Abstract
For centuries, physicians have attempted to use the urine for noninvasive assessment of disease. Today, urinalysis, in particular the measurement of proteinuria, underpins the routine assessment of patients with renal disease. More sophisticated methods for assessing specific urinary protein losses have emerged; however, albumin is still the principal urinary protein measured. Changes in the pattern of urinary protein excretion are not necessarily restricted to nephrourological disease; for instance, the appearance of β-human chorionic gonadotropin in the urine of pregnant women is the basis for all commercially available pregnancy kits. Similarly, microalbuminuria is a clinically important marker not only of early diabetic nephropathy but also of concomitant cardiovascular disease. With the emergence of newer technologies, in particular mass spectrometry, it has become possible to study urinary protein excretion in even more detail. A variety of techniques have been used both to characterize the normal complement of urinary proteins and also to identify proteins and peptides that may facilitate earlier detection of disease, improve assessment of prognosis and allow closer monitoring of response to therapy. Such proteomics-based approaches hold great promise as the basis for new diagnostic tests and as the means to better understand disease pathogenesis. In this review, we summarize the currently available methods for urinary protein analysis and describe the newer approaches being taken to identify urinary biomarkers.
Proteomics is the study of protein expression in a tissue or biological fluid. Comparison of protein patterns in biological fluids between healthy individuals and patients with disease is increasingly being used both to discover biological markers of disease (biomarkers) and to identify biochemical processes important in disease pathogenesis. Although currently available tests for urine proteins measure either the total level of urine protein or the presence of a single protein species, emerging proteomic technologies allow simultaneous examination of the patterns of multiple urinary proteins and their correlation with individual diagnoses, response to treatment or prognosis. Analysis of the various protein constituents of urine may suggest novel, noninvasive diagnostic tests, therapeutic guidance, and prognostic information for patients and clinicians.
In this review, we describe the current practice of urine protein testing and the emerging technologies that are being used for analysis of the urinary proteome.
Background
Normally, the low-molecular-weight proteins and albumin that are filtered from plasma into the early tubular fluid are almost completely reabsorbed and catabolized in the proximal tubules. As a result, daily urinary protein excretion is less than 150 mg/day, of which about 10 mg is albumin. In patients with physiologic proteinuria, the proteins excreted include mucoproteins (mainly Tamm–Horsfall protein), blood-group proteins, albumin, immunoglobulins, mucopolysaccharides and very small amounts of hormones and enzymes. Historically, proteinuria of more than 150 mg/day was regarded as abnormal. However, it is now appreciated that early renal disease is often characterized by low-level albuminuria (between 30 and 300 mg/day).1 This condition is termed microalbuminuria because the concentration of albumin is below the detection limit of traditional assays. Protein or albumin excretion greater than 300 mg/day represents overt proteinuria or macroalbuminuria; at this level, the result of standard urine dipstick testing becomes positive.
Pathological proteinuria can be divided into 3 categories: glomerular proteinuria, tubular proteinuria and overload proteinuria.2 Glomerular proteinuria results from an increase in the permeability of the glomerular capillary wall to macromolecules (particularly albumin) and usually results from glomerular disease. Tubular proteinuria results from reduced reabsorption of proteins that are normally present in the glomerular filtrate or from excretion of proteins derived from injured tubular epithelial cells. It is usually caused by diseases of the tubulointerstitium. Overload proteinuria is due to an excess of low-molecular-weight proteins that are normally reabsorbed by the proximal tubules. These proteins are most often immunoglobulin light chains (in the plasma cell dyscrasias), although lysozyme (in myelomonocytic leukemia), myoglobin (in rhabdomyolysis) or hemoglobin (in intravascular hemolysis) may also be identified.
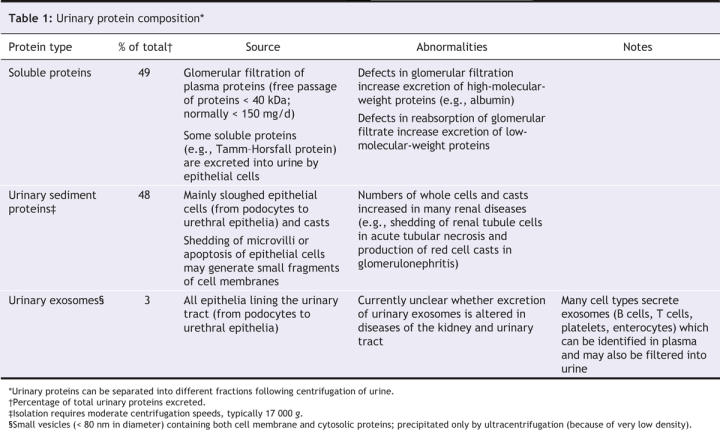
Under normal conditions, urinary proteins exist in different compartments that can be isolated by sequential centrifugation. The resulting fractions contain separate populations of proteins (Table 1).
Table 1
Urine protein analysis: the present
Urine protein testing usually involves a screening test to detect excess protein, a quantitative assay and finally, in certain circumstances, an assay to identify specific proteins.
