Abstract
Background
Myofibroblast differentiation is a key event during wound healing and is triggered primarily by transforming growth factor β (TGFβ). Serum response factor (SRF) is a TGFβ‐inducible transcription factor that is important for wound healing. Injection of SRF expression plasmid into rat gastric ulcers significantly accelerated restoration of epithelium and smooth muscle structures.
Aim
To determine the role of SRF in oesophageal ulcer healing, especially in myofibroblast differentiation.
Subjects
Rats (in vivo), oesophageal epithelial cells (Het1A) and fibroblasts (Rat1‐R12) (in vitro) were used.
Methods
Oesophageal ulcers were induced in rats with acetic acid and subsequently treated by local injection of plasmids expressing either SRF or SRF antisense sequence. Rats were killed at 1, 3, 6, 9 and 14 days after treatment and tissues collected. For in vitro studies, both Het1A and Rat1‐R12 cells were transfected with the plasmids used in ulcer treatment.
Results
Upregulation of SRF increased the myofibroblast population in ulcer granulation tissue; knockdown of SRF suppressed this event. In addition, ulceration induced SRF and TGFβ expression coordinately. In vitro studies showed that overexpression of SRF in either oesophageal epithelial cells or fibroblasts was sufficient to induce myofibroblast phenotype. Furthermore, the TGFβ‐induced myofibroblast phenotype required integrin‐linked kinase (ILK)‐mediated SRF activation, as either knockdown of SRF or inactivation of ILK prevented this action.
Conclusions
SRF is indispensable for myofibroblast differentiation during oesophageal ulcer healing and is required for TGFβ‐induced myofibroblast transition from either epithelial cells or fibroblasts. ILK is a mediator in TGFβ‐induced SRF activation and subsequent myofibroblast differentiation. ILK is associated with SRF, and TGFβ enhances this association.
Keywords: serum response factor, integrin‐linked kinase, transforming growth factor β, cell differentiation, ulcer
Oesophageal ulcers are deep necrotic lesions through squamous mucosae and are most commonly caused by gastro‐oesophageal reflux disease in the US and by diet in the Middle East. Healing of oesophageal ulcers is coordinated by multiple growth factors and transcription factors interacting in a temporally and spatially coordinated manner.
Serum response factor (SRF) is a transcription factor that is inducible by growth factors.1,2 Its activity is greatly enhanced by phosphorylation at serine‐103 (an N‐terminal site immediately adjacent to the DNA‐binding domain).3 About 300 genes are estimated to contain a serum response element (SRE), accounting for 1% of the entire genome.4 Among them, many are immediate early genes (eg, c‐fos and cyr61) which have been shown to be important for wound healing.5,6 Another group is muscle‐specific genes (eg, smooth muscle α‐actin and smoothelin), which encode structural proteins of muscle cells, and some are also shared by myofibroblasts.7,8 SRF functions as a master regulatory platform, which directs cell proliferation,9 migration10 and differentiation,11 the essential processes of wound healing.
Myofibroblasts are contractile cells which can be developed from progenitor cells,12 epithelial cells13 or fibroblasts.14 When the connective tissue has been damaged and denuded of its epithelium during gastrointestinal ulceration, epithelial cells and fibroblasts next to the ulcer area are activated to acquire myofibroblast phenotype and participate in restoration of new epithelial continuity and extracellular matrix. These cells are often distinguished from their ancestors by their expression of smooth muscle α‐actin (SM α‐actin), and from smooth muscle cells by the absence of smoothelin.15,16 Their contractile capability allows them to facilitate contraction of granulation tissue and wound closure.17 Among the many molecules that are able to induce myofibroblast differentiation in vivo or in vitro, transforming growth factor β (TGFβ) appears to be the most potent.18
TGFβ1, a multifunctional polypeptide, plays a pivotal role in various wound healing events; however, its function in oesophageal ulcer healing remains unknown. Results from gastric ulcer studies have been controversial.19 Some studies showed that local injection of TGFβ1 accelerated gastric ulcer healing,20,21 whereas others indicated an opposite effect.22
The induction of SRF by TGFβ has been studied previously,23,24 but its effect on SRF activation is largely unknown. Although several protein kinases, such as casein kinase II and mitogen‐activated protein kinase‐activated protein (MAPKAP) kinase 2, can phosphorylate SRF,3,25,26 none of them is known to have a role in myofibroblast differentiation. Integrin‐linked kinase (ILK), a serine/threonine kinase, is critical for TGFβ1‐induced epithelial–mesenchymal transition (EMT).27 ILK can directly phosphorylate various proteins, including Akt, glycogen synthase kinase 3, smooth muscle myosin and myelin basic protein (MBP);28 however, its role in myofibroblast differentiation and its association with SRF are unclear. A recent study on liver wound healing showed that ILK overexpression in stellate cells upregulates SM α‐actin expression.29
The molecular mechanisms of oesophageal ulcer healing have not been studied in depth. Previous studies showed that acetic acid‐induced oesophageal ulceration in rats triggers expression of multiple growth factors including keratinocyte growth factor, vascular endothelial growth factor and hepatocyte growth factor.30,31,32,33 Although the effect of keratinocyte growth factor or hepatocyte growth factor on SRF remains unknown, our recent study showed that SRF is required for angiogenesis induced by vascular endothelial growth factor.34 Moreover, we found that gastric ulceration activates SRF expression and that gene therapy with SRF promotes restoration of epithelium and smooth muscle during healing.35
Acetic acid‐induced oesophageal ulceration in rat is a validated model for ulcer study.30,31,32,33 Ulcers induced by this method share many morphological similarities, such as basal zone hyperplasia and increased papillae, with human oesophageal ulcers.36 In this study, we used this model to examine the role of SRF in myofibroblast differentiation during oesophageal ulcer healing. We also used oesophageal epithelial and fibroblast cell lines as our in vitro model to explore the mechanisms.
