast responders at diagnosis (P < 0·01, P = 0·02, P < 0·01) and at week 4 (P < 0·01 × 3). Absolute lymphocyte counts did not differ significantly between the two response groups at either time-point. There were significant differences in all cell count types between diagnosis and week 4 in the slow response group. Total WBC counts and absolute monocyte and neutrophil numbers decreased over time (P = 0·02, P = 0·03, P < 0·01), whereas absolute lymphocyte counts increased (P = 0·01). In the fast response group there were no significant differences between any of the cell counts between the two time-points. Lymphocyte counts were lower in community controls than in either of the response groups at both time-points. Community controls had lower counts of total WBC, monocytes and neutrophils than slow responders at both time-points and lower neutrophil counts than fast responders at diagnosis.
Fig. 3.
Peripheral blood white cell counts in community controls and in TB patients with fast and slow responses to TB treatment. Median cell counts, 25th and 75th percentiles and minimum and maximum values of absolute numbers of total white blood cells (WBC) (a), lymphocytes (b), monocytes (c) and neutrophils (d) are shown at one time-point for community controls and at diagnosis and 4 weeks after start of tuberculosis (TB) treatment in fast and slow responders as determined by week 8 sputum smear status. C = community controls; FR = fast responder TB patients [negative Ziehl–Nielsen (ZN)-stained sputum smear after 8 weeks of therapy]; SR = slow responder TB patients. †Statistically significant differences between patient groups and controls; *statistically significant difference between FR and SR groups at a single time-point; #significant differences between diagnosis and the 4-week time-point within a treatment response group.
Forward stepwise discriminant analysis at diagnosis
When forward stepwise discriminant analysis was performed on the diagnosis data set, concentrations of granzyme B and total WBC count were selected as the most informative variables to include in a predictive model for distinction between fast and slow responders to treatment. Using this model, 16 of 17 patients were classified correctly as fast responders (94% correct predictions) and 12 of 18 patients were classified correctly as slow responders (67% correct predictions). One individual from the fast response group was not included in the analysis because peripheral blood count data was not available at the diagnosis time-point. The classification matrix is shown in Table 2a.
Table 2.
Percentage correct predictions of fast and slow treatment responders using forward stepwise discriminant analysis at (a) diagnosis and (b) after 4 weeks of treatment.
| Predicted FR | Predicted SR | % correct predictions* | |
|---|---|---|---|
| (a) All patients | |||
| Observed FR (n = 17) | 16 | 1 | 94 |
| Observed SR (n = 18) | 6 | 12 | 67 |
| Total | 22 | 13 | |
| (b) All patients | Predicted FR | Predicted SR | % correct predictions† |
| Observed FR (n = 18) | 15 | 3 | 83 |
| Observed SR (n = 18) | 5 | 13 | 72 |
| Total | 20 | 16 | |
FR: fast responder; SR: slow responder.
Variables chosen for diagnosis model = Granzyme B and total WBC count.
Variable chosen for week 4 model = absolute monocyte number.
In order to cross-validate the diagnosis model, a training and test set approach was then used. The 17 members of the fast response group were sorted in patient ID order and labelled alternately as either training subjects (n = 8) or test subjects (n = 9). The same process was carried out for the 18 slow responders. Discriminant analysis of the training set resulted in a model which was again based on total WBC count and granzyme B. Application of the model to the training data set resulted in the correct classification of seven of eight fast responders (87% correct predictions) and six of nine slow responders (67% correct predictions). Cross-validation of the model using the test set resulted in the correct classification of seven of nine fast and slow responders (78% correct predictions).
At diagnosis, granzyme B was chosen as one of the two most informative variables for inclusion in a forward stepwise predictive model, yet levels of this cytokine were below the detection limit of the ELISA assay in the serum of 15 of 36 patients. In addition there were no significant differences in concentration of granzyme B between the two response groups at either time-point. For these reasons we repeated discriminant analysis on the diagnosis data using exactly the same method as described above, except that granzyme B was not included as a predictor variable when the analysis was designed. The models constructed for both the complete data set and the training/test set approach incorporated only total WBC count as a predictor variable. Prior to cross-validation, 14 of 17 patients were classified correctly as fast responders (82% correct predictions) and 12 of 18 patients were correctly classified as slow responders (67% correct predictions). The training and test set method of cross-validation was then applied. Seven of eight (87%) fast responders and seven of nine (78%) slow responders were classified correctly in the training set, and six of nine (67%) fast and slow responders were classified correctly in the test set.