Detection of proteinuria
Urine dipstick testing
The urine dipstick test is the basic screening test for proteinuria. With increasing concentrations of urinary protein, a dye indicator (tetrabromophenol blue) undergoes sequential colour changes from pale green to blue.3 The binding of tetrabromophenol blue to proteins is pH dependent: albumin binds at a pH between 5 and 7; other proteins bind only at a pH below 5 and with less affinity than albumin; and Bence-Jones protein does not bind at any pH. Since urinary pH is usually between 5 and 6, urine dipstick testing is essentially albumin specific.
The lower limit of sensitivity for urine dipstick testing is about 250 mg/L, and therefore this method cannot detect microalbuminuria.
Precipitation techniques
Precipitation techniques measure the turbidity that occurs when proteins are precipitated out of solution by sulfosalicylic acid, by trichloroacetic acid or by acetic acid and sodium acetate buffer in the presence of heat. Turbidimetric methods detect essentially all urinary proteins with a sensitivity as low as 2.5 mg/L. They are, however, prone to interference from a variety of exogenous compounds and are therefore rarely used in the clinical setting.4
Quantification of urine total protein
Twenty-four-hour protein excretion
The gold standard for quantification of proteinuria is analysis of a 24-hour urine collection. A number of methods are available for quantifying total urinary protein, but only the biuret method remains in widespread use.
Protein–creatinine ratio
Twenty-four-hour urine collection is time consuming and inconvenient for the patient and is thus subject to important collection errors. The protein–creatinine ratio, which is measured with a single spot urine sample and which corrects for variations in urinary concentration due to hydration, is therefore increasing in popularity.5 There is good correlation between the protein–creatinine ratio and results obtained from a 24-hour urine collection, and many renal guidelines now recommend measurement of urine protein–creatinine ratio over 24-hour urine collections.6–8 The ratio is reported as milligrams of protein per millimole of creatinine. Given that mean daily creatinine excretion is about 10 mmol, 24-hour protein excretion can be estimated by multiplying the ratio value by 10. Therefore, a protein–creatinine ratio of 30 mg/mmol is equivalent to proteinuria of about 300 mg/day, and 300 mg/mmol is equivalent to 3 g/day (i.e., nephrotic-range proteinuria).
Measurement of specific proteins
In particular circumstances, identification and quantification of specific proteins within the urine can be clinically useful.
Albumin and albumin–creatinine ratio
Microalbuminuria is a clinically important marker of early diabetic nephropathy and cardiovascular disease. Albuminuria is specifically measured with nephelometric, turbidimetric, radioimmunoassay and enzyme immunoassay techniques, although some investigators argue for the widespread adoption of techniques based on high-performance liquid chromatography to improve accuracy.9–12 The albumin–creatinine ratio is analogous to the protein–creatinine ratio. An albumin–creatinine ratio of 30 mg/mmol is equivalent to albuminuria of about 300 mg/day and represents the upper limit of the microalbuminuria range.
Immunoglobulin G and transferrin (protein selectivity)
Protein selectivity is traditionally determined by measuring the ratio of the clearances of immunoglobulin G and transferrin. A ratio of less than 0.1 indicates selective proteinuria, which in children with nephrosis implies the presence of minimal change disease and likely responsiveness to corticosteroids.13 However, protein selectivity is now rarely used in the clinical setting.
Proteinuria arising from tubular cell dysfunction
Although not part of routine practice, the identification in the urine of a variety of proteins caused by dysfunction of the tubular epithelial cells has been proposed as a method to diagnose tubulointerstitial disease and to stratify its severity. Proteinuria can arise from tubular cell injury by 2 mechanisms (described in more detail below): release of tubular epithelial cell proteins into the urine and failure to reabsorb filtered low-molecular-weight proteins.
Release of a wide variety of brush-border enzymes from the proximal tubules into the urine has been studied in various disease states; these enzymes include neutral endopeptidase, dipeptidyl aminopeptidase IV, α-glucosidase, trehalase, leucine aminopeptidase, alkaline phosphatase, γ-glutamyltransferase and alanine aminopeptidase.14–17 N-Acetyl-β-D-glucosaminidase, a hydrolytic enzyme present in the lysosomes of proximal tubular epithelial cells, has also been investigated.18 The excretion of all of these proteins is increased by various renal insults, including ischemia, acute interstitial nephritis and the presence of materials such as cisplatin, contrast media, heavy metals and aminoglycoside antibiotics. In addition, the excretion of tubular proteins may predict the outcome of proteinuric glomerular disease.19
A number of filtered proteins, including β2-microglobulin, retinol-binding protein and α1-microglobulin, have been used in the diagnosis and follow-up of tubulointerstitial diseases. Neutrophil gelatinase-associated lipocalin and cystatin C have been identified as markers of ischemia-associated acute tubular injury.20,21 These filtered and tubule-derived proteins are easily measured, but they offer no specificity in terms of the underlying renal insult, and their ability to predict early renal injury is uncertain. Their current clinical utility is therefore limited.