Methods
Experimental oesophageal ulcer model
The animal study was approved by the Subcommittee for Animal Studies of the VA Long Beach Healthcare System. Sixty male Sprague‐Dawley rats (Charles Rivers Labs, Wilmington, Massachusetts, USA) with body weight 225–250 g were fasted for 12 h before operation. The animals were anaesthetised by intraperitoneal injection of pentobarbital (50 mg/kg body weight). Oesophageal ulcers were induced by 3‐min applications of 50 μl acetic acid to the serosal surface of the lower oesophagus through a polyethylene tube (diameter 3.0 mm), as described in the previous studies.30,31,32,33 Ten rats were killed at 1, 3, 6, 9 and 14 days after the intervention, and oesophageal tissue specimens were collected. An additional 10 rats were sham‐operated as controls.
Gene therapy with SRF
The human SRF cDNA (accession number J03161) was cloned into EcoRI/NotI restriction sites of pcDNA3 (Invitrogen, Carlsbad, California, USA) under cytomegalovirus promoter. The C‐terminal myc and His epitope tags were ready for detection. To create an antisense construct, a 287‐bp fragment of SRF cDNA was cloned into the ApaI restriction site of pcDNA3 in antisense orientation. All plasmids were prepared as described in our previous study.34,35 Oesophageal ulcers were induced in 120 rats, which were divided into three groups (40 rats each) and each was injected with pcDNA3 expressing SRF (SRF+), antisense SRF (SRF−) or the plasmid vehicle (control) immediately after ulcer induction. In total, 100 μg plasmid DNA was injected into the submucosa around the ulcer induction site, as described in our previous study.35 Ten rats from each group were killed at each time point (3, 6, 9 and 14 days) to collect tissues.
Detection of SRF transgene expression
After plasmid injection, SRF transgene expression was monitored by both reverse transcription PCR and western blotting using an anti‐His antibody (Invitrogen). To detect antisense SRF expression, we used the primers 5′‐CTCACTATAGGGAGACCCAAG‐3′ (forward) from the T7 promoter region of the plasmid and 5′‐CTGGTGGTATGGCTAGAGG‐3′ (reverse) from the insert sequence. A 319‐bp product was expected. Rat β‐actin was amplified simultaneously using the primers 5′‐TTGTAACCAACTGGGACGATATGG‐3′ (forward) and 5′‐GATCTTGATCTTCATGGTGCTAGG‐3′ (reverse) as an internal control (764‐bp fragment).
Histology and immunohistochemistry
Oesophageal tissue was fixed in 10% formalin and embedded in paraffin. Antigen was retrieved by pepsin digestion. Tissue sections were stained with either H&E for histological evaluation or an antibody against one of the following proteins: SRF (1:200 dilution), smoothelin (1:200) (Santa Cruz Biotechnology, Santa Cruz, California, USA), SM α‐actin (1:400; Abcam, Cambridge, Massachusetts, USA), TGFβ1 (15 μg/ml; R&D System, Minneapolis, Minnesota, USA), ILK (5 μg/ml; Upstate Cell Signaling, Lake Placid, New York, USA) and His (1:5000). A signal was developed using a LSAB+ kit with a biotinylated universal linker and horseradish peroxidase‐conjugated streptavidin, following the manufacturer's protocol (DakoCytomation, Carpentaria, California, USA). AEC Chromogen was used as a substrate (DakoCytomation). The effect of SRF gene therapy on ulcer healing was evaluated by measuring ulcer size under a dissecting microscope. Assessment of myofibroblast differentiation was based on both SM α‐actin expression level (western blot) and the number of SM α‐actin‐positive cells, subtracting the number of smoothelin‐positive cells in three randomly selected microscopic fields in each of five sections (n = 15).
Cell culture
Human oesophageal epithelial cells (Het1A) and rat connective tissue fibroblasts (Rat1‐R12) (American Type Culture Collection, Manassas, Virginia, USA) were maintained in culture in KBM‐2 medium (Cambrex, Rockland, Maine, USA) and Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum, respectively. The Het1A cell line was established and characterized in 1991.37 It retains normal oesophageal epithelial morphology in culture. Rat1‐R12 was derived from the Rat1 cell line, which was established and characterized in 1981.38 It is a Fisher rat 3T3‐like connective tissue fibroblast cell line.
Both cell lines were transfected with either the plasmids that were used in our in vivo study or a pcDNA3 carrying a kinase‐dead human ILK cDNA (with a double point mutation produced by the substitution of GC for CG at nucleotides 631 and 632), using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. The entire mutant ILK cDNA was released by EcoRI digestion from the commercial plasmid pUSEamp (Upstate Cell Signalling) and subcloned into pcDNA3. The detailed sequence information can be found on the manufacturer's website.
To examine the effect of TGFβ on SRF, cells were treated with either a human recombinant TGFβ1 (R&D Systems) or vehicle (control) as indicated. To determine SRF protein stability, cells were pretreated with TGFβ1 at 2.5 ng/ml for 6 h and then treated with either cycloheximide (Sigma‐Aldrich, St Louis, Missouri, USA) or vehicle (control) as indicated.
For immunocytochemistry, cells were cultured on type I collagen‐coated coverslips overnight. After 24 h of serum deprivation followed by the indicated treatment, cells were fixed in 3.7% formaldehyde, permeabilised in cold acetone, and stained as indicated by following our previous protocol.34,35
Northern and western blot analysis
Northern and western blot analyses were carried out as described in our previous studies.34,35,39 The SRF cDNA from the pcDNA3 vector was used as a probe for northern blotting. The signals were quantified with the ImageQuant 5.0 software (GE Healthcare System, Piscataway, New Jersey, USA) based on at least six replicates.
Electrophoretic mobility shift assay
This was performed as described in our previous studies.34,35,39 The consensus and mutant oligonucleotide of SM α‐actin SRE are 5′‐GTTTTACCTAATTAGGAAATGC‐3′ and 5′‐ GTTTTACCTAATTATTAAATGC‐3′, respectively (synthesized by Retrogen, San Diego, California, USA).