Forward stepwise discriminant analysis at week 4
When forward stepwise analysis was carried out on the week 4 data set, absolute monocyte number was selected as the most informative variable for inclusion in a predictive model distinguishing between fast and slow responders. Using this model, 15 of 18 patients were classified correctly as fast responders (83% correct predictions) and 13 of 18 patients were classified correctly as slow responders (72% correct predictions). The classification matrix is shown in Table 2b.
The training and test set method was again employed as described for the diagnosis data for cross-validation of the week 4 model. Discriminant analysis (selecting for cross-validation) of the training data set resulted in a model which was also based on absolute monocyte number, and resulted in the correct classification of eight of nine fast responders (89% correct predictions) and seven of nine slow responders (78% correct predictions). When the model was cross-validated with the test set data, seven of nine fast responders (78% correct predictions) and six of nine slow responders (67% correct predictions) were predicted correctly.
Best subset discriminant analysis
At both diagnosis and week 4 the predictor variables chosen by the best subsets method were total WBC count, granzyme B and sTNF-R1. The percentage of correct predictions after leave-one-out cross-validation for fast and slow responders were 88% and 67% at diagnosis and 78% and 83% at week 4.
Resampling analysis
Using the resampling method, at diagnosis granzyme B, WBC count and absolute neutrophil numbers were thevariables selected most frequently for inclusion in a predictive model, each being chosen in over half of the 500 best subsets models. A discriminant analysis model using only the above three variables yielded a 94% (16 of 17) correct classification of fast responders, and 67% (12 of 18) correct classification of slow responders following leave-one-out cross-validation. Resampling analysis of the week 4 data showed only granzyme B standing out as the most popular variable for inclusion in a predictive model, being chosen in approximately 300 of the 500 best subsets models. A discriminant analysis model which used only granzyme B gave low predictive accuracy for both response groups at week 4. When the discriminant analysis model also included absolute monocyte number and sIL-R2α, the next two most commonly occurring variables in the resampling analysis (being chosen in 200–250 models), the predictive accuracy for fast responders was 78% (14 of 18) and 67% (12 of 18) for slow responders at week 4 after leave-one-out cross-validation.
Support vector machines
Results from the SVM analysis corresponded to a large degree with that of the discriminant analyses. At diagnosis, granzyme B and absolute neutrophil numbers were chosen by all three kernel functions (linear, polynomial and radial). The remaining six variables were chosen by one or no kernel functions. At week 4, only granzyme B was chosen by all three kernel functions and sTNF-R1 and total WBC count were each selected by two kernel functions. The remaining five variables were chosen by one or no kernel functions. The leave-one-out cross-validation of the SVM analysis generally gave more than 80% correct classification for both fast and slow responders.
Discussion
The potential for cytokines measured in sputum and serum to act as early markers for response to anti-TB therapy has been demonstrated previously [35]. In the present study patients were identified as fast or slow responders to standard intensive phase TB treatment. The sustained positive smear status at 8 weeks in the slow response group is not likely to be due to an initially higher mycobacterial load, as the two patient groups were matched closely for sputum smear grade at diagnosis, but may be associated with differences in the immune response in slow compared to fast responders. The two groups were also matched carefully for extent of disease in order to ensure as far as possible that there was not a greater degree of clinical illness in one group than the other at diagnosis. Based on this matching it was hypothesized that the two groups may have similar pretreatment immunological profiles at diagnosis and that differences between fast and slow responders might become apparent only after a period of therapy. There were, however, significant differences observed in levels of sTNF-R1, total WBC count and absolute monocyte and neutrophil numbers prior to initiation of treatment, indicating that these groups were immunologically different at the time of diagnosis. These observed differences may be associated with the subsequent response to the intensive phase of TB treatment. Soluble TNF-R1 was the only factor measured in serum for which we observed higher concentrations at diagnosis in the slow response group. sTNF-R1 binds to TNF-α and inhibits its activity [36]. Our results support previous findings that serum concentrations of sTNF-R1 are increased in patients with active TB [16,37] and extend this observation to suggest that relatively higher concentrations of sTNF-R1 correlate with a slow response to chemotherapy. Concentrations of both TNF receptors had decreased significantly by week 4 in the slow response group, which may reflect dampening of the immune response as treatment begins to take effect. Concentrations of sIL-2Rα also showed significant decreases after 4 weeks of treatment in the slow response group. This receptor has been shown to be a sensitive and specific marker for monitoring disease activity in pulmonary TB [10].