Immunoglobulin light chains
Henry Bence-Jones described the detection of urinary immunoglobulin light chains in 1848. He observed that light chains precipitate when urine is heated to between 40°C and 60°C, dissolve on further heating to 100°C and then reprecipitate on cooling back to 40°C to 60°C.22 Because false-positive results may occur in the presence of excess polyclonal light chains, this technique is no longer in clinical use.23 Urinary light chains are now identified either by urine protein electrophoresis followed by immunofixation or by an immunoassay-based free light chain assay.24 Immunofixation is capable of detecting a monoclonal protein concentration of 0.04 g/L.25
β-Human chorionic gonadotropin
Urine pregnancy tests use an immunochromatographic method to detect β-human chorionic gonadotropin. The hormone interacts with 2 monoclonal antibodies impregnated on the test strip, one of which is conjugated with gold. In the presence of the hormone, the 2 antibodies are approximated and a colour reaction develops. These tests can detect β-human chorionic gonadotropin to a sensitivity of 5 IU/L.26
Leukocyte esterase
Leukocyte esterase testing is readily available as a dipstick test specific for the presence of leukocytes in the urine.27 Neutrophil esterase hydrolyses the dipstick substrate (3-hydroxy-5-phenyl-pyrrole esterified with an indoxyl carbonic acid ester) into 3-hydroxy-5-phenyl-pyrrole and indoxyl, which react with a diazonium salt to yield an azo dye.
Urine biomarker discovery: the future
There are 2 principal approaches to the discovery of new urine biomarkers. The first involves the study of candidate biomarkers (usually tubular proteins, cytokines, growth factors and inflammatory mediators) in specific diseases where laboratory studies have suggested a pathological link. The second involves biomarker discovery studies in which urine is screened for disease-associated proteins using an array of technologies, predominantly based on mass spectrometry.
Techniques for screening the urinary proteome
Currently, the most popular method of identifying new urine biomarkers involves centrifugation of the urine sample to collect either the soluble urine proteins or the urinary exosomes (Table 1), followed by 1 or 2 protein separation steps before visualization and finally identification of potential biomarkers, usually by mass spectrometry28 (Figure 1). As with standard methods of urinalysis, a number of factors strongly influence the interpretation of such studies; these include the timing of collection of the urine sample, the storage conditions used and the need for normalization of urinary biomarker excretion (correction for variations in concentration of urine due to hydration) between different individuals.29 The Human Kidney and Urine Proteome Project (www.hkupp.org/), under the auspices of the World Human Proteome Organisation (www.hupo.org/), is currently addressing these issues and is publishing standardized methods for urinary proteomic studies.
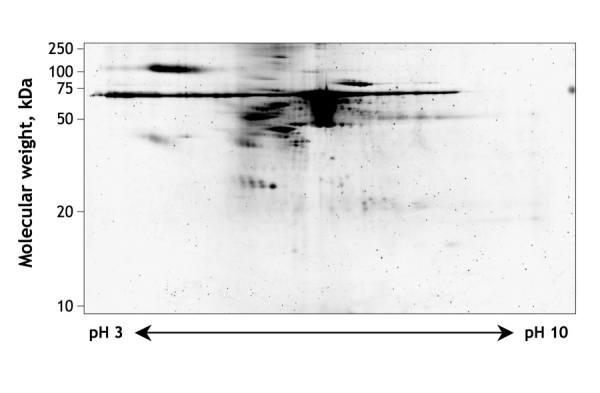
Figure 1: Two-dimensional gel electrophoresis of normal human urine. Proteins are first separated by isoelectric focusing, which separates on the basis of isoelectric point (horizontal separation). This is followed by sodium dodecyl sulfate — polyacrylamide gel electrophoresis, which separates on the basis of molecular weight (vertical separation). Individual spots of interest can be cut from the gel and identified by mass spectrometry. (Gel courtesy of Jan Novak, G. Robinson, H. Kim and Bruce A. Julian, University of Alabama at Birmingham, Birmingham, Ala.).
Until recently, 2-dimensional gel electrophoresis was the most widely used method for separating and visualizing urinary proteins (Box 1).30 Urinary biomarkers can be identified by comparing gels from patients and healthy subjects, and individual biomarkers can be identified by excising protein spots from the gel and subjecting them to amino acid sequencing. However, this technique is time consuming and difficult to integrate with other proteomic techniques, and it has a limited resolution capacity, particularly for small proteins and peptides. These limitations have led to increased use of techniques based on mass spectrometry (Box 1).
Box 1.

Standard mass spectrometry techniques usually generate the spectra of the proteins and peptides within a fractionated urine sample, although they do not in themselves allow identification of potential protein biomarkers (Figure 2). The proteins can be identified with prior enzymatic digestion of individual protein spots or tandem mass spectrometry, which allows specific proteins from an initial spectrum to be selected, isolated and fragmented, the amino acid sequence of the selected protein deduced and the parent protein identified. Surface-enhanced laser desorption and ionization combined with time-of-flight mass spectrometry is another commonly used technique in which sample prefractionation occurs on specially designed protein chips that capture specific proteins and peptides, which are then visualized by mass spectrometry (Figure 2).31 This technique has the advantage of facilitating the screening of large numbers of urine samples for potential biomarkers; unlike tandem mass spectrometry, however, it does not allow identification of any potential biomarkers.