Immunoprecipitation and kinase assay
Exact 200 μg total protein lysates from either the cultured cells or oesophageal tissues were incubated with 2 μg of either the anti‐SRF or anti‐ILK antibody (Upstate) for 2 h and with protein A–agarose beads (Upstate) for an additional 2 h at 4°C with agitation. The beads were washed twice with the lysis buffer. For ILK assay, the beads were washed twice more with a kinase buffer containing 25 mM Tris (pH 7.5), 5 mM β‐glycerophosphate, 2 mM dithiothreitol, 0.1 mM Na3VO4, and 10 mM MgCl2, then incubated in 50 μl kinase buffer containing 0.2 mM ATP and 1 μg MBP as substrate at 30°C for 30 min. The proteins were eventually dissociated from the beads by boiling in SDS protein sample buffer for 5 min, separated by western blotting, and probed with antibodies that detect total SRF and ILK or phosphorylated SRF (Cell Signaling, Danvers, Massachusetts, USA) and MBP (Upstate).
Statistical analysis
All numerical data are expressed as mean (SD) and analysed by single‐classification one‐way analysis of variance. p<0.05 was considered significant.
Results
Local overexpression of SRF promotes myofibroblast differentiation during ulcer healing
Our previous study showed that local upregulation of SRF by gene therapy accelerates gastric ulcer healing by promoting restoration of epithelium and smooth muscle components.35 These promising results inspired us to apply the same strategy to oesophageal ulcer healing.
Plasmid expression in ulcers was confirmed by reverse transcription PCR after injection. A single product (∼800 bp) was detected only in the tissue samples collected at days 3 and 6 from the rats that received (SRF+) plasmid (fig 1A). Likewise, a band expected at 300 bp was generated only from the tissue samples collected from the antisense (SRF−) plasmid‐treated rats (fig 1A).
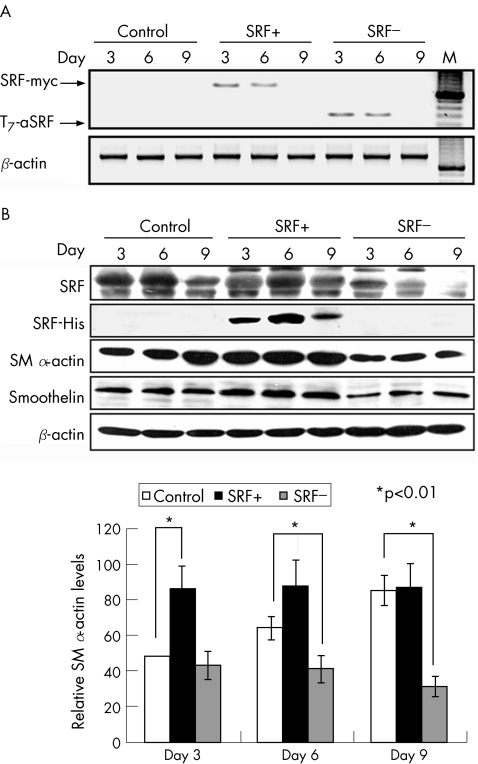
Figure 1 Serum response factor (SRF) gene therapy promotes expression of SM α‐actin in oesophageal ulcers. (A) Reverse transcription PCR analyses showing specific transient expression of SRF transgene (SRF‐myc) in the rats that were injected with the SRF expression plasmid (SRF+) and specific expression of antisense SRF fragment (T7‐aSRF) in the rats that received antisense SRF (SRF−) plasmid. β‐Actin was used as an internal control. M, Molecular marker. (B) Western blot analyses showing a His‐tagged SRF protein product in the rats that were injected with the SRF+ plasmid in parallel with the increased total SRF protein concentrations in these rats, reduced local SRF protein concentrations in the rats that received SRF− plasmid, and their corresponding effects on expressions of smooth muscle (SM) α‐actin and smoothelin. (C) Quantification of SM α‐actin protein expression after gene therapy based on 10 replicates. *p<0.01. Error bars represent SD.
Transgene expression was also examined at protein level by both western blotting and immunohistochemistry. Both techniques detected a His‐tagged protein product in the tissue samples collected at days 3, 6 and 9 from the rats that received SRF+ plasmid (fig 1B; 2H), together indicating a specific local and transient expression of the plasmids. In parallel with the expression of the SRF+ plasmid, total SRF protein concentrations in these tissue samples were significantly increased by 29.5 (4.9)% and 127.3 (10.6)% (both p<0.01) at days 6 and 9, respectively, compared with the control group (fig 1B). Expression of antisense SRF significantly reduced local SRF protein contents by 28.2 (4.7)%, 71.0 (11.8)% and 81.8 (14.5)% (all p<0.01) at days 3, 6 and 9, respectively, compared with the control group (fig 1B).
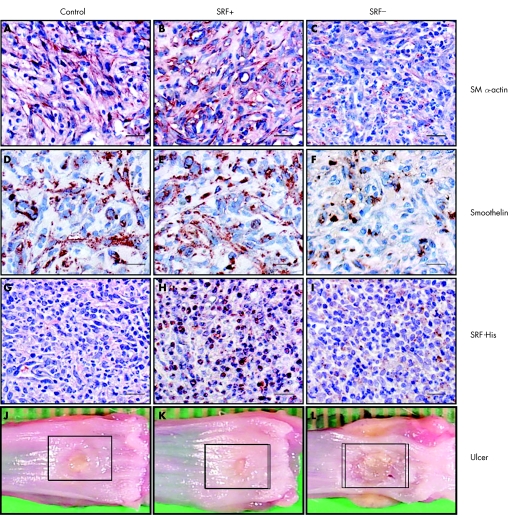
In addition to the improvements in re‐epithelialisation and restoration of smooth muscle structures as observed in gastric ulcers, local upregulation of SRF in oesophageal ulcers significantly increased SM α‐actin expression by 79.5 (8.1)% and 38.0 (6.8)% at days 3 and 6, respectively; knockdown of SRF reduced it by 35.4 (0.2)% and 63.9 (3.9)% correspondingly compared with controls (both p<0.01; fig 1B). Immunohistochemistry showed that the number of SM α‐actin‐positive cells was increased by 55 (12)% in ulcer granulation tissue at day 6 as the result of upregulation of SRF (fig 2B) and decreased by 78 (17)% as the result of knockdown of SRF (fig 2C).