Rapid consumption of IFN-γ and compartmentalized production of nitrotyrosine could explain the paucity of these markers in the serum of our patients. As IFN-γ is released by T lymphocytes following antigenic stimulation, analysis of supernatants after antigen-stimulated culture of whole blood [38] or peripheral blood mononuclear cells [39] may be a more suitable means of investigating this cytokine as a potential surrogate marker of treatment response in the future.
Higher absolute monocyte and neutrophil numbers as well as total WBC count were observed in the slow responders at diagnosis and week 4. Lower cell numbers at either time-point are therefore associated with becoming smear negative by week 8. An increased WBC count (normal range 3·11–13·8 × 109/l) would suggest acute infection and inflammation. Neutrophils are the most abundant granulocyte and one of the main defences against bacteria. High numbers of these cells in the slow responder group may be suggestive of an ongoing innate response against a background of an inadequate adaptive immune response against mycobacteria. This suggests that the quality of the cellular immune response is a more important determinant of outcome than the number of circulating cells, as has been shown in studies of HIV infection [40,41].
The concept of biomarkers is not novel [42] and they have proved to be beneficial in other clinical disciplines [43,44]. In the present study discriminant analysis of the cell count and serum data consistently gave 67% and between 83% and 94% predictive accuracy in classification of slow and fast responders, respectively, at the diagnosis time-point. Forward stepwise, best subsets and resampling analyses all selected total white blood cell count and granzyme B for inclusion in models predicting slow or fast response at this time-point. Support vector machines (SVM) analysis also suggested that granzyme B data were important for differentiating between response types at diagnosis. The majority of the models based on the week 4 data also included granzyme B. It was surprising that granzyme B was prioritized in predictive models at both time-points, as this serum factor was undetectable in the serum of 15 of the 36 patients and showed no significant difference between the slow response and fast response groups at either diagnosis or week 4. Excluding granzyme B from the forward stepwise analysis decreased the predictive ability of the diagnosis data; however, the effect was not strong, indicating that the total white blood cell count was the main contributor to the model. Based on the resampling result, including only granzyme B in discriminant analysis of the week 4 data resulted in a low percentage of correct predictions of slow response; however, adding absolute monocyte number and sIL-2Rα boosted the predictive accuracy of the model to 67%. These findings suggest that either granzyme B data need to be used in conjunction with other biomarker data in order to be useful in discriminant analysis, or that granzyme B needs to be replaced with an immune marker which is more easily detectable in serum and therefore more informative for use in future discriminant analysis. All the variables selected for inclusion in the various models presented here could play a role in the classification of patients into fast and slow response categories.
It was encouraging that discriminant analysis was able to differentiate between patients who had been categorized according to smear status, as not all diagnostic laboratories that carry out microscopy for TB surveillance purposes also have access to reliable culture facilities. Additionally, the models relied on only one or two predictor variables that can be measured with basic laboratory equipment. In order for predictive models to be applicable in the field it would be important that data analysis required a small number of predictor variables that are technically straightforward to obtain. For example, the full blood count test can be performed in most developing countries, where the burden of TB is highest.
We propose that discriminant analysis of simple immunological data collected at diagnosis could be used to predict treatment response during the intensive phase. This would improve monitoring of patients found to be at risk of poor response to treatment and could expedite clinical trials of new drugs by allowing stratification of patients into distinct response phenotype groups to enhance statistical power and improve evaluation of the drug. The statistical models created from this study are based on a small sample size and need to be validated with a larger group of patients. The use of technologies such as luminescent multiplex cytokine systems [45], proteomic analysis [46] of sera and culture supernatants and gene expression profiling [47,48] could identify alternative or additional surrogate markers of treatment response which may improve the frequency of correct predictions reported here. In conclusion, the results from this proof-of-concept study indicate that an approach that incorporates combinations of different immune markers holds promise for prediction of early treatment outcome. This approach should act as a guide for future expansive searches for the optimal set of surrogate markers.
Acknowledgments
The authors would like to thank the Desmond Tutu TB Centre (Stellenbosch University), Wena Moelich, Makkie van de Merwe, the data team and the nursing sisters of the Desmond Tutu TB Centre for the recruitment of TB patients, the communities for their support and willingness to participate in the research, the clinic staff at Ravensmead, Uitsig, Adriaanse and Elsiesrivier and the Department of Health and the City of Cape Town for their support. We would like to acknowledge Ilse Steyn and Siobhan Harnett for technical assistance in the laboratory, Robert Gie for interpretation of the chest radiographs, the Department of Haematology at Tygerberg Hospital for conducting full blood count analysis and Pieter Uys for statistical input. This work was supported by the GlaxoSmithKline Action TB Initiative and the European and Developing Countries Clinical Trials Partnership.