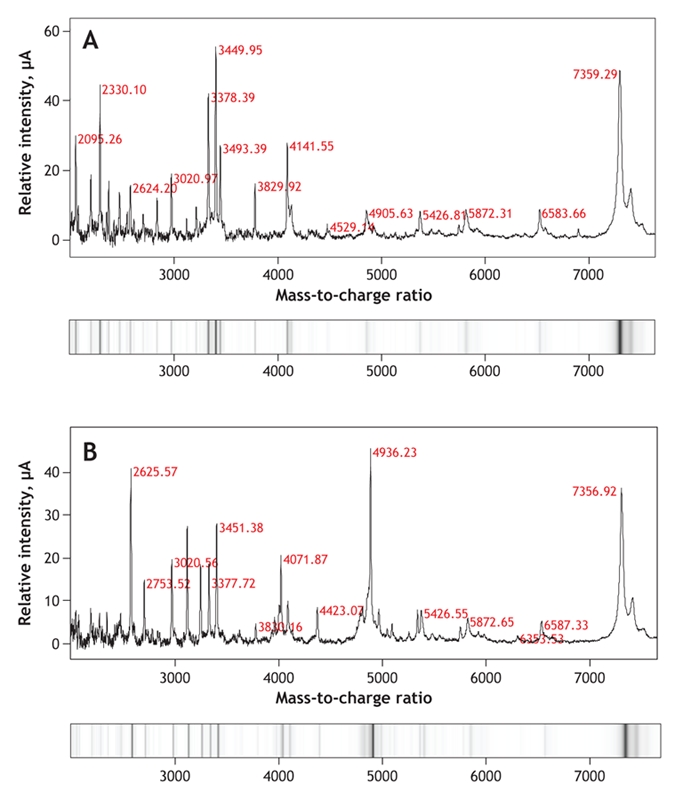
Figure 2: Spectra and virtual gels generated by surface-enhanced laser desorption and ionization, followed by time-of-flight mass spectrometry of urine samples from a healthy subject (A) and a patient with glomerular disease (B). Mass-to-charge values for selected peptides and proteins are shown in red. Some proteins and peptides are present in the urine of both subjects (e.g., mass-to-charge ratios 7359.29 and 7356.92, 5426.81 and 5426.55, and 2624.20 and 2625.57). Other proteins are apparent only in the urine of the patient with glomerular disease (e.g., mass-to-charge ratio 4936.23).
Proteomic profiling of normal human urine through mass spectrometry methods has identified at least 1000 different protein gene products and many more peptide fragments of larger proteins. A urinary exosome protein database derived from mass spectrometry data is now available (http://dir.nhlbi.nih.gov/papers/lkem/exosome/index.htm), and it is hoped that this will be expanded over time.
Current biomarker candidates
The number of published urinary biomarker studies is increasing rapidly.32 Most studies have looked at the soluble urine protein fraction, focusing on the identification of potential biomarkers in renal disease and diseases of the urogenital tract. They include studies of acute kidney injury,33,34 acute renal allograft rejection,35–38 glomerular disease39–46 and carcinoma of the kidney, bladder and prostate.47–49 In many cases the reported biomarkers remain unidentified, although some studies have proceeded to identification of the biomarker proteins.35,50
Acute kidney injury
Acute kidney injury is a common clinical problem associated with high morbidity and mortality. Routinely used measures of renal function change only after substantial kidney injury has occurred and then with a delay. Sensitive and specific biomarkers are needed to detect early kidney injury and to guide therapeutic strategies that might avert irreversible renal damage. A number of candidate urinary proteins, mainly renal tubular proteins, have been evaluated as biomarkers of renal injury, and some of the most promising are listed in Table 2. However, problems have been identified in the reliable use of individual molecules to identify and monitor kidney injury in animal models and in human subjects, and none of the molecules has yet made the transition from bench to bedside. It has been suggested that a “urine panel” consisting of interleukin-18, kidney injury molecule-1 and neutrophil gelatinase-associated lipocalin may prove more useful, and clinical trials with this panel are planned.33
Table 2
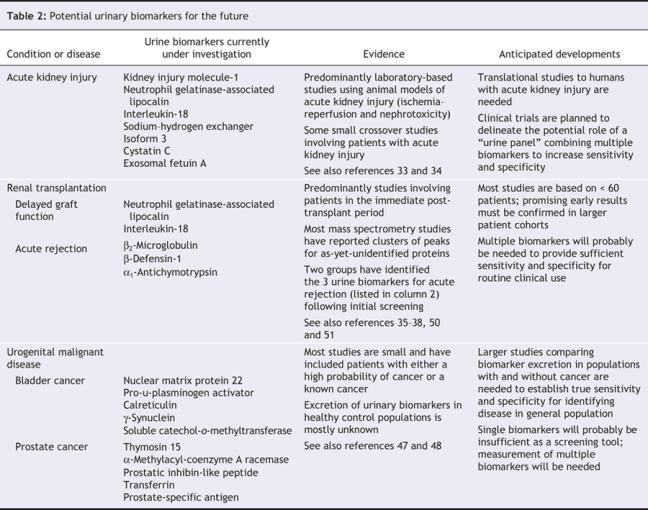
Urinary exosomes contain a number of proteins that may be altered in association with renal disease and that are free from much of the interference of the most abundant urinary proteins. Exosomal fetuin-A has recently been identified as a potential urinary biomarker of acute kidney injury in both animal models and patients in the intensive care setting, and further work on this biomarker is awaited with interest.34
Renal transplantation
Renal tubular damage is a principal feature of both ischemia–reperfusion injury at the time of surgery and acute allograft rejection. Early detection of acute tubular injury is essential if therapeutic interventions are to be effective in preserving allograft function. In parallel with work in acute kidney injury, there has been interest in the use of renal tubular proteins as biomarkers of ischemia–reperfusion injury and delayed graft function (Table 2). Screening studies using mass spectrometry have identified biomarkers predictive of acute allograft rejection, but for the most part these biomarkers are protein peak clusters that remain unidentified (as in Figure 2).35–38,50,51 Two studies have now reported the identification of biomarkers after initial urine screening (Table 2).35,50 The use of these urine biomarkers for noninvasive monitoring of renal allografts remains a little way off; however, work is now under way to examine their utility, along with that of other urine biomarkers of tubular injury, in assessing renal allograft function.
Glomerular disease
