Short abstract
Mucosa‐associated E Coli are reported to be increased in Crohn's disease
There is a widespread assumption that both of the major inflammatory bowel diseases, Crohn's disease and ulcerative colitis, arise as a result of a host response to intestinal bacteria. Evidence to support this includes the involvement of non‐pathogenic bacteria in the development of colitis in genetically altered animals, knowledge that the Crohn's disease‐associated gene NOD2/CARD15 is a receptor for bacterial cell‐wall peptidoglycan, the common presence of circulating anti‐bacterial antibodies in Crohn's disease, and the role of known pathogens in precipitating relapse in ulcerative colitis.1 We still need to know which bacteria are the culprits, where they are (intraluminal, intramucosal, intracellular) and whether there is an abnormal host response to “commensal” bacteria or whether the bacteria themselves have pathogenic features.
Study of the gut microbiota is not easy. The human gut contains 500–1000 bacterial species2 and around 80% of these have yet to be cultured.3 There is growing evidence that the bacteria that are closely related to or adherent to the mucosa may be more relevant to mucosal inflammation than those in the faeces. Studies of the mucosa‐associated bacteria are, however, affected by the method used. The colonic mucosa, unlike the small intestine, has a near‐continuous mucus coat4 and bacteria adherent to the surface of this coat will differ in number and nature from those underneath the mucus. Moreover, the surface of the mucus layer, like the faeces, is likely to suit the growth predominantly of anaerobic bacteria, whereas mucus represents a significant barrier to oxygen diffusion,5 so the sub‐mucus niche may be relatively well oxygenated by the underlying mucosa and more suitable for micro‐aerophilic bacteria. We have found that aerobic culture of colonoscopic biopsies after removal of the mucus layer with dithiothreitol is often sterile in control colons, whereas the colon in Crohn's disease and colon cancer contains increased bacterial numbers in this sub‐mucus niche, more than half of which are E coli,6 even though these organisms account for less than 1% of the faecal microbiota.
At least five independent groups have reported that mucosa‐associated E coli are increased in Crohn's disease6,7,8,9,10 and the study by Kotlowski et al in this issue11 (see page 669) makes this six. Two of the studies7,11 have also shown an increase in mucosa‐associated E coli in ulcerative colitis. One group has reported selective growth of the E coli from distal ileal mucosal samples in Crohn's disease,8 whilst the others have reported on colonic mucosal samples. The present study adds particular weight to the potential importance of E coli because of the initial screen that was used, which should have been able to pick up any bacteria that were overexpressed in the inflammatory bowel disease tissue samples. The study used ribosomal intergenic spacer analysis to identify DNA segments that were more commonly present (in about 70%) in Crohn's disease and ulcerative colitis mucosal biopsies than controls (about 30%). Five of these segments were then sequenced and all were found to contain E coli DNA. Subsequent study focused on E coli and again confirmed increased culture of E coli from Crohn's disease samples and, to a lesser extent, from ulcerative colitis samples, compared with controls. There was a poor correlation between site of inflammation and presence of E coli, in agreement with other studies that have tended to show that the same organisms can be identified from various sites within the same colon.6,7 This is compatible with the organisms having a causative role in the inflammation rather than merely colonising inflamed mucosa.
Where do the E coli end up? In Crohn's disease but not ulcerative colitis there is good evidence of bacterial invasion into tissues and this is supported by the tendency for abscess and fistula formation. Some of the bacteria undoubtedly invade after a breach in the mucosa has already occurred and these are likely to be a cross‐spectrum of the faecal microbiota.12,13 Nevertheless, an immunohistochemical study using a broad range of anti‐microbial antibodies showed a selective presence in Crohn's disease tissue of E coli, Listeria and Streptococci, all within macrophages.14 Subsequent studies of Listeria have been contradictory15 but E coli DNA has also been identified within 12/15 Crohn's disease granulomas dissected out from tissue sections by laser‐dissecting microscopy, compared with 1/10 control granulomas, although the same group also identified M paratuberculosis within Crohn's disease granulomas using the same technique.16 The possibility that the E coli may end up chronically replicating within tissue macrophages is supported by evidence that Crohn's disease E coli isolates are particularly capable of replicating within macrophage vesicles in vitro when compared with laboratory control E coli strains.17
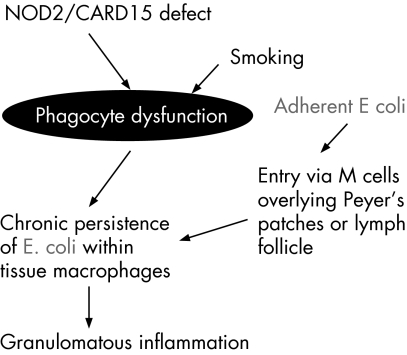
What is the site of entry? Crohn's disease mucosal E coli isolates have been shown to be better able than laboratory strains to adhere to and invade colon epithelial cell lines in culture.6,8 However, this phenomenon is cell‐line specific6 and there is little evidence that they are able to invade the normal colonic epithelial cells in vivo. Moreover, there is good evidence that bona fide invasive pathogens, such as Salmonella and Shigella, are unable to invade via normal colon epithelial cells and instead make their initial entry via the specialised M cells within the dome epithelium overlying Peyer's patches in the distal ileum and lymphoid follicles in the colon.18,19 There is no mucus layer overlying the dome epithelium since it possesses no goblet cells. Moreover, the M cells lack the “fuzzy” glycocalyx that other small intestinal and colonic epithelial cells possess.20 If unequivocal pathogenic organisms need M cells as a portal, it seems likely that the Crohn's E coli, which lack conventional pathogenicity genes, will also have to enter via the M cells. This fits well with evidence suggesting that the earliest lesions in Crohn's disease occur at Peyer's patches in the ileum and lymphoid follicles in the colon21 (fig 1).