References
- 1.Abramo C, Meijgaarden KE, Garcia D, et al. Monokine induced by interferon gamma and IFN-gamma response to a fusion protein of Mycobacterium tuberculosis ESAT-6 and CFP-10 in Brazilian tuberculosis patients. Microbes Infect. 2006;8:45–51. doi: 10.1016/j.micinf.2005.05.019. [DOI] [PubMed] [Google Scholar]
- 2.Tobias HJ, Schafer MP, Pitesky M, et al. Bioaerosol mass spectrometry for rapid detection of individual airborne Mycobacterium tuberculosis H37Ra particles. Appl Environ Microbiol. 2005;71:6086–95. doi: 10.1128/AEM.71.10.6086-6095.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benator D, Bhattacharya M, Bozeman L, et al. Rifapentine and isoniazid once a week versus rifampicin and isoniazid twice a week for treatment of drug-susceptible pulmonary tuberculosis in HIV-negative patients: a randomised clinical trial. Lancet. 2002;360:528–34. doi: 10.1016/s0140-6736(02)09742-8. [DOI] [PubMed] [Google Scholar]
- 4.Enarson DE, Rieder HL, Arnadottir T, Trebucq A. France: Fifth International Union Against Tuberculosis and Lung Disease; Management of tuberculosis: a guide for low income countries. [pamphlet]. [Google Scholar]
- 5.The Provincial Administration of The Western Cape Metropole Region and City Health. City of Cape Town/Metropole Region TB Control Programme Progress Report 1997–2002. Available at: http://www.capegateway.gov.za/Text/2003/12/cct_tb_report_1997to2002.pdf. 2002. (Last accessed 2002)
- 6.Kaufmann SH, Cole ST, Mizrahi V, Rubin E, Nathan C. Mycobacterium tuberculosis and the host response. J Exp Med. 2005;201:1693–7. doi: 10.1084/jem.20050842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murray HW. Gamma interferon, cytokine-induced macrophage activation, and antimicrobial host defense. In vitro, in animal models, and in humans. Diagn Microbiol Infect Dis. 1990;13:411–21. doi: 10.1016/0732-8893(90)90012-k. [DOI] [PubMed] [Google Scholar]
- 8.Serbina NV, Lazarevic V, Flynn JL. CD4(+) T cells are required for the development of cytotoxic CD8(+) T cells during Mycobacterium tuberculosis infection. J Immunol. 2001;167:6991–7000. doi: 10.4049/jimmunol.167.12.6991. [DOI] [PubMed] [Google Scholar]
- 9.Newport MJ, Huxley CM, Huston S, et al. A mutation in the interferon-gamma-receptor gene and susceptibility to mycobacterial infection. N Engl J Med. 1996;335:1941–9. doi: 10.1056/NEJM199612263352602. [DOI] [PubMed] [Google Scholar]
- 10.Rajalingam R, Mehra NK, Pande JN, Jain RC, Singla R. Correlation of serum interleukin-2 receptor-alpha levels with clinical manifestations in pulmonary tuberculosis. Tuber Lung Dis. 1996;77:374–9. doi: 10.1016/s0962-8479(96)90105-7. [DOI] [PubMed] [Google Scholar]
- 11.Shikama Y, Kobayashi K, Kasahara K, et al. Granuloma formation by artificial microparticles in vitro. Macrophages and monokines play a critical role in granuloma formation. Am J Pathol. 1989;134:1189–99. [PMC free article] [PubMed] [Google Scholar]
- 12.Bozkurt B, Kribbs SB, Clubb FJ, Jr, et al. Pathophysiologically relevant concentrations of tumor necrosis factor-alpha promote progressive left ventricular dysfunction and remodeling in rats. Circulation. 1998;97:1382–91. doi: 10.1161/01.cir.97.14.1382. [DOI] [PubMed] [Google Scholar]
- 13.Peetre C, Thysell H, Grubb A, Olsson I. A tumor necrosis factor binding protein is present in human biological fluids. Eur J Haematol. 1988;41:414–19. doi: 10.1111/j.1600-0609.1988.tb00220.x. [DOI] [PubMed] [Google Scholar]
- 14.Cope AP, Aderka D, Doherty M, et al. Increased levels of soluble tumor necrosis factor receptors in the sera and synovial fluid of patients with rheumatic diseases. Arthritis Rheum. 1992;35:1160–9. doi: 10.1002/art.1780351008. [DOI] [PubMed] [Google Scholar]