Figure 2 Immunohistochemistry and macroscopic appearances of oesophageal ulcers 6 days after serum response factor (SRF) gene therapy. Scale bar, 100 μm. (A–C) Granulation tissues stained with the antibody against smooth muscle (SM) α‐actin showing an increase in the number of SM α‐actin expression cells due to local upregulation of SRF (SRF+) and a decrease due to local knockdown of SRF (SRF−). (D–F) Granulation tissues stained with the antibody against smoothelin showing an increase in the number of smooth muscle cells due to local upregulation of SRF (SRF+) and a decrease due to local knockdown of SRF (SRF−). (G–I) Granulation tissues stained with the antibody recognising His tag showing specific nuclear staining in the rats that received SRF+ plasmid injection. (J–L) Macroscopic pictures of dissected oesophagus showing reduced ulcer size by local upregulation of SRF (SRF+) and delayed healing due to SRF deficiency (SRF−).
As both myofibroblasts and smooth muscle cells express SM α‐actin, to assess net myofibroblast population, we examined protein expression of smoothelin, a specific marker for smooth muscle cells, in the same tissue samples. Upregulation of SRF increased smoothelin concentration marginally, whereas knockdown of SRF reduced it by 17.9 (5.0)% and 32.3 (4.0)% at days 6 and 9 respectively (fig 1B; both p<0.01). Staining for smoothelin showed similar results (fig 2E,F).
After subtracting the number of smoothelin‐positive cells from the number of SM α‐actin‐positive cells, as a result of gene therapy, upregulation of SRF had increased the number of myofibroblasts in granulation tissue by 74 (8)% at day 6 and knockdown of SRF had reduced it by 86 (10)% (both p<0.005).
Ulcer closure results from the combined effort of proliferating/migrating epithelial cells and contracting myofibroblasts. Measurement of ulcer area at day 6 indicated that ulcer size was reduced by 54 (9)% in SRF(+) rats (fig 2K), whereas it was 108 (12)% larger in SRF(−) rats (fig 2L) compared with controls (fig 2J) (both p<0.01).
Overexpression of SRF in either oesophageal epithelial cells or fibroblasts can induce myofibroblast phenotype
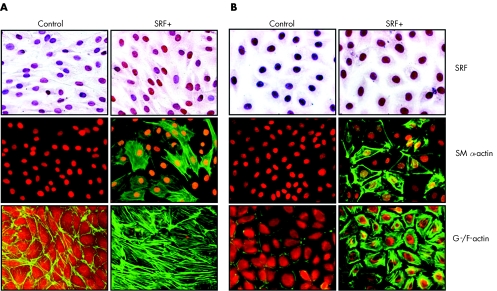
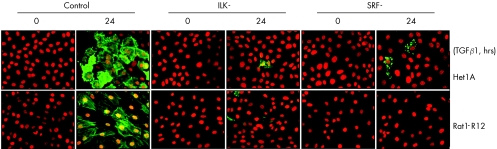
The origin of myofibroblasts in wound tissue has not been fully elucidated; however, it is generally accepted that their main sources are local fibroblasts and epithelial cells. The latter can become either fibroblasts or myofibroblasts through EMT. To investigate the role of SRF in myofibroblast differentiation, we transfected an oesophageal epithelial cell line (Het1A) and a rat connective tissue fibroblast cell line (Rat1‐R12) with either the SRF expressing plasmid or vehicle (control) used in our in vivo study, and looked for evidence of myofibroblast phenotype: expression of SM α‐actin and formation of actin stress fibres. The controls neither expressed SM α‐actin nor formed actin stress fibres. However, forced overexpression of SRF (SRF+) induced SM α‐actin expression and extensive actin stress fibre formation in both cells (fig 3), suggesting that upregulation of SRF in either epithelial cells or fibroblasts can transform these cells into myofibroblasts.
Figure 3 Overexpression of serum response factor (SRF) in either oesophageal epithelial cells or fibroblasts is sufficient to induce myofibroblast phenotype. Both Het1A and Rat1‐R12 cells were transfected with either SRF+ plasmid or plasmid vehicle (control) and stained for SRF (counterstained with haematoxylin), smooth muscle (SM) α‐actin (counterstained with propidium iodide) or G‐/F‐actin (Oregon Green‐conjugated phalloidin and Texas Red‐conjugated deoxynuclease I). Overexpression of SRF (SRF+) induced SM α‐actin expression and actin stress fibre formation in both (A) Rat1‐R12 cells and (B) Het1A cells.
SRF upregulation during ulcer healing is coordinated with TGFβ
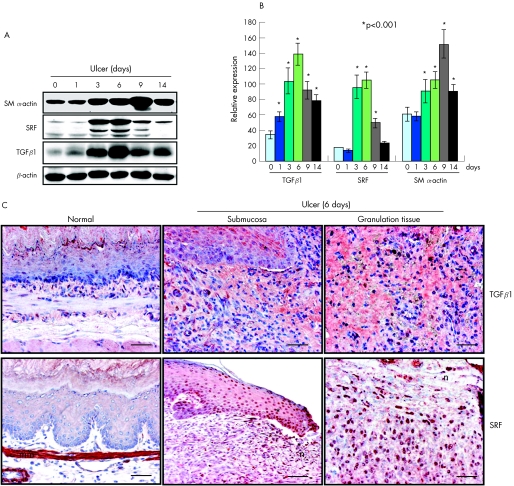
Wound healing requires activation of multiple growth factors and transcription factors around the healing area in a coordinated manner. TGFβ is widely recognised to be the most potent inducer of myofibroblast differentiation both in vivo and in vitro; however, the role of TGFβ in oesophageal ulcer healing has never been investigated. Therefore, we examined TGFβ1 expression in oesophageal tissue after ulcer induction. TGFβ1 was significantly upregulated through the entire 14‐day monitoring period, peaking with a 38‐fold increase at day 6 (p<0.001; fig 4A). Immunohistochemical staining showed a strong TGFβ1 signal in both ulcer margin and granulation tissue (fig 4C). A similar dynamic expression and localisation were also found for SRF (fig 4A–C), suggesting possible coordination between these two molecules.