Figure 1 Models for involvement of E coli in IBD pathogenesis: 1. Crohn's disease.
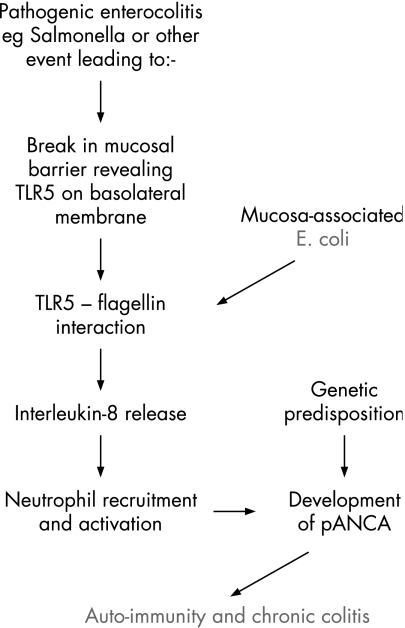
What role could these E coli have in ulcerative colitis? The study by Kotlowski and colleagues published here11 and also that by Swidsinski and colleagues7 both show increased mucosa‐associated E coli in ulcerative colitis, possibly to a slightly lesser extent than in Crohn's disease. The lack of association reported by others6 may reflect differences in sampling technique, such as removal of the mucus layer prior to culture. There is no good evidence for mucosal invasion by E coli in ulcerative colitis and the typically continuous inflammation does not suggest any specific focus of inflammation around lymphoid follicles. It seems more plausible that, in ulcerative colitis, the mucosa‐associated E coli may interact with the surface epithelial cells without invasion. This would fit with the superficial nature of mild colitis and also with evidence of surface epithelial NFkappaB activation as a very early event.22 Another likely result of such interaction would be release from the epithelial cells of interleukin‐8 (IL‐8), a potent chemotactant for neutrophils. So far, most evidence suggests that bacteria‐induced IL‐8 release from colon epithelial cells is usually a consequence of flagellin‐Toll receptor 5 (TLR5) interaction and it is notable that this can result from interaction even with E coli strains that are conventionally regarded as “commensal”.23 Since TLR5 is located on the basolateral aspect of the epithelial cells, this would require prior weakening of the mucosal barrier, as occurs experimentally with dextran sulphate, to allow flagellin to pass through the tight junctions and reach TLR5.24 This represents a plausible model for ulcerative colitis,25 perhaps with inflammation perpetuated by an auto‐immune mechanism (fig 2).
Figure 2 Models for involvement of E coli in IBD pathogenesis: 2. Ulcerative colitis.
Further work is needed to characterise the inflammatory bowel disease mucosa‐associated E coli and their mechanisms of interaction with epithelial cells. In many ways their adherence and invasion of epithelial cells in the absence of any of the typical pathogenicity genes, other than adhesins, is similar to the properties of uropathogenic E coli;26,27 indeed they may be the same organisms. There is already a considerable literature on how uropathogenic E coli interact with epithelial cells26,28 and much of this may be relevant in the colon. This may include interaction with members of the carcinoembryonic antigen‐cell adhesion molecule (CEACAM) family29,30,31 that are present in the glycocalyx, the “fuzzy coat” that underlies the mucus layer and overlies colon and small intestinal epithelial cells. The study by Kotlowski and colleagues takes this a step forward with the demonstration that the Crohn's disease E coli isolates are more likely to be in phylogenetic groups B2 and D; groups that characteristically contain E coli that are pathogenic at extra‐intestinal sites and that tend to adhere to epithelial cells.32,33 The inflammatory bowel disease isolates are also shown commonly to possess serine protease autotransporters (SPATEs), some of which possess mucinase activity.34 This could explain why these organisms seem typically to have been found within35 or beneath6 the colonic mucus layer.