- 15.Tartaglia LA, Goeddel DV. Two TNF receptors. Immunol Today. 1992;13:151–3. doi: 10.1016/0167-5699(92)90116-O. [DOI] [PubMed] [Google Scholar]
- 16.Lawn SD, Rudolph D, Wiktor S, Coulibaly D, Ackah A, Lal RB. Tuberculosis (TB) and HIV infection are independently associated with elevated serum concentrations of tumour necrosis factor receptor type 1 and beta2-microglobulin, respectively. Clin Exp Immunol. 2000;122:79–84. doi: 10.1046/j.1365-2249.2000.01341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bartholdy C, Nansen A, Marker O, Thomsen AR. Soluble tumour necrosis factor (TNF)-receptor levels in serum as markers of anti-viral host reactivity. Clin Exp Immunol. 1999;116:299–306. doi: 10.1046/j.1365-2249.1999.00903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grotzke JE, Lewinsohn DM. Role of CD8+ T lymphocytes in control of Mycobacterium tuberculosis infection. Microbes Infect. 2005;7:776–88. doi: 10.1016/j.micinf.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 19.Podack ER, Young JD, Cohn ZA. Isolation and biochemical and functional characterization of perforin 1 from cytolytic T-cell granules. Proc Natl Acad Sci USA. 1985;82:8629–33. doi: 10.1073/pnas.82.24.8629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hudig D, Ewoldt GR, Woodard SL. Proteases and lymphocyte cytotoxic killing mechanisms. Curr Opin Immunol. 1993;5:90–6. doi: 10.1016/0952-7915(93)90086-8. [DOI] [PubMed] [Google Scholar]
- 21.Dennert G, Podack ER. Cytolysis by H-2-specific T killer cells. Assembly of tubular complexes on target membranes. J Exp Med. 1983;157:1483–95. doi: 10.1084/jem.157.5.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang CH, Lin HC, Liu CY, et al. Upregulation of inducible nitric oxide synthase and cytokine secretion in peripheral blood monocytes from pulmonary tuberculosis patients. Int J Tuberc Lung Dis. 2001;5:283–91. [PubMed] [Google Scholar]
- 23.Tunctan B, Okur H, Calisir CH, et al. Comparison of nitric oxide production by monocyte/macrophages in healthy subjects and patients with active pulmonary tuberculosis. Pharmacol Res. 1998;37:219–26. doi: 10.1006/phrs.1997.0284. [DOI] [PubMed] [Google Scholar]
- 24.Nicholson S, Bonecini-Almeida MG, Lapa e Silva JR, et al. Inducible nitric oxide synthase in pulmonary alveolar macrophages from patients with tuberculosis. J Exp Med. 1996;183:2293–302. doi: 10.1084/jem.183.5.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonecini-Almeida MG, Chitale S, Boutsikakis I, et al. Induction of in vitro human macrophage anti-Mycobacterium tuberculosis activity: requirement for IFN-gamma and primed lymphocytes. J Immunol. 1998;160:4490–9. [PubMed] [Google Scholar]
- 26.Kamijo R, Gerecitano J, Shapiro D, et al. Generation of nitric oxide and clearance of interferon-gamma after BCG infection are impaired in mice that lack the interferon-gamma receptor. J Inflamm. 1995;46:23–31. [PubMed] [Google Scholar]
- 27.Radi R. Nitric oxide, oxidants, and protein tyrosine nitration. Proc Natl Acad Sci USA. 2004;101:4003–8. doi: 10.1073/pnas.0307446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brahmbhatt S, Carroll N, Beyers N, van Helden P, Lukey P, Walzl G. Surrogate markers for tuberculosis. SA Resp J. 2003;9:105. [abstract]. [Google Scholar]
- 29.Walzl G, Beyers N, van Helden P. TB: a partnership for the benefit of research and community. Trans R Soc Trop Med Hyg. 2005;99(Suppl. 1):S15–19. doi: 10.1016/j.trstmh.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 30.Den Boon S, van Lill SW, Borgdorff MW, et al. Association between smoking and tuberculosis infection: a population survey in a high tuberculosis incidence area. Thorax. 2005;60:555–7. doi: 10.1136/thx.2004.030924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crowther JR. Stages in ELISAThe ELISA guidebook. New Jersey: Humana Press; 2001. [Google Scholar]