Figure 4 Oesophageal ulceration induces expressions of transforming growth factor β1 (TGFβ1) and serum response factor (SRF) in a temporally and spatially coordinated manner. (A) Western blot analyses showing the dynamic expressions of TGFβ1, smooth muscle (SM) α‐actin and SRF during ulcer healing. (B) Quantitative analysis based on 10 replicates. *p<0.001. Error bars represent SD. (C) Immunohistochemistry showing extensive expression of TGFβ1 at the ulcer margin (?) and in granulation tissue 6 days after ulcer induction, compared with normal oesophagus. Ulceration also induced SRF expression in similar locations. mm, Muscularis mucosa; n, necrotic tissue. Scale bar, 100 μm.
TGFβ1 can activate SRF in both oesophageal epithelial cells and fibroblasts in an ILK‐dependent manner
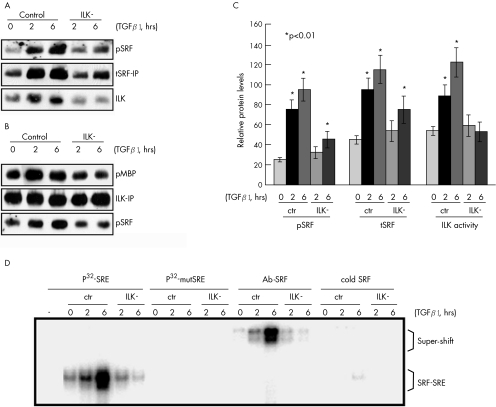
The effect of TGFβ1 on SRF expression was previously documented in other cell types.23,24 However, little is known about its effect on SRF activation. To select the most appropriate dose for our study, we treated both Het1A and Rat1‐R12 cells with a human recombinant TGFβ1 for 6 h at the following concentrations: 1, 2.5, 5.0 and 10 ng/ml. TGFβ1 clearly increased SRF expression levels by a similar amount at 2.5, 5.0 and 10 ng/ml. Therefore, we chose to use 2.5 ng/ml for the rest of the study. To determine whether TGFβ1 induces SRF activation in oesophageal epithelial cells or fibroblasts, both Het1A and Rat1‐R12 cells were treated with TGFβ1 for 2 and 6 h. TGFβ1 significantly increased SRF protein concentration in Het1A cells by 171.4 (25)% and 228.6 (39)% at 2 and 6 h, respectively (fig 5A,C) (both p<0.01). More interestingly, almost all of the SRF protein induced by TGFβ1 was also phosphorylated at serine‐103 (fig 5A), which is known to promote SRF interaction with DNA.3 Similar results were obtained in Rat1‐R12 cells.
Figure 5 Transforming growth factor β1 (TGFβ1) induces serum response factor (SRF)‐binding activity to smooth muscle (SM) α‐actin serum response element (SRE) in an integrin‐linked kinase (ILK)‐dependent manner. (A) Immunoprecipitation with an anti‐SRF antibody (tSRF‐IP) showing that SRF is associated with ILK and inactivation of ILK (ILK−) prevents both TGFβ1‐induced SRF phosphorylation (pSRF) and total SRF protein upregulation (tSRF). (B) Immunoprecipitation with an anti‐ILK antibody (ILK‐IP) followed by a kinase assay using myelin basic protein (MBP) as a substrate confirming that SRF phosphorylation is associated with ILK activation. (C) Quantitative analysis based on 10 replicates. *p<0.01. Error bars represent SD. ctr, Control. (D) Electrophoretic mobility shift assay showing that TGFβ1 treatment increases SRF protein binding to smooth muscle α‐actin SRE in an ILK‐dependent manner. 10 μg protein lysate was incubated with 5 ng of either 32P‐labelled consensus smooth muscle α‐actin SRE oligonucleotide (P32‐SRE) or 32P‐labelled mutant SRE (P32‐mutSRE). For super‐shift assays, 2 μg of the anti‐SRF antibody (Ab‐SRF) was also included in the reaction. To confirm the specificity of the binding products, a 50‐fold ( = 250 ng) excess of unlabelled consensus SRE oligonucleotide (cold SRE) was added to the reaction to compete with the 32P‐labelled SRE for SRF binding.
Several protein kinases have been reported to phosphorylate SRF, but none is known to be involved in EMT.3,25,26 ILK is required for TGFβ‐induced EMT in several other cell types.27 A recent study showed that ILK overexpression in stellate cells dramatically upregulates SM α‐actin expression.29 On the basis of the literature, it was reasonable to speculate that ILK may have been involved in TGFβ‐induced SRF phosphorylation. To test this speculation, we transfected both Het1A and Rat1‐R12 cells with a kinase‐dead ILK vector (ILK−). ILK activity was monitored by kinase assay using MBP as substrate (fig 5B). Inactivation of ILK dramatically suppressed TGFβ1‐induced increases in both SRF phosphorylation and total SRF protein content (fig 5A), suggesting that SRF phosphorylation might have increased SRF protein stability. Moreover, immunoprecipitation of either SRF or ILK demonstrated a direct association between these two proteins (fig 5A,B). Meanwhile, inactivation of ILK also clearly suppressed TGFβ1‐activated SRF binding to sm α‐actin SRE (fig 5D), indicating that ILK is a mediator of this action.
Inactivation of ILK destabilises SRF protein
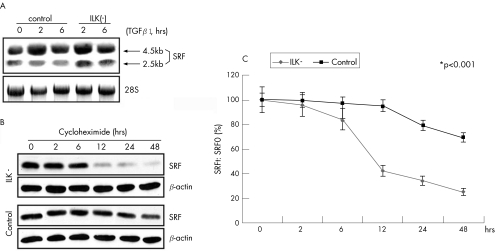
To determine whether SRF protein stability depends on ILK activation, we first examined SRF mRNA concentration in the ILK(−) cells. TGFβ1 treatment significantly increased SRF mRNA expression in Het1A cells by 72.9 (9.3)% and 45.3 (5.2)% at 2 and 6 h, respectively (both p<0.01; fig 6A), regardless of ILK activity. Secondly, we pretreated both control and ILK(−) cells with TGFβ1 at 2.5 ng/ml for 6 h and then treated them with cycloheximide (a protein synthesis inhibitor) at 100 μg/ml for 2, 6, 12, 24 and 48 h. Western blot analysis showed that SRF protein content fell by 50% in ILK(−) Het1A cells within 12 h of cycloheximide treatment, whereas no significant change was observed in control cells (fig 6B,C), indicating that inactivation of ILK destabilises SRF protein. Similar results were obtained in Rat1‐R12 cells.