Evidence is thus accumulating that E coli, probably “mildly pathogenic” by virtue of their possession of adhesion mechanisms and secretion of proteases, may have a causative role in inflammatory bowel disease. In Crohn's disease they probably invade the tissue, perhaps as a result of defective clearance by neutrophils,36 and chronically infect macrophages, whereas they may have a more superficial relationship with the mucosal surface in ulcerative colitis. Koch's postulates have yet to be fulfilled but it is intriguing that E coli with a similar adherent and invasive phenotype have been shown to be associated with granulomatous colitis in boxer dogs.37 I think we now have a new therapeutic target.
Footnotes
Competing interests: JMR is a past/present member of advisory boards for Procter and Gamble, Schering‐Plough, Chiesi, Falk, and Celltech/UCB, and, with the University of Liverpool and Provexis UK, holds a patent for use of a soluble fibre preparation as maintenance therapy for Crohn's disease.
References
- 1.Subramanian S, Campbell B J, Rhodes J M. Bacteria in the pathogenesis of inflammatory bowel disease. Curr Opin Infect Dis 200619475–484. [DOI] [PubMed] [Google Scholar]
- 2.Egert M, de Graaf A A, Smidt H.et al Beyond diversity: functional microbiomics of the human colon. Trends Microbiol 20061486–91. [DOI] [PubMed] [Google Scholar]
- 3.Eckburg P B, Bik E M, Bernstein C N.et al Diversity of the human intestinal microbial flora. Science 20053081635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atuma C, Strugala V, Allen A.et al The adherent gastrointestinal mucus gel layer: thickness and physical state in vivo. Am J Physiol Gastrointest Liver Physiol 2001280G922–G929. [DOI] [PubMed] [Google Scholar]
- 5.Saldena T A, Saravi F D, Hwang H J.et al Oxygen diffusive barriers of rat distal colon: role of subepithelial tissue, mucosa, and mucus gel layer. Dig Dis Sci 2000452108–2114. [DOI] [PubMed] [Google Scholar]
- 6.Martin H M, Campbell B J, Hart C A.et al Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology 200412780–93. [DOI] [PubMed] [Google Scholar]
- 7.Swidsinski A, Ladhoff A, Pernthaler A.et al Mucosal flora in inflammatory bowel disease. Gastroenterology 200212244–54. [DOI] [PubMed] [Google Scholar]
- 8.Darfeuille‐Michaud A, Boudeau J, Bulois P.et al High prevalence of adherent‐invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 2004127412–421. [DOI] [PubMed] [Google Scholar]
- 9.Mylonaki M, Rayment N B, Rampton D S.et al Molecular characterization of rectal mucosa‐associated bacterial flora in inflammatory bowel disease. Inflamm Bowel Dis 200511481–487. [DOI] [PubMed] [Google Scholar]
- 10.Alpern J, Sasaki M, Sitaraman S.et al Invasive E coli strains are increased in Crohn's disease. Gastroenterology 2006130A362 [Google Scholar]
- 11.Kotlowski R, Bernstein C N, Sepehri S.et al High prevalence of Escherichia coli belonging to the B2 and D phylogenetic groups in inflammatory bowel disease. Gut 200756669–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takesue Y, Ohge H, Uemura K.et al Bacterial translocation in patients with Crohn's disease undergoing surgery. Dis Colon Rectum 2002451665–1671. [DOI] [PubMed] [Google Scholar]
- 13.Ambrose N S, Johnson M, Burdon D W.et al Incidence of pathogenic bacteria from mesenteric lymph nodes and ileal serosa during Crohn's disease surgery. Br J Surg 198471623–625. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, van Kruiningen H J, West A B.et al Immunocytochemical evidence of Listeria, Escherichia coli, and Streptococcus antigens in Crohn's disease. Gastroenterology 19951081396–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen W, Li D, Paulus B.et al Detection of Listeria monocytogenes by polymerase chain reaction in intestinal mucosal biopsies from patients with inflammatory bowel disease and controls. J Gastroenterol Hepatol 2000151145–1150. [DOI] [PubMed] [Google Scholar]
- 16.Ryan P, Kelly R G, Lee G.et al Bacterial DNA within granulomas of patients with Crohn's disease – detection by laser capture microdissection and PCR. Am J Gastroenterol 2004991539–1543. [DOI] [PubMed] [Google Scholar]