- 32.Daniel WW. Applied nonparametric statistics. Boston: Houghton Mifflin Company; 1978. [Google Scholar]
- 33.Hastie T, Tibrishani R, Friedman J. The elements of statistical learning. New York: Springer Series In Statistics; 2001. [Google Scholar]
- 34.Efron B, Tibrishani R. An introduction to the bootstrap. New York: Chapman & Hall/CRC; 1993. [Google Scholar]
- 35.Ribeiro-Rodrigues R, Resende CT, Johnson JL, et al. Sputum cytokine levels in patients with pulmonary tuberculosis as early markers of mycobacterial clearance. Clin Diagn Lab Immunol. 2002;9:818–23. doi: 10.1128/CDLI.9.4.818-823.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aderka D, Engelmann H, Maor Y, Brakebusch C, Wallach D. Stabilization of the bioactivity of tumor necrosis factor by its soluble receptors. J Exp Med. 1992;175:323–9. doi: 10.1084/jem.175.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Juffermans NP, Verbon A, van Deventer SJ, van Deutekom H, Speelman P, van der Poll T. Tumor necrosis factor and interleukin-1 inhibitors as markers of disease activity of tuberculosis. Am J Respir Crit Care Med. 1998;157:1328–31. doi: 10.1164/ajrccm.157.4.9709126. [DOI] [PubMed] [Google Scholar]
- 38.Weir RE, Brennan PJ, Butlin CR, Dockrell HM. Use of a whole blood assay to evaluate in vitro T cell responses to new leprosy skin test antigens in leprosy patients and healthy subjects. Clin Exp Immunol. 1999;116:263–9. doi: 10.1046/j.1365-2249.1999.00892.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mustafa AS, Amoudy HA, Wiker HG, et al. Comparison of antigen-specific T-cell responses of tuberculosis patients using complex or single antigens of Mycobacterium tuberculosis. Scand J Immunol. 1998;48:535–43. doi: 10.1046/j.1365-3083.1998.00419.x. [DOI] [PubMed] [Google Scholar]
- 40.Alimonti JB, Koesters SA, Kimani J, et al. CD4+ T cell responses in HIV-exposed seronegative women are qualitatively distinct from those in HIV-infected women. J Infect Dis. 2005;191:20–4. doi: 10.1086/425998. [DOI] [PubMed] [Google Scholar]
- 41.Migueles SA, Laborico AC, Shupert WL, et al. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat Immunol. 2002;3:1061–8. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- 42.Baker M. In biomarkers we trust? Nat Biotechnol. 2005;23:297–304. doi: 10.1038/nbt0305-297. [DOI] [PubMed] [Google Scholar]
- 43.Milunsky A, Nebiolo L. Maternal serum triple analyte screening and adverse pregnancy outcome. Fetal Diagn Ther. 1996;11:249–53. doi: 10.1159/000264310. [DOI] [PubMed] [Google Scholar]
- 44.Rosenberg AL. Recent innovations in intensive care unit risk-prediction models. Curr Opin Crit Care. 2002;8:321–30. doi: 10.1097/00075198-200208000-00009. [DOI] [PubMed] [Google Scholar]
- 45.de Jager W, te Velhuis H, Prakken BJ, Kuis W, Rijkers GT. Simultaneous detection of 15 human cytokines in a single sample of stimulated peripheral blood mononuclear cells. Clin Diagn Lab Immunol. 2003;10:133–9. doi: 10.1128/CDLI.10.1.133-139.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang CG, Chromy BA, McCutchen-Maloney SL. Host–pathogen interactions: a proteomic view. Expert Rev Proteomics. 2005;2:187–202. doi: 10.1586/14789450.2.2.187. [DOI] [PubMed] [Google Scholar]
- 47.Hakonarson H, Bjornsdottir US, Halapi E, et al. Profiling of genes expressed in peripheral blood mononuclear cells predicts glucocorticoid sensitivity in asthma patients. Proc Natl Acad Sci USA. 2005;102:14789–94. doi: 10.1073/pnas.0409904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodgers A, Whitmore KM, Walker KB. Potential correlates of BCG induced protection against tuberculosis detected in a mouse aerosol model using gene expression profiling. Tuberculosis (Edinb) 2006;86:255–62. doi: 10.1016/j.tube.2006.01.020. [DOI] [PubMed] [Google Scholar]