Glomerulonephritis is a major cause of renal failure and an important cause of morbidity and mortality in multisystem diseases. The diagnosis of glomerular disease combines assessment of renal function, quantification of proteinuria and analysis of urine by microscopy and dipstick testing. These tests are useful in assessing renal injury, but they are poor indicators of ongoing inflammatory activity. A number of candidate biomarkers, including cytokines, chemokines and growth factors, have been studied, and more recently mass spectrometry has been used to screen for urine biomarkers in glomerulonephritis both in animal models and in patients with various levels of disease activity.52 Urine biomarkers have been described that discriminate active from inactive lupus nephritis and allow detection of relapse and remission earlier than traditional clinical markers.44 Protein peak clusters revealed by mass spectrometry have been reported but not yet identified. Similarly, a number of urine biomarkers identified in immunoglobulin A nephropathy may discriminate this pattern of glomerulonephritis from other proteinuric glomerular diseases.39,41 Interestingly, changes were observed in the excretion of these protein peak clusters with increasing use of antihypertensive agents, which suggests that urine biomarkers may be used in the future to monitor the clinical effectiveness of treatment in glomerulonephritis.39 This possibility was supported by the results of a separate study of diabetic nephropathy, in which it was found that commencement of candesartan significantly reduced the excretion of biomarkers toward the levels seen in healthy control urine.46
Diabetic nephropathy is an important cause of morbidity and mortality in patients with both type 1 and type 2 diabetes mellitus. Microalbuminuria is an important marker of early diabetic nephropathy, but by the time this problem becomes apparent, significant glomerular injury has already occurred. A number of studies using mass spectrometry have identified urinary polypeptide patterns specific to patients with normoalbuminuria in diabetes, which suggests that it may be possible in the future to identify patients at risk of renal injury before nephropathy becomes established.42,43,46
Malignancy of the urogenital tract
Many nephrourological tumour-associated molecules are secreted into the urine and it is therefore not surprising that several studies have evaluated urine biomarkers for the early diagnosis and surveillance of renal cell, bladder and prostate cancer (Table 2).47,49,53 Most of these studies have been based on small patient cohorts, and the reported sensitivity and specificity of urine biomarkers are wide ranging. With only a few studies published on each set of biomarkers, it is difficult at present to assess their potential clinical impact in the future.
Non-nephrourological disease
Because urine is a filtrate of blood, it contains protein components similar to those found in the blood. Changes in the urinary proteome may therefore be used to detect not only abnormalities within the kidney and the urogenital tract but also systemic disease associated with small circulating protein and peptide markers that can pass through the glomerular filter. Recent reports have identified urinary biomarkers predictive of acute pancreatitis,54 obstructive sleep apnea,55 early ovarian cancer56 and non-small-cell lung cancer.57 It is likely that urine testing will be used in the future to screen for a plethora of systemic disorders, the majority of which will have no or limited renal involvement.
Conclusions
Urinary proteomics offers a great opportunity for the development of novel, noninvasive assays for the diagnosis and monitoring of both renal and systemic diseases. To date, most biomarker studies have been conducted on small patient cohorts, with few having been validated by independent research groups. It is unlikely that any single urine biomarker will have sufficient sensitivity and specificity for clinical use, and it seems more probable that any new biomarker will be used as part of a “biomarker panel” or will be integrated into a clinical scoring algorithm to refine existing diagnostic scoring systems. As more data become available, it will be imperative that urine biomarkers with potential clinical utility are independently validated before investment is made into producing a marketed diagnostic test.