Figure 6 Serum response factor (SRF) protein stability depends on integrin‐linked kinase (ILK) activation in Het1A cells. (A) Northern blot analyses showing that transforming growth factor β1 (TGFβ1) treatment upregulates SRF transcription regardless of ILK activity. (B) Western blot analysis showing the time course of SRF protein stability: cells were pretreated with TGFβ1 for 6 h, cycloheximide was then added at 100 μg/ml, and total protein was isolated at the times indicated. (C) Plot of SRF decay in ILK(−) cells versus controls: SRF protein content at each time point (SRFt) divided by SRF content at time 0 (SRF0) based on six replicates. *p<0.001. Error bars represent SD.
ILK‐mediated SRF activation is essential for TGFβ‐induced myofibroblast differentiation from either oesophageal epithelial cells or fibroblasts
As TGFβ1‐activated ILK mediates SRF transcriptional activity, we examined whether TGFβ1 is still able to induce myofibroblast phenotype without functional ILK or SRF. We transfected both Het1A and Rat1‐R12 cells with either SRF(−) or ILK(−) plasmid or vehicle (control). Knockdown of SRF (SRF−) completely prevented TGFβ‐induced SM α‐actin expression in both cell types (fig 7). Likewise, inactivation of ILK (ILK−) also greatly suppressed the induction of SM α‐actin. This indicates an essential role for SRF in TGFβ1‐induced myofibroblast differentiation.
Figure 7 Knockdown of serum response factor (SRF) in either Het1A or Rat1‐R12 cells prevents transforming growth factor β1 (TGFβ1)‐induced smooth muscle (SM) α‐actin expression. Both Het1A and Rat1‐R12 cells were transfected with (SRF−) plasmid, mutant integrin‐linked kinase (ILK−) or vehicle (control), treated with the recombinant TGFβ1 at 2.5 ng/ml for 24 h, and stained for SM α‐actin. TGFβ1 induced SM α‐actin expression in control cells, but not in ILK(−) or SRF(−) cells.
Oesophageal ulceration induces ILK expression and activation
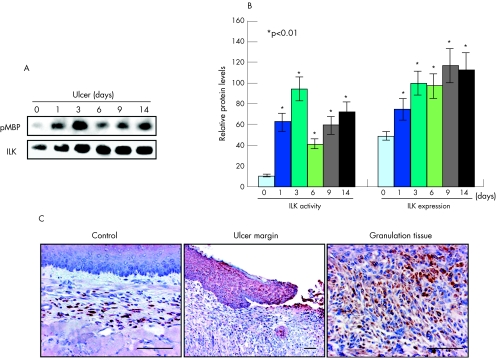
On the basis of our in vitro study, which indicated that ILK is a critical requirement for TGFβ‐induced SRF binding to sm α‐actin SRE and thereafter myofibroblast differentiation, we examined whether ILK is activated by oesophageal ulceration. Immunoprecipitation and ILK assay showed significant increases in both ILK protein expression (peaking at day 9 with a ∼1.5‐fold increase) and activity (peaking at day 3 with a ∼8‐fold increase) (p<0.01; fig 8A,B) after ulcer induction. ILK was localised to both ulcer margin and granulation tissue (fig 8C).
Figure 8 Integrin‐linked kinase (ILK) is activated during oesophageal ulcer healing. (A) Immunoprecipitation followed by a kinase assay using myelin basic protein (MBP) as substrate showing that not only the expression of ILK is upregulated but also its activity during ulcer healing. (B) Quantitative analysis based on 10 replicates each. *p<0.01. Error bars represent SD. (C) Immunohistochemistry showing that ILK is upregulated in both ulcer margin and granulation tissue. Scale bar, 100 μm.
Discussion
In summary, this study shows for the first time that: (a) SRF is critical for myofibroblast differentiation during ulcer healing; (b) overexpression of SRF in either epithelial cells or fibroblasts can induce myofibroblast phenotype; (c) TGFβ‐induced myofibroblast differentiation is mediated by ILK‐regulated SRF phosphorylation; (d) ILK is associated with SRF, and TGFβ enhances this association.
It is well established that myofibroblasts are a key component of wound healing. However, their role in ulcer healing remains largely unknown. Our previous study using SRF gene therapy to treat experimental gastric ulcers showed that overexpression of SRF can promote smooth muscle cell proliferation and migration, which contributes to restoration of muscularis mucosa damaged by ulceration. Myofibroblasts share several features (such as expression of SM α‐actin and contractility) with smooth muscle cells. In this study, we focused on the effect of SRF gene therapy on myofibroblast differentiation during experimental oesophageal ulcer healing and showed that both oesophageal epithelial cells and fibroblasts can be induced to develop a myofibroblast phenotype by overexpression of SRF.
SM α‐actin is a common marker used to identify differentiated epithelial cells or fibroblasts. The SM α‐actin gene has three SREs, which are completely conserved in all species in which the SM α‐actin gene promoter has been cloned.40 A study on cardiac development demonstrated that inactivation of SRF inhibits SM α‐actin expression, thus preventing transdifferentiation from proepicardial cells to smooth muscle cells.41 Here we show that knockdown of SRF in vivo suppresses myofibroblast differentiation in ulcer granulation tissue, and knockdown of SRF in vitro prevents TGFβ1‐induced myofibroblast phenotype in both oesophageal epithelial cells and fibroblasts, which together suggest that SRF is a downstream regulator of myofibroblast differentiation.