- 17.Bringer M A, Glasser A L, Tung C H.et al The Crohn's disease‐associated adherent‐invasive Escherichia coli strain LF82 replicates in mature phagolysosomes within J774 macrophages. Cell Microbiol 20068471–484. [DOI] [PubMed] [Google Scholar]
- 18.Niedergang F, Kraehenbuhl J P. Much ado about M cells. Trends Cell Biol 200010137–141. [DOI] [PubMed] [Google Scholar]
- 19.Sansonetti P J, Phalipon A. M cells as ports of entry for enteroinvasive pathogens: mechanisms of interaction, consequences for the disease process. Semin Immunol 199911193–203. [DOI] [PubMed] [Google Scholar]
- 20.Neutra M R, Mantis N J, Frey A.et al The composition and function of M cell apical membranes: implications for microbial pathogenesis. Semin Immunol 199911171–181. [DOI] [PubMed] [Google Scholar]
- 21.Fujimura Y, Kamoi R, Iida M. Pathogenesis of aphthoid ulcers in Crohn's disease: correlative findings by magnifying colonoscopy, electron microscopy, and immunohistochemistry. Gut 199638724–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bodger K, Halfvarson J, Dodson A R.et al Altered colonic glycoprotein expression in unaffected monozygotic twins of inflammatory bowel disease patients. Gut 200655973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bambou J C, Giraud A, Menard S.et alIn vitro and ex vivo activation of the TLR5 signaling pathway in intestinal epithelial cells by a commensal Escherichia coli strain. J Biol Chem 200427942984–42992. [DOI] [PubMed] [Google Scholar]
- 24.Rhee S H, Im E, Riegler M.et al Pathophysiological role of Toll‐like receptor 5 engagement by bacterial flagellin in colonic inflammation. Proc Natl Acad Sci U S A 200510213610–13615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gitter A H, Wullstein F, Fromm M.et al Epithelial barrier defects in ulcerative colitis: characterization and quantification by electrophysiological imaging. Gastroenterology 20011211320–1328. [DOI] [PubMed] [Google Scholar]
- 26.Servin A L. Pathogenesis of Afa/Dr diffusely adhering Escherichia coli. Clin Microbiol Rev 200518264–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marrs C F, Zhang L, Foxman B.Escherichia coli mediated urinary tract infections: Are there distinct uropathogenic E coli (UPEC) pathotypes? FEMS Microbiol Letts 2005252183–190. [DOI] [PubMed] [Google Scholar]
- 28.Mulvey M A. Adhesion and entry of uropathogenic Escherichia coli. Cell Microbiol 20024257–271. [DOI] [PubMed] [Google Scholar]
- 29.Berger C N, Billker O, Meyer T F.et al Differential recognition of members of the carcinoembryonic antigen family by Afa/Dr adhesins of diffusely adhering Escherichia coli (Afa/Dr DAEC). Mol Microbiol 200452963–983. [DOI] [PubMed] [Google Scholar]
- 30.Frangsmyr L, Baranov V, Hammarstrom S. Four carcinoembryonic antigen subfamily members, CEA, NCA, BGP and CGM2, selectively expressed in the normal human colonic epithelium, are integral components of the fuzzy coat. Tumour Biol 199920277–292. [DOI] [PubMed] [Google Scholar]
- 31.Kuespert K, Pils S, Hauck C R. CEACAMs: their role in physiology and pathophysiology. Curr Opin Cell Biol 200618565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. App Environ Microbiol 2000664555–4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nowrouzian F L, Adlerberth I, Wold A E. Enhanced persistence in the colonic microbiota of Escherichia coli strains belonging to phylogenetic group B2: role of virulence factors and adherence to colonic cells. Microbes Infection 20068834–840. [DOI] [PubMed] [Google Scholar]
- 34.Parham N J, Srinavasan U, Desvaux M.et al PicU, a second serine protease autotransporter of uropathogenic Escherichia coli. FEMS Microbiol Letts 200423073–83. [DOI] [PubMed] [Google Scholar]
- 35.Schultsz C, Van Den Berg F M, Ten Kate F W.et al The intestinal mucus layer from patients with inflammatory bowel disease harbors high numbers of bacteria compared with controls. Gastroenterology 19991171089–1097. [DOI] [PubMed] [Google Scholar]
- 36.Marks D J, Harbord M W, MacAllister R.et al Defective acute inflammation in Crohn's disease: a clinical investigation. Lancet 2006367668–678. [DOI] [PubMed] [Google Scholar]
- 37.Simpson K W, Dogan B, Rishniw M.et al Adherent and invasive Escherichia coli is associated with granulomatous colitis in boxer dogs. Infect Immunity 2006744778–4792. [DOI] [PMC free article] [PubMed] [Google Scholar]