Future studies should therefore focus on evaluating the sensitivity and specificity of biomarker panels in large patient populations and should be managed in the same way as any other clinical trial. It will be important to have a well-defined “gold standard” diagnostic test against which to compare each biomarker. Given that the excretion of urine biomarkers varies with sex, age, concomitant renal disease, hypertension and current medications, urine biomarkers validated for one patient group may not be diagnostic in another; therefore, inclusion and exclusion criteria will have to be clearly defined. Furthermore, widespread implementation of any new biomarker assay is more likely if the assay is an extension of current practice. Therefore, biomarker identification is likely to be an important step, as it will be necessary to generate biomarker-specific antibodies for development of an appropriate assay.
We remain some way from integrating any of the new urine biomarkers into clinical practice; urine protein screening with urine dipsticks and quantification using the protein–creatinine ratio or albumin–creatinine ratio will remain the focus of urine diagnostics for the near future.
Key points
• For patients with intrinsic renal disease, urinary protein excretion provides important diagnostic and prognostic information.
• Current methods of assessing urinary protein excretion are crude and do not discriminate between different forms of renal and extrarenal disease.
• Innovative urine proteomic studies are identifying increasing numbers of novel urine proteins that may prove useful for the diagnosis and monitoring of renal and systemic diseases.
• Integration of urine biomarkers in routine clinical practice has been slow; it is likely that multiple biomarkers will be needed to provide the sensitivity and specificity necessary for their use in clinical decision-making.
Acknowledgments
We thank Drs. Jan Novak, G. Robinson, H. Kim and Bruce A. Julian, University of Alabama at Birmingham, Birmingham, Ala., for supplying the 2-dimensional gel electrophoresis of human urine.
Footnotes
Une version française de ce résumé est disponible à l'adresse www.cmaj.ca/cgi/content/full/177/4/361/DC1
This article has been peer reviewed.
Contributors: Both authors contributed to the literature review and to drafting and critically revising the article. Both approved the final version to be published.
Competing interests: None declared.
Correspondence to: Dr. Jonathan Barratt, Department of Infection, Immunity and Inflammation, John Walls Renal Unit, Leicester General Hospital, Gwendolen Road, Leicester UK LE5 4PW; fax 0116 258 4764; jb81@le.ac.uk
REFERENCES
- 1.Viberti GC, Hill RD, Jarrett RJ, et al. Microalbuminuria as a predictor of clinical nephropathy in insulin-dependent diabetes mellitus. Lancet 1982;1:1430-2. [DOI] [PubMed]
- 2.Abuelo JG. Proteinuria: diagnostic principles and procedures. Ann Intern Med 1983;98:186-91. [DOI] [PubMed]
- 3.Free AH, Rupe CO, Metzler I. Studies with a new colorimetric test for proteinuria. Clin Chem 1957;3:716-27. [PubMed]
- 4.McElderry LA, Tarbit IF, Cassells-Smith AJ. Six methods for urinary protein compared. Clin Chem 1982;28:356-60. [PubMed]
- 5.Ginsberg JM, Chang BS, Matarese RA, et al. Use of single voided urine samples to estimate quantitative proteinuria. N Engl J Med 1983;309:1543-6. [DOI] [PubMed]
- 6.Chitalia VC, Kothari J, Wells EJ, et al. Cost–benefit analysis and prediction of 24-hour proteinuria from the spot urine protein–creatinine ratio. Clin Nephrol 2001;55:436-47. [PubMed]
- 7.Schwab SJ, Christensen RL, Dougherty K, et al. Quantitation of proteinuria by the use of protein-to-creatinine ratios in single urine samples. Arch Intern Med 1987;147:943-4. [PubMed]
- 8.Steinhauslin F, Wauters JP. Quantitation of proteinuria in kidney transplant patients: accuracy of the urinary protein/creatinine ratio. Clin Nephrol 1995;43:110-5. [PubMed]
- 9.Comper WD, Jerums G, Osicka TM. Differences in urinary albumin detected by four immunoassays and high-performance liquid chromatography. Clin Biochem 2004;37:105-11. [DOI] [PubMed]
- 10.Giampietro O, Penno G, Clerico A, et al. Which method for quantifying “microalbuminuria” in diabetics? Comparison of several immunological methods (immunoturbidimetric assay, immunonephelometric assay, radioimmunoassay and two semiquantitative tests) for measurement of albumin in urine. Acta Diabetol 1992;28:239-45. [DOI] [PubMed]