TGFβ1 is the most potent inducer of myofibroblast differentiation both in vivo and in vitro. In this study, we revealed a dramatic increase (38‐fold) in TGFβ1 expression in granulation tissue 6 days after wounding. Although TGFβ1 is known to induce SRF expression,23,24 little is known about how it affects SRF activity. The classic signalling cascade downstream of TGFβ1 involves activation of the Smads family of transcription factors. For example, Smad7 (an inhibitor of TGFβ signalling) has been found in association with SRF in unstimulated airway smooth muscle cells, and TGFβ weakens this interaction.42 However, alternative pathways, such as ILK activation, have also recently been linked to TGFβ1 signalling.27 Our data show that TGFβ1 induces SRF phosphorylation at serine‐103 and binding activity to SM α‐actin SRE in an ILK‐dependent manner. Moreover, ILK activation seemed to increase SRF protein stability, because inactivation of ILK greatly shortened the half‐life of SRF. This notion was also supported by co‐immunoprecipitation, which indicated that SRF is associated with ILK, and TGFβ1 treatment enhances this association. However, owing to a lack of purified SRF protein, at this point we were unable to confirm whether SRF is a direct target or a downstream target of ILK phosphorylation.
SRF was discovered during treatment of fibroblasts with serum 20 years ago.1 When the same experiment was examined again using microarray technology, a wound healing programme was uncovered.43 It appears that cells are programmed to interpret the abrupt exposure to serum (which normally comes into direct contact with cells in vivo only after tissue injury) not only as a general mitogenic stimulus, but also as a specific physiological signal, signifying a wound. On the basis of this notion, the features of a cell that is responding to serum are comparable to some aspects of the wound healing process. Therefore, SRF activation during oesophageal ulcer healing shown in this study can be taken as an essential component of the whole healing programme, which applies not only to fibroblasts but also to epithelial, endothelial and other cells involved.
Acetic acid‐induced experimental oesophageal ulcers in rats share many morphological similarities with human oesophageal ulcers.36 Therefore, this study provides new insight into common molecular mechanisms of ulcer healing. Local gene therapy, by introducing vectors that transiently express desirable genes (in this case, SRF) or suppress undesirable gene expression, appears to be a feasible treatment for the promotion of ulcer healing without affecting other organs.
Acknowledgements
This study was supported by both the Merit Review Entry Program of the Department of Veterans Affairs of the USA and the Grant‐in‐Aid of the American Heart Association awarded to JC.
Abbreviations
EMT - epithelial–mesenchymal transition
ILK - integrin‐linked kinase
ILK(−) - inactivation of ILK
MBP - myelin basic protein
SM α‐actin - smooth muscle α‐actin
SRE - serum response element
SRF - serum response factor
SRF(+) - overexpression of SRF
SRF(−) - knockdown of SRF
TGFβ1 - transforming growth factor β1
Footnotes
Competing interests: None.
References
- 1.Treisman R. Identification of a protein‐binding site that mediates transcriptional response of the c‐fos gene to serum factors. Cell 198646567–574. [DOI] [PubMed] [Google Scholar]
- 2.Chai J, Tarnawski A S. Serum response factor: discovery, biochemistry, biological roles and implications for tissue injury healing. J Physiol Pharmacol 200253147–157. [PubMed] [Google Scholar]
- 3.Rivera V M, Miranti C K, Misra R P.et al A growth factor‐induced kinase phosphorylates the serum response factor at a site that regulates its DNA‐binding activity. Mol Cell Biol 1993136260–6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orr L. Scientists align billion‐year‐old protein with embryonic heart defects. Medical News Today 2004
- 5.Wang J Y, Johnson L R. Expression of protooncogenes c‐fos and c‐myc in healing of gastric mucosal stress ulcers. Am J Physiol 1994266G878–G886. [DOI] [PubMed] [Google Scholar]
- 6.Schober J M, Lau L F, Ugarova T P.et al Identification of a novel integrin alphaMbeta2 binding site in CCN1 (CYR61), a matricellular protein expressed in healing wounds and atherosclerotic lesions. J Biol Chem 200327825808–25815. [DOI] [PubMed] [Google Scholar]
- 7.Mack C P, Thompson M M, Lawrenz‐Smith S.et al Smooth muscle alpha‐actin CArG elements coordinate formation of a smooth muscle cell‐selective, serum response factor‐containing activation complex. Circ Res 200086221–232. [DOI] [PubMed] [Google Scholar]
- 8.Rensen S S, Niessen P M, Long X.et al Contribution of serum response factor and myocardin to transcriptional regulation of smoothelins. Cardiovasc Res 200670136–145. [DOI] [PubMed] [Google Scholar]
- 9.Poser S, Impey S, Trinh K.et al SRF‐dependent gene expression is required for PI3‐kinase‐regulated cell proliferation. EMBO J 2000194955–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schratt G, Philippar U, Berger J.et al Serum response factor is crucial for actin cytoskeletal organization and focal adhesion assembly in embryonic stem cells. J Cell Biol 2002156737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang Y, Zhe X, Phan S H.et al Involvement of srf isoforms in myofibroblast differentiation during bleomycin‐induced lung injury. Am J Respir Cell Mol Biol 200329583–590. [DOI] [PubMed] [Google Scholar]
- 12.Brittan M, Chance V, Elia G.et al A regenerative role for bone marrow following experimental colitis: contribution to neovasculogenesis and myofibroblasts. Gastroenterology 20051281984–1995. [DOI] [PubMed] [Google Scholar]
- 13.Yang J, Liu Y. Dissection of key events in tubular epithelial to myofibroblast transition and its implications in renal interstitial fibrosis. Am J Pathol 20011591465–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans R A, Tian Y C, Steadman R.et al TGF‐beta1‐mediated fibroblast‐myofibroblast terminal differentiation‐the role of Smad proteins. Exp Cell Res 200328290–100. [DOI] [PubMed] [Google Scholar]