- 11.Polkinghorne KR, Su Q, Chadban SJ, et al. Population prevalence of albuminuria in the Australian Diabetes, Obesity, and Lifestyle (AusDiab) study: immunonephelometry compared with high-performance liquid chromatography. Am J Kidney Dis 2006;47:604-13. [DOI] [PubMed]
- 12.Roberts WL, Calcote CB, Cook CB, et al. Comparison of four commercial urinary albumin (microalbumin) methods: implications for detecting diabetic nephropathy using random urine specimens. Clin Chim Acta 1998;273:21-33. [DOI] [PubMed]
- 13.Cameron JS, Blandford G. The simple assessment of selectivity in heavy proteinuria. Lancet 1966;2:242-7. [DOI] [PubMed]
- 14.Cavaliere G, Arrigo G, D'Amico G, et al. Tubular nephrotoxicity after intravenous urography with ionic high-osmolal and nonionic low-osmolal contrast media in patients with chronic renal insufficiency. Nephron 1987;46:128-33. [DOI] [PubMed]
- 15.Hartmann HG, Braedel HE, Jutzler GA. Detection of renal tubular lesions after abdominal aortography and selective renal arteriography by quantitative measurements of brush-border enzymes in the urine. Nephron 1985;39:95-101. [DOI] [PubMed]
- 16.Jung K, Pergande M, Schimke E, et al. Urinary enzymes and low-molecular-mass proteins as indicators of diabetic nephropathy. Clin Chem 1988;34:544-7. [PubMed]
- 17.Marchewka Z, Kuzniar J, Dlugosz A. Enzymuria and beta2-mikroglobulinuria in the assessment of the influence of proteinuria on the progression of glomerulopathies. Int Urol Nephrol 2001;33:673-6. [DOI] [PubMed]
- 18.Sherman RL, Drayer DE, Leyland-Jones BR, et al. N-Acetyl-beta-glucosaminidase and beta2-microglobulin. Their urinary excretion in patients with renal parenchymal disease. Arch Intern Med 1983;143:1183-5. [DOI] [PubMed]
- 19.Bazzi C, Petrini C, Rizza V, et al. Urinary N-acetyl-beta-glucosaminidase excretion is a marker of tubular cell dysfunction and a predictor of outcome in primary glomerulonephritis. Nephrol Dial Transplant 2002;17:1890-6. [DOI] [PubMed]
- 20.Herget-Rosenthal S, Marggraf G, Husing J, et al. Early detection of acute renal failure by serum cystatin C. Kidney Int 2004;66:1115-22. [DOI] [PubMed]
- 21.Mishra J, Dent C, Tarabishi R, et al. Neutrophil gelatinase-associated lipocalin (NGAL) as a biomarker for acute renal injury after cardiac surgery. Lancet 2005;365:1231-8. [DOI] [PubMed]
- 22.Putnam FW, Easley CW, Lynn LT, et al. The heat precipitation of Bence-Jones proteins. I. Optimum conditions. Arch Biochem Biophys 1959;83:115-30. [DOI] [PubMed]
- 23.Perry MC, Kyle RA. The clinical significance of Bence Jones proteinuria. Mayo Clin Proc 1975;50:234-8. [PubMed]
- 24.Mayo MM, Johns GS. Serum free light chains in the diagnosis and monitoring of patients with plasma cell dyscrasias. Contrib Nephrol 2007;153:44-65. [DOI] [PubMed]
- 25.International Myeloma Working Group. Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: a report of the International Myeloma Working Group. Br J Haematol 2003;121:749-57. [PubMed]
- 26.Cole LA, Khanlian SA, Sutton JM, et al. Accuracy of home pregnancy tests at the time of missed menses. Am J Obstet Gynecol 2004;190:100-5. [DOI] [PubMed]
- 27.Scheer WD. The detection of leukocyte esterase activity in urine with a new reagent strip. Am J Clin Pathol 1987;87:86-93. [DOI] [PubMed]
- 28.Hortin GL, Jortani SA, Ritchie JC Jr, et al. Proteomics: a new diagnostic frontier. Clin Chem 2006;52:1218-22. [DOI] [PubMed]
- 29.Pisitkun T, Johnstone R, Knepper MA. Discovery of urinary biomarkers. Mol Cell Proteomics 2006;5:1760-71. [DOI] [PubMed]
- 30.Gorg A, Weiss W, Dunn MJ. Current two-dimensional electrophoresis technology for proteomics. Proteomics 2004;4:3665-85. [DOI] [PubMed]
- 31.Xiao Z, Prieto D, Conrads TP, et al. Proteomic patterns: their potential for disease diagnosis. Mol Cell Endocrinol 2005;230:95-106. [DOI] [PubMed]
- 32.Janech MG, Raymond JR, Arthur JM. Proteomics in renal research. Am J Physiol Renal Physiol 2007;292:F501-12. [DOI] [PubMed]
- 33.Nguyen MT, Devarajan P. Biomarkers for the early detection of acute kidney injury. Pediatr Nephrol 2007 Mar 30; [Epub ahead of print]. DOI 10.1007/s00467-007-0470-x. [DOI] [PMC free article] [PubMed]
- 34.Zhou H, Pisitkun T, Aponte A, et al. Exosomal fetuin-A identified by proteomics: a novel urinary biomarker for detecting acute kidney injury. Kidney Int 2006;70:1847-57. [DOI] [PMC free article] [PubMed]
- 35.Schaub S, Wilkins JA, Antonovici M, et al. Proteomic-based identification of cleaved urinary beta2-microglobulin as a potential marker for acute tubular injury in renal allografts. Am J Transplant 2005;5:729-38. [DOI] [PubMed]
- 36.Schaub S, Rush D, Wilkins J, et al. Proteomic-based detection of urine proteins associated with acute renal allograft rejection. J Am Soc Nephrol 2004;15:219-27. [DOI] [PubMed]
- 37.O'Riordan E, Orlova TN, Mei JJ, et al. Bioinformatic analysis of the urine proteome of acute allograft rejection. J Am Soc Nephrol 2004;15:3240-8. [DOI] [PubMed]
- 38.Clarke W, Silverman BC, Zhang Z, et al. Characterization of renal allograft rejection by urinary proteomic analysis. Ann Surg 2003;237:660-4; discussion 664-5. [DOI] [PMC free article] [PubMed]
- 39.Haubitz M, Wittke S, Weissinger EM, et al. Urine protein patterns can serve as diagnostic tools in patients with IgA nephropathy. Kidney Int 2005;67:2313-20. [DOI] [PubMed]
- 40.Candiano G, Musante L, Bruschi M, et al. Repetitive fragmentation products of albumin and alpha1-antitrypsin in glomerular diseases associated with nephrotic syndrome. J Am Soc Nephrol 2006;17:3139-48. [DOI] [PubMed]
- 41.Park MR, Wang EH, Jin DC, et al. Establishment of a 2-D human urinary proteomic map in IgA nephropathy. Proteomics 2006;6:1066-76. [DOI] [PubMed]
- 42.Meier M, Kaiser T, Herrmann A, et al. Identification of urinary protein pattern in type 1 diabetic adolescents with early diabetic nephropathy by a novel combined proteome analysis. J Diabetes Complications 2005;19:223-32. [DOI] [PubMed]
- 43.Mischak H, Kaiser T, Walden M, et al. Proteomic analysis for the assessment of diabetic renal damage in humans. Clin Sci (Lond) 2004;107:485-95. [DOI] [PubMed]
- 44.Mosley K, Tam FW, Edwards RJ, et al. Urinary proteomic profiles distinguish between active and inactive lupus nephritis. Rheumatology (Oxford) 2006;45:1497-504. [DOI] [PubMed]
- 45.Ngai HH, Sit WH, Jiang PP, et al. Serial changes in urinary proteome profile of membranous nephropathy: implications for pathophysiology and biomarker discovery. J Proteome Res 2006;5:3038-47. [DOI] [PubMed]
- 46.Rossing K, Mischak H, Parving HH, et al. Impact of diabetic nephropathy and angiotensin II receptor blockade on urinary polypeptide patterns. Kidney Int 2005;68:193-205. [DOI] [PubMed]
- 47.van Rhijn BW, van der Poel HG, van der Kwast TH. Urine markers for bladder cancer surveillance: a systematic review. Eur Urol 2005;47:736-48. [DOI] [PubMed]
- 48.M'Koma AE, Blum DL, Norris JL, et al. Detection of pre-neoplastic and neoplastic prostate disease by MALDI profiling of urine. Biochem Biophys Res Commun 2007;353:829-34. [DOI] [PMC free article] [PubMed]
- 49.Perroud B, Lee J, Valkova N, et al. Pathway analysis of kidney cancer using proteomics and metabolic profiling. Mol Cancer 2006;5:64. [DOI] [PMC free article] [PubMed]
- 50.O'Riordan E, Orlova TN, Podust VN, et al. Characterization of urinary peptide biomarkers of acute rejection in renal allografts. Am J Transplant 2007;7:930-40. [DOI] [PubMed]
- 51.Wittke S, Haubitz M, Walden M, et al. Detection of acute tubulointerstitial rejection by proteomic analysis of urinary samples in renal transplant recipients. Am J Transplant 2005;5:2479-88. [DOI] [PubMed]
- 52.Boumpas DT, Kuroiwa T. Urine proteomics for the early diagnosis of acute glomerulonephritis: Has the time come? Arthritis Rheum 2007;56:705-9. [DOI] [PubMed]
- 53.Downes MR, Byrne JC, Pennington SR, et al. Urinary markers for prostate cancer. BJU Int 2007;99:263-8. [DOI] [PubMed]
- 54.Comte B, Franceschi C, Sadoulet MO, et al. Detection of bile salt-dependent lipase, a 110 kDa pancreatic protein, in urines of healthy subjects. Kidney Int 2006;69:1048-55. [DOI] [PubMed]
- 55.Polotsky VY, O'Donnell CP. Genomics of sleep-disordered breathing. Proc Am Thorac Soc 2007;4:121-6. [DOI] [PubMed]
- 56.Chambers AF, Vanderhyden BC. Ovarian cancer biomarkers in urine. Clin Cancer Res 2006;12:323-7. [DOI] [PubMed]
- 57.Tantipaiboonwong P, Sinchaikul S, Sriyam S, et al. Different techniques for urinary protein analysis of normal and lung cancer patients. Proteomics 2005;5:1140-9. [DOI] [PubMed]