- 15.Tomasek J J, Gabbiani G, Hinz B.et al Myofibroblasts and mechano‐regulation of connective tissue remodelling. Nat Rev Mol Cell Biol 20023349–363. [DOI] [PubMed] [Google Scholar]
- 16.Powell D W, Mifflin R C, Valentich J D.et al Myofibroblasts. I. Paracrine cells important in health and disease. Am J Physiol 1999277C1–C9. [DOI] [PubMed] [Google Scholar]
- 17.Feugate J E, Li Q, Wong L.et al The cxc chemokine cCAF stimulates differentiation of fibroblasts into myofibroblasts and accelerates wound closure. J Cell Biol 2002156161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Desmouliere A, Geinoz A, Gabbiani F.et al Transforming growth factor‐beta 1 induces alpha‐smooth muscle actin expression in granulation tissue myofibroblasts and in quiescent and growing cultured fibroblasts. J Cell Biol 1993122103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanigawa T, Pai R, Arakawa T.et al TGF‐beta signaling pathway: its role in gastrointestinal pathophysiology and modulation of ulcer healing. J Physiol Pharmacol 2005563–13. [PubMed] [Google Scholar]
- 20.Tominaga K, Arakawa T, Kim S.et al Increased expression of transforming growth factor‐beta1 during gastric ulcer healing in rats. Dig Dis Sci 199742616–625. [DOI] [PubMed] [Google Scholar]
- 21.Ernst H, Konturek P C, Brzozowski T.et al Subserosal application of transforming growth factor‐beta 1 in rats with chronic gastric ulcers: effect on gastric ulcer healing and blood flow. J Physiol Pharmacol 199647443–454. [PubMed] [Google Scholar]
- 22.Ernst H, Konturek P, Hahn E G.et al Acceleration of wound healing in gastric ulcers by local injection of neutralising antibody to transforming growth factor beta 1. Gut 199639172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ueyama T, Kawashima S, Sakoda T.et al Transforming growth factor‐beta1 and protein kinase C synergistically activate the c‐fos serum response element in myocardial cells. J Mol Cell Cardiol 199830551–562. [DOI] [PubMed] [Google Scholar]
- 24.Qiu P, Feng X H, Li L. Interaction of Smad3 and SRF‐associated complex mediates TGF‐beta1 signals to regulate SM22 transcription during myofibroblast differentiation. J Mol Cell Cardiol 2003351407–1420. [DOI] [PubMed] [Google Scholar]
- 25.Marais R M, Hsuan J J, McGuigan C.et al Casein kinase II phosphorylation increases the rate of serum response factor‐binding site exchange. EMBO J 19921197–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heidenreich O, Neininger A, Schratt G.et al MAPKAP kinase 2 phosphorylates serum response factor in vitro and in vivo. J Biol Chem 199927414434–14443. [DOI] [PubMed] [Google Scholar]
- 27.Lee Y I, Kwon Y J, Joo C K. Integrin‐linked kinase function is required for transforming growth factor beta‐mediated epithelial to mesenchymal transition. Biochem Biophys Res Commun 2004316997–1001. [DOI] [PubMed] [Google Scholar]
- 28.Legate K R, Montanez E, Kudlacek O.et al ILK, PINCH and parvin: the tIPP of integrin signalling. Nat Rev Mol Cell Biol 2006720–31. [DOI] [PubMed] [Google Scholar]
- 29.Shafiei M, Rockey D C. The role of integrin linked kinase in liver wound healing. J Biol Chem 200628124863–24872. [DOI] [PubMed] [Google Scholar]
- 30.Baatar D, Kawanaka H, Szabo I L.et al Esophageal ulceration activates keratinocyte growth factor and its receptor in rats: implications for ulcer healing. Gastroenterology 2002122458–468. [DOI] [PubMed] [Google Scholar]
- 31.Baatar D, Jones M K, Tsugawa K.et al Esophageal ulceration triggers expression of hypoxia‐inducible factor‐1 alpha and activates vascular endothelial growth factor gene: implications for angiogenesis and ulcer healing. Am J Pathol 20021611449–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basson M D. Gut mucosal healing: is the science relevant? Am J Pathol 20021611101–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baatar D, Jones M K, Pai R.et al Selective cyclooxygenase‐2 blocker delays healing of esophageal ulcers in rats and inhibits ulceration‐triggered c‐Met/hepatocyte growth factor receptor induction and extracellular signal‐regulated kinase 2 activation. Am J Pathol 2002160963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chai J, Jones M K, Tarnawski A S. Serum response factor is a critical requirement for VEGF signaling in endothelial cells and VEGF‐induced angiogenesis. FASEB J 2004181264–1266. [DOI] [PubMed] [Google Scholar]
- 35.Chai J, Baatar D, Tarnawski A. Serum response factor promotes re‐epithelialization and muscular structure restoration during gastric ulcer healing. Gastroenterology 20041261809–1818. [DOI] [PubMed] [Google Scholar]
- 36.Livstone E M, Sheahan D G, Behar J. Studies of esophageal epithelial cell proliferation in patients with reflux esophagitis. Gastroenterology 1977731315–1319. [PubMed] [Google Scholar]
- 37.Stoner G D, Kaighn M E, Reddel R R.et al Establishment and characterization of SV40 T‐antigen immortalized human esophageal epithelial cells. Cancer Res 199151365–371. [PubMed] [Google Scholar]
- 38.Topp W C. Normal rat cell lines deficient in nuclear thymidine kinase. Virology . 1981;113408–411. [DOI] [PubMed]
- 39.Zhang X, Chai J, Azhar G.et al Early postnatal cardiac changes and premature death in transgenic mice overexpressing a mutant form of serum response factor. J Biol Chem 200127640033–40040. [DOI] [PubMed] [Google Scholar]
- 40.Tomasek J J, McRae J, Owens G K.et al Regulation of alpha‐smooth muscle actin expression in granulation tissue myofibroblasts is dependent on the intronic CArG element and the transforming growth factor‐beta1 control element. Am J Pathol 20051661343–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landerholm T E, Dong X R, Lu J.et al A role for serum response factor in coronary smooth muscle differentiation from proepicardial cells. Development 19991262053–2062. [DOI] [PubMed] [Google Scholar]
- 42.Camoretti‐Mercado B, Fernandes D J, Dewundara S.et al Inhibition of transforming growth factor beta‐enhanced serum response factor‐dependent transcription by SMAD7. J Biol Chem 200628120383–20392. [DOI] [PubMed] [Google Scholar]
- 43.Iyer V R, Eisen M B, Ross D T.et al The transcriptional program in the response of human fibroblasts to serum. Science 199928383–87. [DOI] [PubMed] [Google Scholar]